Phage-Based Biosensing for Rapid and Specific Detection of Staphylococcus aureus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical and Materials
2.2. Bacterial Strains and Culture Condition
2.3. Isolation and Purification of Phage LSA2302
2.4. Host Range Determination
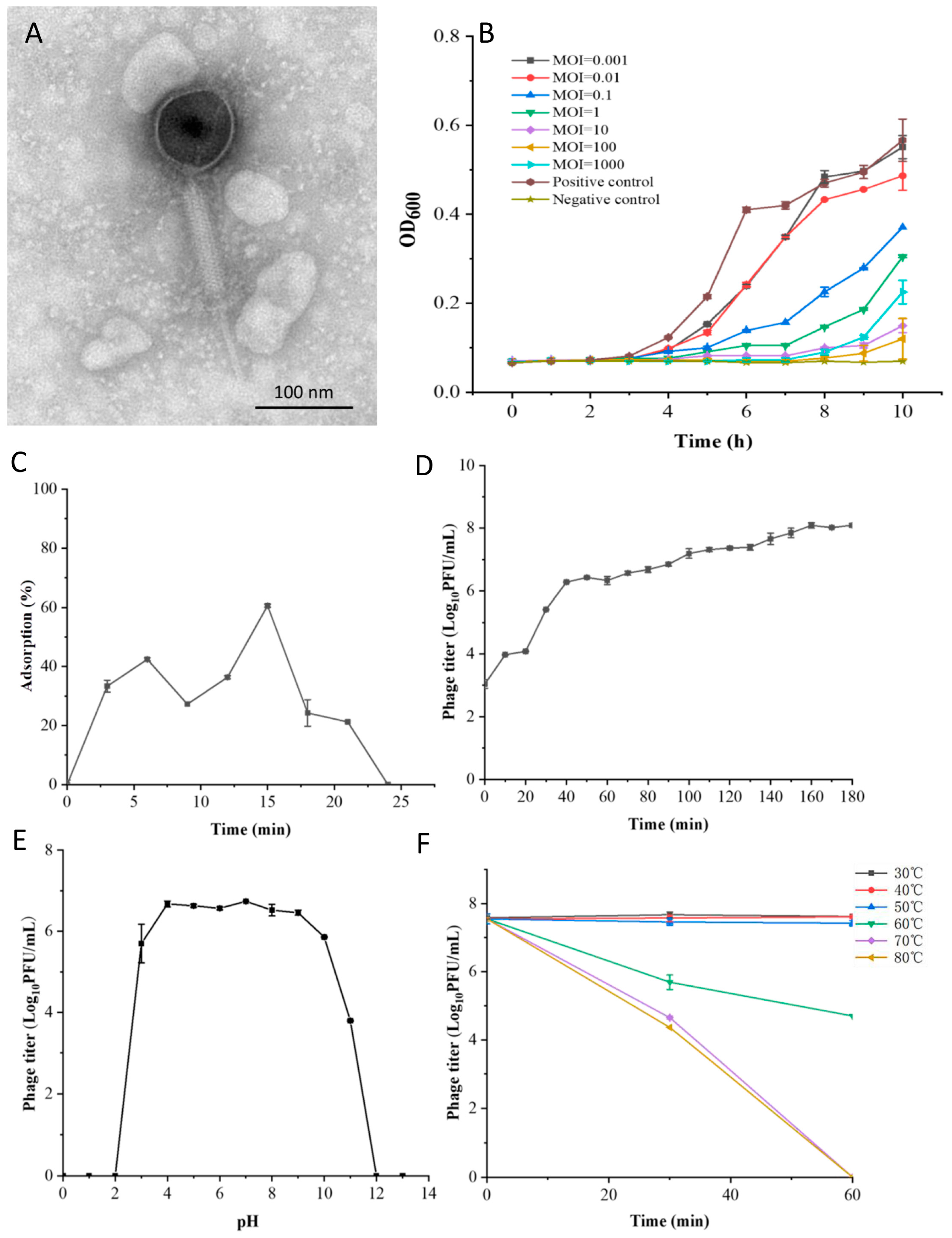
2.5. Morphological and Structural Protein Analysis
2.6. The Optimal Multiplicity of Infection
2.7. Adsorption Rate
2.8. One-Step Growth Curve
2.9. Thermal and pH Stability
2.10. Lytic Curve
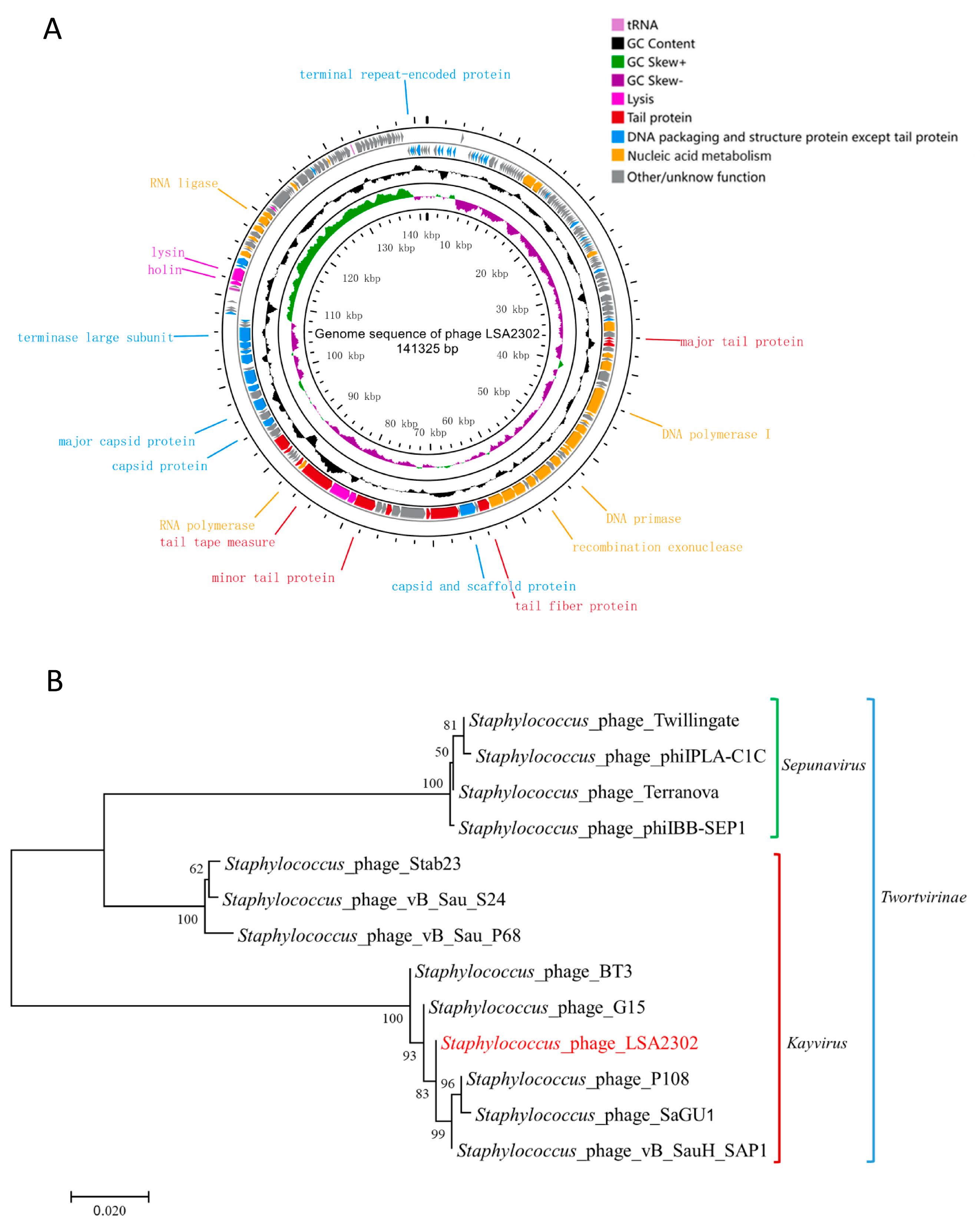
2.11. Genomic Analysis
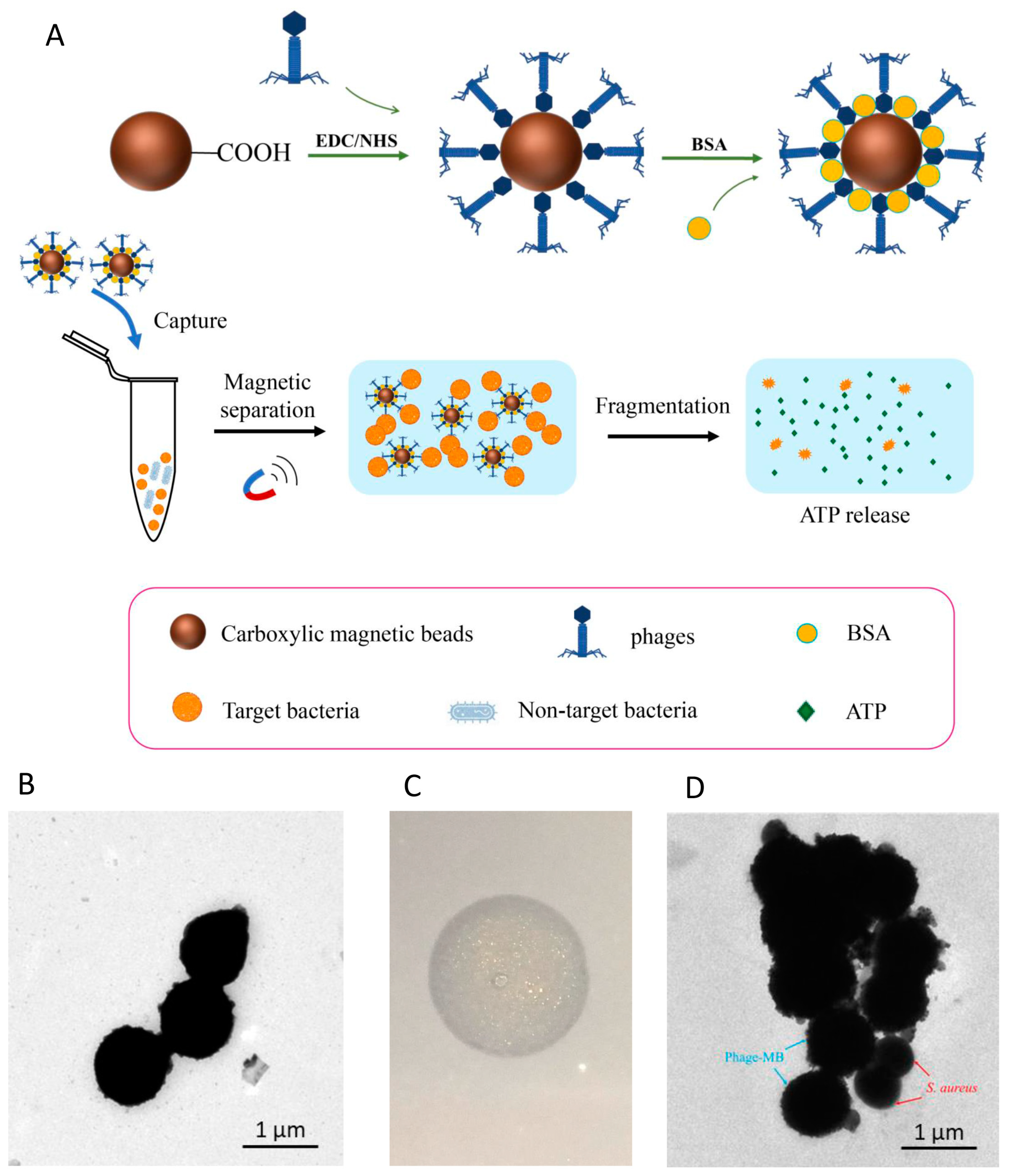
2.12. Functionalization of Magnetic Beads by Phage LSA2302
2.13. Detection of S. aureus by pMB Combined with ATP Bioluminescence
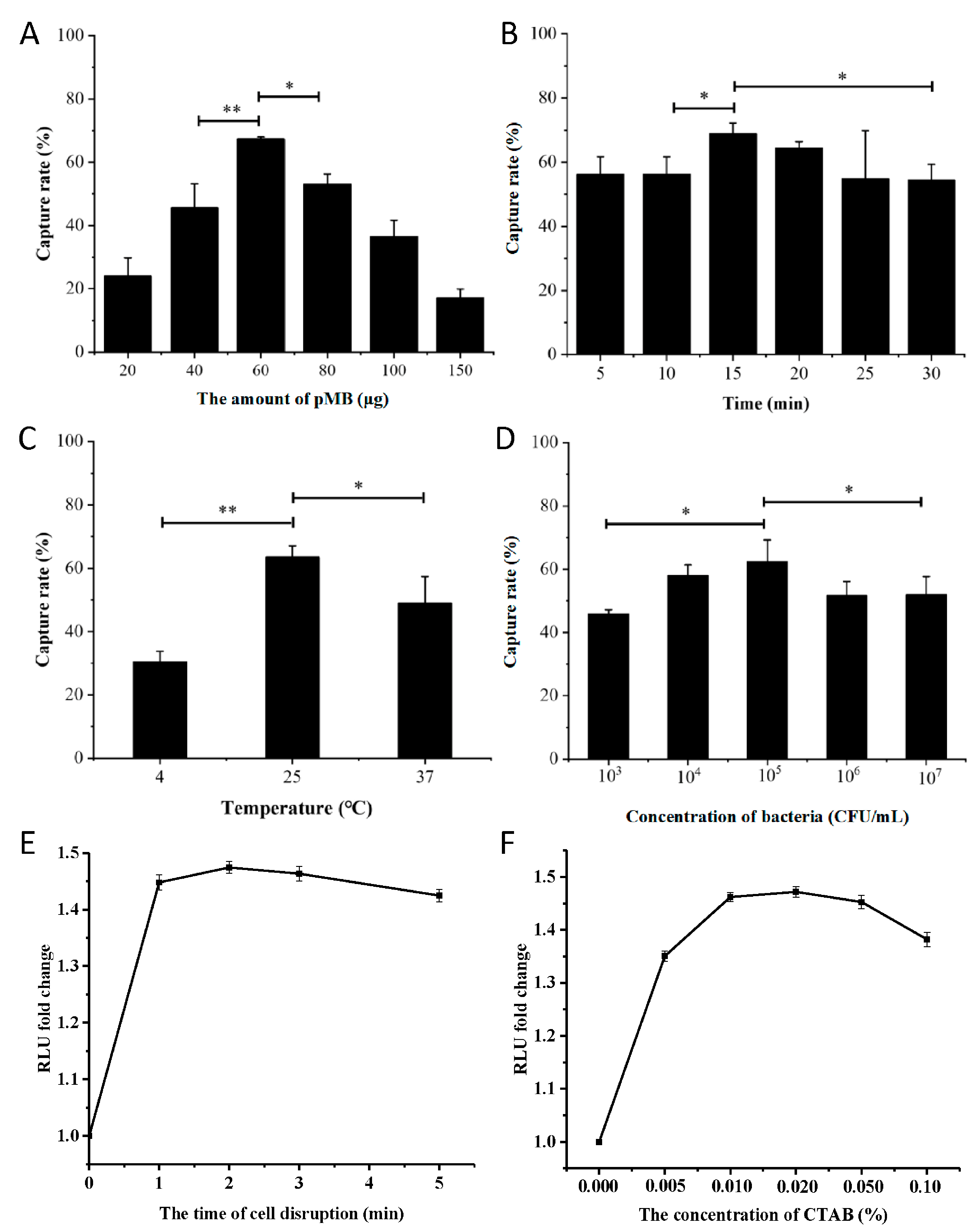
2.14. Detection Performance of pMB under Different Conditions
2.15. Analytical Sensitivity and Specificity of pMB-Based Detection of S. aureus
2.16. Detection of S. aureus in Spiked Samples
2.17. Statistical Analysis
3. Results
3.1. Biological Characteristics of Phage LSA2302
3.2. Genomic Features of Phage LSA2302
3.3. Assembly and Characterization of Phage-Functionalized Magnetic Beads
3.4. Optimization of Conditions for pMB-Based Detection of S. aureus
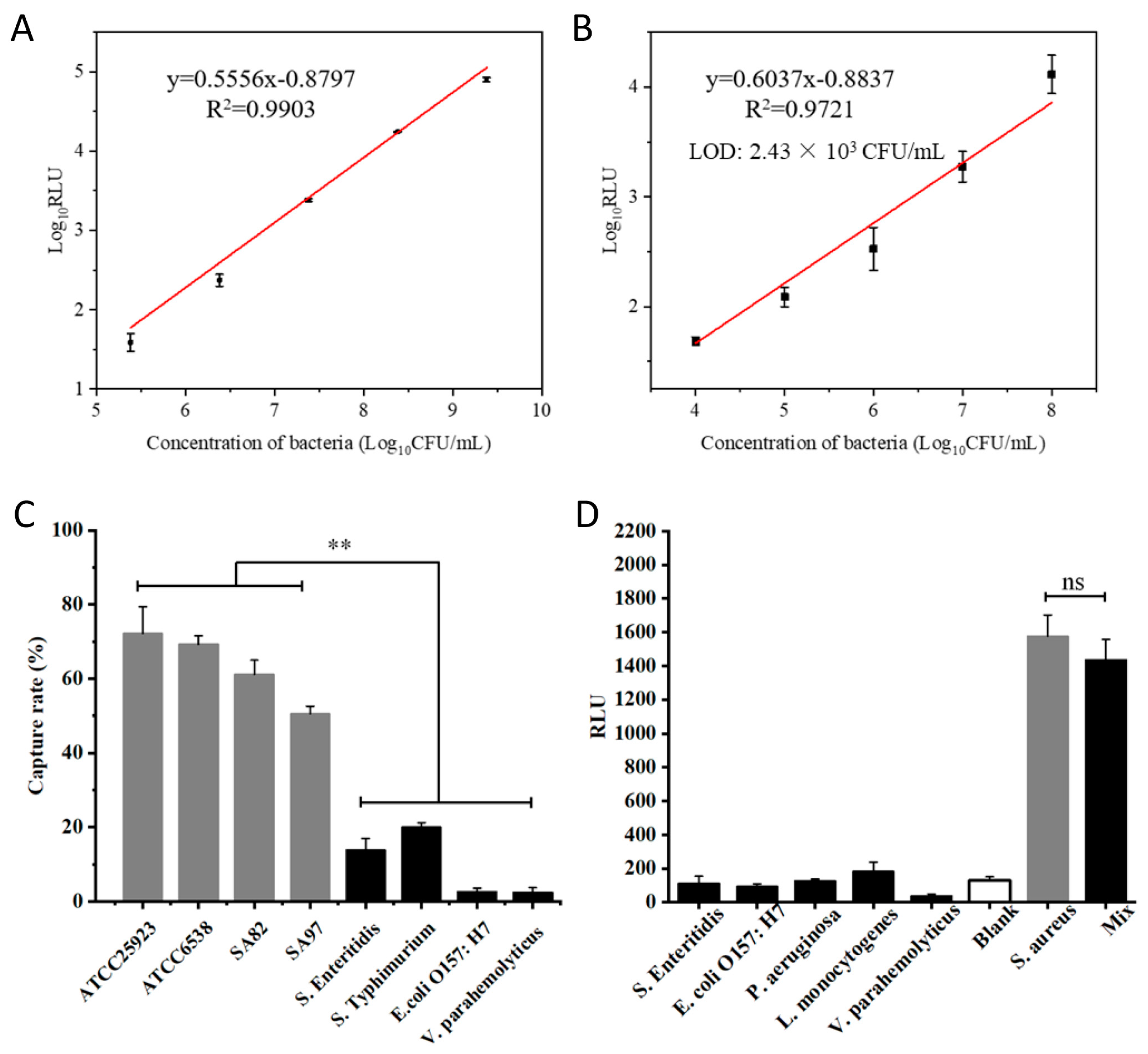
3.5. Analytical Sensitivity and Specificity of pMB-Based Detection of S. aureus
3.6. Evaluation of pMB-Based Detection of S. aureus in Spiked Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Hennekinne, J.-A.; De Buyser, M.-L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef]
- Samani, S.S.; Khojastehnezhad, A.; Ramezani, M.; Alibolandi, M.; Yazdi, F.T.; Mortazavi, S.A.; Khoshbin, Z.; Abnous, K.; Taghdisi, S.M. Ultrasensitive detection of micrococcal nuclease activity and Staphylococcus aureus contamination using optical biosensor technology-A review. Talanta 2021, 226, 122168. [Google Scholar] [CrossRef]
- Li, H.; Li, W.; Dai, Y.; Jiang, Y.; Liang, J.; Wang, S.; Zhuang, M.; Huang, Z.; Xu, L.; Xue, B. Characteristics of Settings and Etiologic Agents of Foodborne Disease Outbreaks—China, 2020. China CDC Wkly. 2021, 3, 889. [Google Scholar] [CrossRef]
- Seaton, R.; Johal, S.; Coia, J.; Reid, N.; Cooper, S.; Jones, B. Economic evaluation of treatment for MRSA complicated skin and soft tissue infections in Glasgow hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 305–311. [Google Scholar] [CrossRef]
- Bhunia, A.K. One day to one hour: How quickly can foodborne pathogens be detected? Fut. Microbiol. 2014, 9, 935–946. [Google Scholar] [CrossRef]
- Foddai, A.C.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Wei, S.; Oh, D.-H. A highly selective enrichment broth combined with real-time PCR for detection of Staphylococcus aureus in food samples. LWT 2018, 94, 103–110. [Google Scholar] [CrossRef]
- Kim, S.-O.; Kim, S.-S. Bacterial pathogen detection by conventional culture-based and recent alternative (polymerase chain reaction, isothermal amplification, enzyme linked immunosorbent assay, bacteriophage amplification, and gold nanoparticle aggregation) methods in food samples: A review. J. Food Saf. 2021, 41, e12870. [Google Scholar] [CrossRef]
- Aliakbar Ahovan, Z.; Hashemi, A.; De Plano, L.M.; Gholipourmalekabadi, M.; Seifalian, A. Bacteriophage Based Biosensors: Trends, Outcomes and Challenges. Nanomaterials 2020, 10, 501. [Google Scholar] [CrossRef]
- Brüssow, H.; Hendrix, R.W. Phage genomics: Small is beautiful. Cell 2002, 108, 13–16. [Google Scholar] [CrossRef]
- Lomakina, G.Y.; Modestova, Y.A.; Ugarova, N.N. Bioluminescence Assay for Cell Viability. Biochemistry 2015, 80, 701–713. [Google Scholar] [CrossRef]
- Li, Z.; Li, W.; Ma, W.; Ding, Y.; Zhang, Y.; Yang, Q.; Wang, J.; Wang, X. Characterization and Application of a Lytic Phage D10 against Multidrug-Resistant Salmonella. Viruses 2021, 13, 1626. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Ding, Y.; Huang, C.; Wang, J.; Wang, J.; Wang, X. Genomic characterization of a novel bacteriophage STP55 revealed its prominent capacity in disrupting the dual-species biofilm formed by Salmonella Typhimurium and Escherichia coli O157: H7 strains. Arch. Microbiol. 2022, 204, 597. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ding, Y.; Li, W.; Zhu, W.; Wang, J.; Wang, X. Application of a Novel Lytic Podoviridae Phage Pu20 for Biological Control of Drug-Resistant Salmonella in Liquid Eggs. Pathogens 2021, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Cheng, M.; Zhai, S.; Xi, H.; Cai, R.; Wang, Z.; Zhang, H.; Wang, X.; Xue, Y.; Li, X.; et al. Preventive effect of the phage VB-SavM-JYL01 on rabbit necrotizing pneumonia caused by Staphylococcus aureus. Vet. Microbiol. 2018, 229, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.; Kropinski, A.M.; Lavigne, R. Bacteriophages; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Sliusarenko, O.; Heinritz, J.; Emonet, T.; Jacobs-Wagner, C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 2011, 80, 612–627. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Stevens, R. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2013, 42, D206–D214. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA Genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016, 45, gkw1004. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Laube, T.; Cortés, P.; Llagostera, M.; Alegret, S.; Pividori, M.I. Phagomagnetic immunoassay for the rapid detection of Salmonella. Appl. Microbiol. Biotechnol. 2014, 98, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Wang, D.; Nugen, S.R.; Chen, J. An Engineered Reporter Phage for the Fluorometric Detection of Escherichia coli in Ground Beef. Microorganisms 2021, 9, 436. [Google Scholar] [CrossRef] [PubMed]
- Jakub, B.; François, E.; Dutilh, B.E.; Margo, B.S.; Edwards, R.A.; Annika, G.; Jochen, K.; Petar, K.; Mart, K.; Kuhn, J.H. Analysis of Spounaviruses as a Case Study for the Overdue Reclassification of Tailed Phages. Syst. Biol. 2020, 69, 110–123. [Google Scholar]
- Wang, S.I.N. Bacteriophage Adsorption Rate and Optimal Lysis Time. Genetics 2008, 180, 471. [Google Scholar]
- Cui, Z.; Feng, T.; Gu, F.; Li, Q.; Dong, K.; Zhang, Y.; Zhu, Y.; Han, L.; Qin, J.; Guo, X. Characterization and complete genome of the virulent Myoviridae phage JD007 active against a variety of Staphylococcus aureus isolates from different hospitals in Shanghai, China. Virol. J. 2017, 14, 26. [Google Scholar] [CrossRef]
- Feng, T.; Leptihn, S.; Dong, K.; Loh, B.; Cui, Z. JD419, a Staphylococcus aureus Phage With a Unique Morphology and Broad Host Range. Front. Microbiol. 2021, 12, 602902. [Google Scholar] [CrossRef]
- Reineke, K.; Mathys, A.; Knorr, D. Shift of pH-Value During Thermal Treatments in Buffer Solutions and Selected Foods. Int. J. Food Prop. 2011, 14, 870–881. [Google Scholar] [CrossRef]
- Ali, A.A.; Altemimi, A.B.; Alhelfi, N.; Ibrahim, S.A. Application of Biosensors for Detection of Pathogenic Food Bacteria: A Review. Biosensors 2020, 10, 58. [Google Scholar] [CrossRef]
- Bal, B.; Nayak, S.; Das, A. Recent advances in molecular techniques for the diagnosis of foodborne diseases. Nanotechnol. Appl. Food 2017, 267–285. [Google Scholar] [CrossRef]
- Paczesny, J.; Richter, U.; Hoyst, R. Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review. Viruses 2020, 12, 845. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Mahboubat, B.Y.; Ding, Y.; Yang, Q.; Wang, J.; Zhou, M.; Wang, X. Development of a rapid Salmonella detection method via phage-conjugated magnetic bead separation coupled with real-time PCR quantification. LWT 2021, 142, 111075. [Google Scholar] [CrossRef]
- Lee, J.; Park, C.; Kim, Y.; Park, S. Signal enhancement in ATP bioluminescence to detect bacterial pathogens via heat treatment. BioChip J. 2017, 11, 287–293. [Google Scholar] [CrossRef]
- Rahman, M.A.; Rahman, A.A.; Islam, M.A.; Alam, M.M. Multi–drug resistant Staphylococcus aureus isolated from milk, chicken meat, beef and egg in Bangladesh. Res. Agric. Livest. Fish. 2018, 5, 175. [Google Scholar] [CrossRef]
- Sadat, A.; Shata, R.R.; Farag, A.M.M.; Ramadan, H.; Alkhedaide, A.; Soliman, M.M.; Elbadawy, M.; Abugomaa, A.; Awad, A. Prevalence and Characterization of PVL-Positive Staphylococcus aureus Isolated from Raw Cow’s Milk. Toxins 2022, 14, 97. [Google Scholar] [CrossRef]
- Yue, X.; Huang, Y.; Zhang, Y.; Ouyang, H.; Xie, J.; Fu, Z. Mycobacteriophage SWU1-Functionalized magnetic particles for facile bioluminescent detection of Mycobacterium smegmatis. Anal. Chim. Acta 2021, 1145, 17–25. [Google Scholar] [CrossRef]
- He, Y.; Wang, M.; Fan, E.; Ouyang, H.; Yue, H.; Su, X.; Liao, G.; Wang, L.; Lu, S.; Fu, Z. Highly Specific Bacteriophage-Affinity Strategy for Rapid Separation and Sensitive Detection of Viable Pseudomonas aeruginosa. Anal. Chem. 2017, 89, 1916–1921. [Google Scholar] [CrossRef]
| Sample | Standard Method | pMB | Recovery (%) | RSD a (%) |
|---|---|---|---|---|
| Skim milk | 1.20 × 107 | 1.19 × 107 | 99.17 | 1.93 |
| 5.60 × 106 | 4.54 × 106 | 81.07 | 1.13 | |
| 4.10 × 105 | 3.68 × 105 | 89.76 | 2.06 | |
| Chicken | 1.70 × 107 | 1.60 × 107 | 94.12 | 2.85 |
| 5.30 × 106 | 4.61 × 106 | 86.98 | 1.64 | |
| 1.30 × 105 | 1.36 × 105 | 104.62 | 3.48 | |
| pre-cultivation b | 2.10 × 105 | 2.07 × 105 c | 98.57 | 4.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Li, Z.; Huang, C.; Ding, Y.; Wang, J.; Wang, X. Phage-Based Biosensing for Rapid and Specific Detection of Staphylococcus aureus. Microorganisms 2023, 11, 2098. https://doi.org/10.3390/microorganisms11082098
Li R, Li Z, Huang C, Ding Y, Wang J, Wang X. Phage-Based Biosensing for Rapid and Specific Detection of Staphylococcus aureus. Microorganisms. 2023; 11(8):2098. https://doi.org/10.3390/microorganisms11082098
Chicago/Turabian StyleLi, Ruining, Zhiwei Li, Chenxi Huang, Yifeng Ding, Jia Wang, and Xiaohong Wang. 2023. "Phage-Based Biosensing for Rapid and Specific Detection of Staphylococcus aureus" Microorganisms 11, no. 8: 2098. https://doi.org/10.3390/microorganisms11082098
APA StyleLi, R., Li, Z., Huang, C., Ding, Y., Wang, J., & Wang, X. (2023). Phage-Based Biosensing for Rapid and Specific Detection of Staphylococcus aureus. Microorganisms, 11(8), 2098. https://doi.org/10.3390/microorganisms11082098









