An Overview of Antimicrobial Resistance in Saudi Arabia (2013–2023) and the Need for National Surveillance
Abstract
1. Introduction
2. Search Strategy
3. Resistant Gram-Positive Pathogens
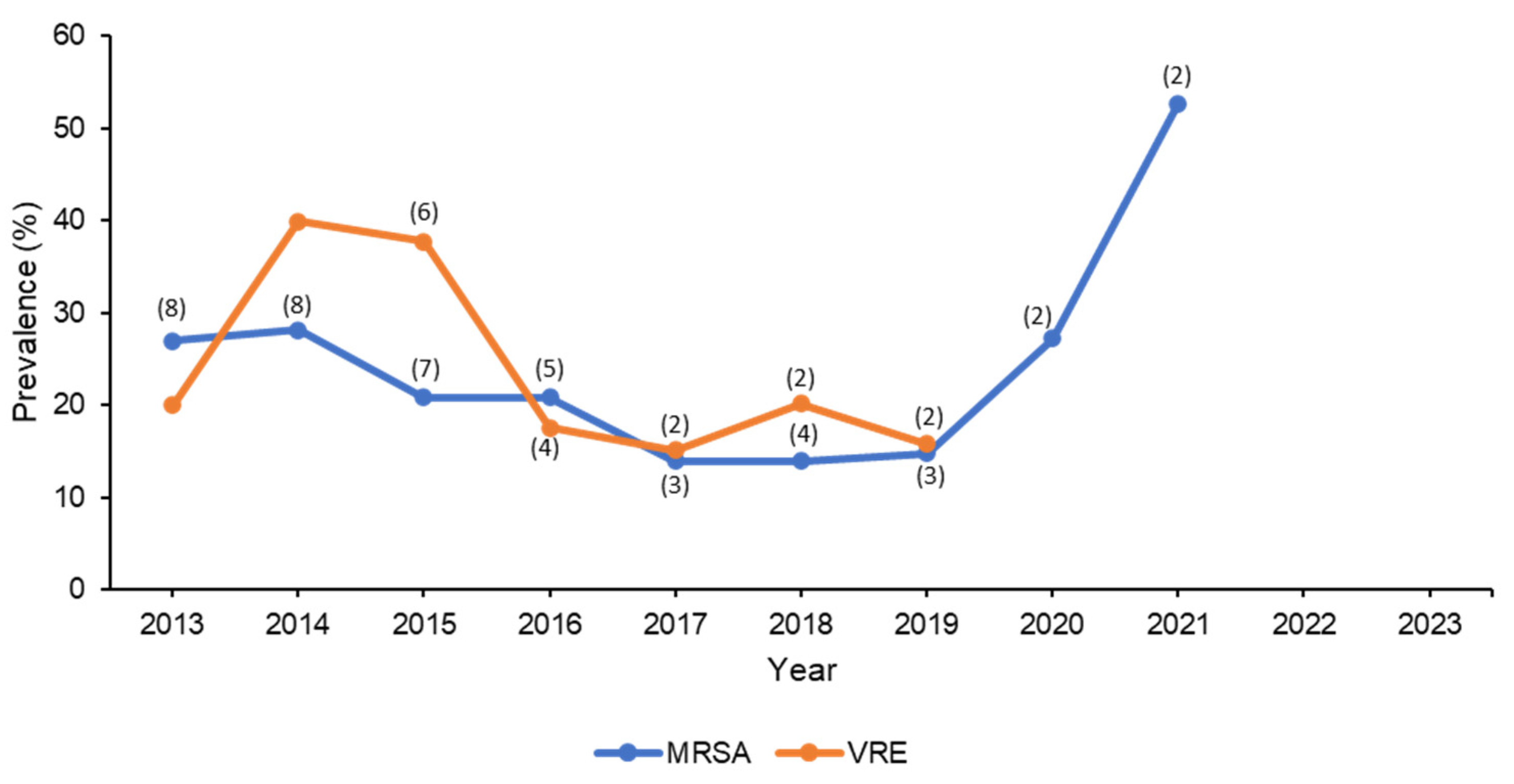
3.1. Methicillin- and Vancomycin-Resistant Staphylococcus aureus
3.2. Vancomycin-Resistant Enterococcus spp.
4. Resistant Gram-Negative Pathogens
4.1. Enterobacterales
4.2. Extended-Spectrum β-Lactamases Producing Enterobacterales
4.3. Carbapenem-Resistant Enterobacterales
4.4. Pseudomonas aeruginosa
4.5. Acinetobacter baumannii
5. Future Directions
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Thabit, A.K.; Crandon, J.L.; Nicolau, D.P. Antimicrobial resistance: Impact on clinical and economic outcomes and the need for new antimicrobials. Expert Opin. Pharmacother. 2015, 16, 159–177. [Google Scholar] [CrossRef]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 25 May 2023).
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically [Document M07-A10]; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2015. [Google Scholar]
- Masterton, R. The importance and future of antimicrobial surveillance studies. Clin. Infect. Dis. 2008, 47 (Suppl. S1), S21–S31. [Google Scholar] [CrossRef]
- Critchley, I.A.; Karlowsky, J.A. Optimal use of antibiotic resistance surveillance systems. Clin. Microbiol. Infect. 2004, 10, 502–511. [Google Scholar] [CrossRef]
- Engelkirk, P.E.; Janet, L. Duben-Engelkirk. Laboratory Diagnosis of Infectious Diseases: Essentials of Diagnostic Microbiology; Lippincott Williams & Wilkins: Baltimore, MD, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Thertieth Informational Supplement [Document M100-S33]; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2023. [Google Scholar]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, ix–xv. [Google Scholar] [CrossRef]
- Riedel, S.; Neoh, K.M.; Eisinger, S.W.; Dam, L.M.; Tekle, T.; Carroll, K.C. Comparison of commercial antimicrobial susceptibility test methods for testing of Staphylococcus aureus and Enterococci against vancomycin, daptomycin, and linezolid. J. Clin. Microbiol. 2014, 52, 2216–2222. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration (Center for Devices and Radiological Health). Guidance for Industry and FDA. Class II Special Controls Guidance Document: Antimicrobial Susceptibility Test (AST) Systems; Silver Spring: Rockville, MD, USA, 2009. [Google Scholar]
- Sader, H.S.; Farrell, D.J.; Flamm, R.K.; Jones, R.N. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn. Microbiol. Infect. Dis. 2014, 78, 443–448. [Google Scholar] [CrossRef]
- Farrell, D.J.; Flamm, R.K.; Sader, H.S.; Jones, R.N. Antimicrobial activity of ceftolozane-tazobactam tested against Enterobacteriaceae and Pseudomonas aeruginosa with various resistance patterns isolated in U.S. Hospitals (2011–2012). Antimicrob. Agents Chemother. 2013, 57, 6305–6310. [Google Scholar] [CrossRef]
- Flamm, R.K.; Mendes, R.E.; Ross, J.E.; Sader, H.S.; Jones, R.N. Linezolid surveillance results for the United States: LEADER surveillance program 2011. Antimicrob. Agents Chemother. 2013, 57, 1077–1081. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Farrell, D.J.; Sader, H.S.; Jones, R.N. AWARE Ceftaroline Surveillance Program (2008–2010): Trends in resistance patterns among Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States. Clin. Infect. Dis. 2012, 55, S187–S193. [Google Scholar] [CrossRef]
- Sader, H.S.; Jones, R.N. Antimicrobial susceptibility of Gram-positive bacteria isolated from US medical centers: Results of the Daptomycin Surveillance Program (2007–2008). Diagn. Microbiol. Infect. Dis. 2009, 65, 158–162. [Google Scholar] [CrossRef]
- Sutherland, C.A.; Verastegui, J.E.; Nicolau, D.P. In vitro potency of amikacin and comparators against E. coli, K. pneumoniae and P. aeruginosa respiratory and blood isolates. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Housman, S.T.; Sutherland, C.A.; Nicolau, D.P. Pharmacodynamic profile of commonly utilised parenteral therapies against meticillin-susceptible and meticillin-resistant Staphylococcus aureus collected from US hospitals. Int. J. Antimicrob. Agents 2014, 44, 235–241. [Google Scholar] [CrossRef]
- Bertrand, X.; Dowzicky, M.J. Antimicrobial susceptibility among gram-negative isolates collected from intensive care units in North America, Europe, the Asia-Pacific Rim, Latin America, the Middle East, and Africa between 2004 and 2009 as part of the Tigecycline Evaluation and Surveillance Trial. Clin. Ther. 2012, 34, 124–137. [Google Scholar]
- Gemmell, C.G. Susceptibility of a variety of clinical isolates to linezolid: A European inter-country comparison. J. Antimicrob. Chemother. 2001, 48, 47–52. [Google Scholar] [CrossRef]
- Queenan, A.M.; Pillar, C.M.; Deane, J.; Sahm, D.F.; Lynch, A.S.; Flamm, R.K.; Peterson, J.; Davies, T.A. Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: Results from CAPITAL Surveillance 2010. Diagn. Microbiol. Infect. Dis. 2012, 73, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Muraki, Y.; Kitamura, M.; Maeda, Y.; Kitahara, T.; Mori, T.; Ikeue, H.; Tsugita, M.; Tadano, K.; Takada, K.; Akamatsu, T.; et al. Nationwide surveillance of antimicrobial consumption and resistance to Pseudomonas aeruginosa isolates at 203 Japanese hospitals in 2010. Infection 2013, 41, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Denton, M.; O’Connell, B.; Bernard, P.; Jarlier, V.; Williams, Z.; Henriksen, A.S. The EPISA study: Antimicrobial susceptibility of Staphylococcus aureus causing primary or secondary skin and soft tissue infections in the community in France, the UK and Ireland. J. Antimicrob. Chemother. 2008, 61, 586–588. [Google Scholar] [CrossRef]
- Sievert, D.M.; Ricks, P.; Edwards, J.R.; Schneider, A.; Patel, J.; Srinivasan, A.; Kallen, A.; Limbago, B.; Fridkin, S.; National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013, 34, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Alhumaid, S.; Al Mutair, A.; Al Alawi, Z.; Alzahrani, A.J.; Tobaiqy, M.; Alresasi, A.M.; Bu-Shehab, I.; Al-Hadary, I.; Alhmeed, N.; Alismail, M. Antimicrobial susceptibility of gram-positive and gram-negative bacteria: A 5-year retrospective analysis at a multi-hospital healthcare system in Saudi Arabia. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 43. [Google Scholar] [CrossRef]
- Farah, S.M.; Alshehri, M.A.; Alfawaz, T.S.; Alasmeri, F.A.; Alageel, A.A.; Alshahrani, D.A. Trends in antimicrobial susceptibility patterns in King Fahad Medical City, Riyadh, Saudi Arabia. Saudi Med. J. 2019, 40, 252. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Al-Motair, K.A.; Alenezi, M.M.; Altheban, A.S.; Hammam, S.A.; Al-Mogbel, M.S. Nosocomial Pathogens-A Single Center Study in Saudi Arabia. J. Pure Appl. Microbiol. 2018, 12, 1411–1416. [Google Scholar] [CrossRef]
- Alam, M.Z.; Alam, Q.; Jiman-Fatani, A.A.; Shukri, H.A.; Haque, A. A surveillance study on the prevalence and antimicrobial resistance pattern among different groups of bacteria isolated from Western province of Saudi Arabia. Biomed. Res. 2017, 28, 898–906. [Google Scholar]
- Al Yousef, S.A. Surveillance of antibiotic-resistant bacteria in King Khalid Hospital, Hafr Al-Batin, Saudi Arabia, during 2013. Jundishapur J. Microbiol. 2016, 9, e19552. [Google Scholar] [CrossRef] [PubMed]
- Al Musawi, S.; Alkhaleefa, Q.; Alnassri, S.; Alamri, A.M.; Alnimr, A. Eleven-Year surveillance of methicillin-resistant Staphylococcus aureus infections at an Academic Health Centre. J. Prev. Med. Hyg. 2022, 63, E132–E138. [Google Scholar] [CrossRef]
- Alarjani, K.M.; Almutairi, A.M.; AlQahtany, F.S.; Soundharrajan, I. Methicillin and multidrug resistant pathogenic Staphylococcus aureus associated sepsis in hospitalized neonatal infections and antibiotic susceptibility. J. Infect. Public Health 2021, 14, 1630–1634. [Google Scholar] [CrossRef] [PubMed]
- Alhussaini, M.S. Methicillin-resistant Staphylococcus aureus nasal carriage among patients admitted at Shaqra general Hospital in Saudi Arabia. Pak. J. Biol. Sci. 2016, 19, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Bazaid, A.S.; Saeed, A.; Alrashidi, A.; Alrashidi, A.; Alshaghdali, K.; Hammam, S.A.; Alreshidi, T.; Alshammary, M.; Alarfaj, A.; Thallab, R. Antimicrobial surveillance for bacterial uropathogens in Ha’il, Saudi Arabia: A Five-year multicenter retrospective study. Infect. Drug Resist. 2021, 14, 1455–1465. [Google Scholar] [CrossRef]
- Eed, E.M.; Ghonaim, M.M.; Hussein, Y.M.; Saber, T.M.; Khalifa, A.S. Phenotypic and molecular characterization of HA-MRSA in Taif hospitals, Saudi Arabia. J. Infect. Dev. Ctries. 2015, 9, 298–303. [Google Scholar] [CrossRef]
- Aljohani, S.; Layqah, L.; Masuadi, E.; Al Alwan, B.; Baharoon, W.; Gramish, J.; Baharoon, S. Occurrence of vancomycin MIC creep in methicillin resistant isolates in Saudi Arabia. J. Infect. Public Health 2020, 13, 1576–1579. [Google Scholar] [CrossRef]
- El-Saed, A.; Balkhy, H.H.; Alshamrani, M.M.; Aljohani, S.; Alsaedi, A.; Al Nasser, W.; El Gammal, A.; Almohrij, S.A.; Alyousef, Z.; Almunif, S. High contribution and impact of resistant gram negative pathogens causing surgical site infections at a multi-hospital healthcare system in Saudi Arabia, 2007–2016. BMC Infect. Dis. 2020, 20, 275. [Google Scholar] [CrossRef] [PubMed]
- Bazzi, A.M.; Rabaan, A.A.; Fawarah, M.M.; Al-Tawfiq, J.A. Prevalence of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus infections in a Saudi Arabian hospital. J. Infect. Public Health 2015, 8, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.E.; Al-Ruwaili, N.M.; El-Masry, E.A.; Saad, A.E.; Taher, I.A. MRSA as an indicator of infection control measures in Turaif General Hospital, Northern Area-Saudi Arabia. J. Infect. Dev. Ctries. 2022, 16, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Al Mutair, A.; Alhumaid, S.; Al Alawi, Z.; Zaidi, A.R.Z.; Alzahrani, A.J.; Al-Tawfiq, J.A.; Al-Shammari, H.; Rabaan, A.A.; Khojah, O.; Al-Omari, A. Five-year resistance trends in pathogens causing healthcare-associated infections at a multi-hospital healthcare system in Saudi Arabia, 2015–2019. J. Glob. Antimicrob. Resist. 2021, 25, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Balkhy, H.H.; El-Saed, A.; Alshamrani, M.M.; Alsaedi, A.; Al Nasser, W.; El Gammal, A.; Aljohany, S.M.; Almunif, S.; Arabi, Y.; Alqahtani, S. Ten-year resistance trends in pathogens causing healthcare-associated infections; reflection of infection control interventions at a multi-hospital healthcare system in Saudi Arabia, 2007–2016. Antimicrob. Resist. Infect. Control 2020, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Said, K.B.; AlGhasab, N.S.; Alharbi, M.S.; Alsolami, A.; Bashir, A.I.; Saleem, M.; Syed Khaja, A.S.; Aldakheel, D.F.; Rakha, E.; Alshamri, J.A. A Sequalae of Lineage Divergence in Staphylococcus aureus from Community-Acquired Patterns in Youth to Hospital-Associated Profiles in Seniors Implied Age-Specific Host-Selection from a Common Ancestor. Diagnostics 2023, 13, 819. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Kelesidis, T.; Tsiodras, S.; Hindler, J.; Humphries, R.M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 2012, 68, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Stosor, V.; Peterson, L.R.; Postelnick, M.; Noskin, G.A. Enterococcus faecium bacteremia: Does vancomycin resistance make a difference? Arch. Intern. Med. 1998, 158, 522–527. [Google Scholar] [CrossRef]
- Farman, M.; Yasir, M.; Al-Hindi, R.R.; Farraj, S.A.; Jiman-Fatani, A.A.; Alawi, M.; Azhar, E.I. Genomic analysis of multidrug-resistant clinical Enterococcus faecalis isolates for antimicrobial resistance genes and virulence factors from the western region of Saudi Arabia. Antimicrob. Resist. Infect. Control 2019, 8, 55. [Google Scholar] [CrossRef]
- Somily, A.M.; Al-Mohizea, M.M.; Absar, M.M.; Fatani, A.J.; Ridha, A.M.; Al-Ahdal, M.N.; Senok, A.C.; Al-Qahtani, A.A. Molecular epidemiology of vancomycin resistant enterococci in a tertiary care hospital in Saudi Arabia. Microb. Pathog. 2016, 97, 79–83. [Google Scholar] [CrossRef]
- Alamri, A.; Hamid, M.E.; Abid, M.; Alwahhabi, A.M.; Alqahtani, K.M.; Alqarni, M.S.; Abomughaid, M. Trend analysis of bacterial uropathogens and their susceptibility pattern: A 4-year (2013–2016) study from Aseer region, Saudi Arabia. Urol. Ann. 2018, 10, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Al Wutayd, O.; Al Nafeesah, A.; Adam, I.; Babikir, I. The antibiotic susceptibility patterns of uropathogens isolated in Qassim, Saudi Arabia. J. Infect. Dev. Ctries. 2018, 12, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Said, K.B.; Alsolami, A.; Khalifa, A.M.; Khalil, N.A.; Moursi, S.; Osman, A.; Fahad, D.; Rakha, E.; Rashidi, M.; Moussa, S.; et al. A multi-point surveillance for antimicrobial resistance profiles among clinical isolates of Gram-negative bacteria recovered from major Ha’il hospitals, Saudi Arabia. Microorganisms 2021, 9, 2024. [Google Scholar] [CrossRef]
- Wani, F.A.; Bandy, A.; Alenzi, M.J.S.; Alzarea, A.I.; Alanazi, A.S.; Sayeed, M.U.; Thirunavukkarasu, A.; Tantry, B.; Dar, M. Resistance patterns of Gram-negative bacteria recovered from clinical specimens of intensive care patients. Microorganisms 2021, 9, 2246. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.E. High antimicrobial resistant rates among Gram-negative pathogens in intensive care units. A retrospective study at a tertiary care hospital in Southwest Saudi Arabia. Saudi Med. J. 2018, 39, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Knothe, H.; Shah, P.; Krcmery, V.; Antal, M.; Mitsuhashi, S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 1983, 11, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Alqasim, A.; Abu Jaffal, A.; Alyousef, A.A. Prevalence of multidrug resistance and extended-spectrum beta-lactamase carriage of clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. Int. J. Microbiol. 2018, 2018, 3026851. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Munoz-Price, L.S. The new β-lactamases. N. Engl. J. Med. 2005, 352, 380–391. [Google Scholar] [CrossRef]
- Bandy, A.; Almaeen, A.H. Pathogenic spectrum of blood stream infections and resistance pattern in Gram-negative bacteria from Aljouf region of Saudi Arabia. PLoS ONE 2020, 15, e0233704. [Google Scholar] [CrossRef]
- Alotaibi, B.S.; Tantry, B.A.; Farhana, A.; Alammar, M.A.; Shah, N.N.; Mohammed, A.H.; Wani, F.; Bandy, A. Resistance pattern in mostly Gram-negative bacteria causing urinary tract infections. Infect. Disord. Drug Targets 2023, 23, 56–64. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; Aljallal, A.; Radwan, H.H.; Shibl, A.M. Characterization of carbapenemases, ESBLs, and plasmid-mediated quinolone determinants in carbapenem-insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J. Infect. Public Health 2018, 11, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Badger-Emeka, L.I.; Al-Sultan, A.A.; Bohol, M.F.F.; Al-Anazi, M.R.; Al-Qahtani, A.A. Genetic analysis, population structure, and characterisation of multidrug-resistant Klebsiella pneumoniae from the Al-Hofuf region of Saudi Arabia. Pathogens 2021, 10, 1097. [Google Scholar] [CrossRef] [PubMed]
- Badger-Emeka, L.I.; Kausar, N.; Estrella, E.; Angeles, G.B. A three-year look at the phylogenetic profile, antimicrobial resistance, and associated virulence genes of uropathogenic Escherichia coli. Pathogens 2022, 11, 631. [Google Scholar] [CrossRef] [PubMed]
- Ejaz, H. Analysis of diverse beta-lactamases presenting high-level resistance in association with OmpK35 and OmpK36 porins in ESBL-producing Klebsiella pneumoniae. Saudi J. Biol. Sci. 2022, 29, 3440–3447. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Rabaan, A.A.; Saunar, J.V.; Bazzi, A.M. Antimicrobial resistance of Gram-negative bacteria: A six-year longitudinal study in a hospital in Saudi Arabia. J. Infect. Public Health 2020, 13, 737–745. [Google Scholar] [CrossRef]
- Alasmary, M.Y. Antimicrobial resistance patterns and ESBL of uropathogens isolated from adult females in Najran region of Saudi Arabia. Clin. Pract. 2021, 11, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Aldrazi, F.A.; Rabaan, A.A.; Alsuliman, S.A.; Aldrazi, H.A.; Alabdalslam, M.J.; Alsadiq, S.A.; Alhani, H.M.; Bueid, A.S. ESBL expression and antibiotic resistance patterns in a hospital in Saudi Arabia: Do healthcare staff have the whole picture? J. Infect. Public Health 2020, 13, 759–766. [Google Scholar] [CrossRef]
- Kabrah, A. Extended-spectrum beta-lactamase and carbapenem-resistant Gram-negative pathogens in Makkah, Saudi Arabia. Ethiop. J. Health Sci. 2022, 32, 1221–1230. [Google Scholar] [CrossRef]
- Yasir, M.; Ajlan, A.M.; Shakil, S.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Farman, M.; Baazeem, Z.M.; Baabdullah, R.; Alawi, M.; Al-Abdullah, N.; et al. Molecular characterization, antimicrobial resistance and clinico-bioinformatics approaches to address the problem of extended-spectrum beta-lactamase-producing Escherichia coli in western Saudi Arabia. Sci. Rep. 2018, 8, 14847. [Google Scholar] [CrossRef]
- Al-Garni, S.M.; Ghonaim, M.M.; Ahmed, M.M.M.; Al-Ghamdi, A.S.; Ganai, F.A. Risk factors and molecular features of extended-spectrum beta-lactamase producing bacteria at southwest of Saudi Arabia. Saudi. Med. J. 2018, 39, 1186–1194. [Google Scholar] [CrossRef]
- Taher, I.; Almaeen, A.; Aljourfi, H.; Bohassan, E.; Helmy, A.; El-Masry, E.; Saleh, B.; Aljaber, N. Surveillance of antibiotic resistance among uropathogens in Aljouf region northern Saudi Arabia. Iran J. Microbiol. 2019, 11, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Abalkhail, A.; AlYami, A.S.; Alrashedi, S.F.; Almushayqih, K.M.; Alslamah, T.; Alsalamah, Y.A.; Elbehiry, A. The prevalence of multidrug-resistant Escherichia coli producing ESBL among male and female patients with urinary tract infections in Riyadh Region, Saudi Arabia. Healthcare 2022, 10, 1778. [Google Scholar] [CrossRef] [PubMed]
- Mashwal, F.A.; El Safi, S.H.; George, S.K.; Adam, A.A.; Jebakumar, A.Z. Incidence and molecular characterization of the extended spectrum beta lactamase-producing Escherichia coli isolated from urinary tract infections in Eastern Saudi Arabia. Saudi Med. J. 2017, 38, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Abdel Azim, N.S.; Al-Harbi, M.A.; Al-Zaban, M.I.; Nofal, M.Y.; Somily, A.M. Prevalence and antibiotic susceptibility among Gram-negative bacteria isolated from intensive care units at a tertiary care hospital in Riyadh, Saudi Arabia. J. Pure Appl. Microbiol. 2019, 13, 201–208. [Google Scholar] [CrossRef]
- Aldawsari, A.; Tawfik, K.; Al-Zaagi, I., Sr. Antimicrobial-resistant bacteria and prescription of antibiotics at a tertiary care hospital in Riyadh, Saudi Arabia. Cureus 2020, 12, e12098. [Google Scholar] [CrossRef] [PubMed]
- Shami, A.; Al-Mijalli, S.; Somily, A.; Almasri, R.; Alsalem, R.; Abdurahim, S.A. Incidence and susceptibility patterns of urine bacterial flora in young Saudi females. J. Pure Appl. Microbiol. 2022, 16, 2791–2801. [Google Scholar] [CrossRef]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm. Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Eltahlawi, R.A.; Jiman-Fatani, A.; Gad, N.M.; Ahmed, S.H.; Al-Rabia, M.W.; Zakai, S.; Kharaba, A.; El-Hossary, D. Detection of carbapenem-resistance in CRE by comparative assessment of RAPIDEC((R)) CARBA NP and XpertCarba-R assay. Infect. Drug Resist. 2023, 16, 1123–1131. [Google Scholar] [CrossRef]
- Khan, M.A.; Mohamed, A.M.; Faiz, A.; Ahmad, J. Enterobacterial infection in Saudi Arabia: First record of Klebsiella pneumoniae with triple carbapenemase genes resistance. J. Infect. Dev. Ctries. 2019, 13, 334–341. [Google Scholar] [CrossRef]
- Abd El Ghany, M.; Sharaf, H.; Al-Agamy, M.H.; Shibl, A.; Hill-Cawthorne, G.A.; Hong, P.Y. Genomic characterization of NDM-1 and 5, and OXA-181 carbapenemases in uropathogenic Escherichia coli isolates from Riyadh, Saudi Arabia. PLoS ONE 2018, 13, e0201613. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Rabaan, A.A.; Saunar, J.V.; Bazzi, A.M. Genotypes and prevalence of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa in a hospital in Saudi Arabia. Trans. R Soc. Trop. Med. Hyg. 2022, 116, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdely, H.; AlHababi, R.; Dada, H.M.; Roushdy, H.; Alanazi, M.M.; Alessa, A.A.; Gad, N.M.; Alasmari, A.M.; Radwan, E.E.; Al-Dughmani, H.; et al. Molecular characterization of carbapenem-resistant Enterobacterales in thirteen tertiary care hospitals in Saudi Arabia. Ann. Saudi Med. 2021, 41, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Taha, R.; Mowallad, A.; Mufti, A.; Althaqafi, A.; Jiman-Fatani, A.A.; El-Hossary, D.; Ossenkopp, J.; AlhajHussein, B.; Kaaki, M.; Jawi, N.; et al. Prevalence of carbapenem-resistant Enterobacteriaceae in western Saudi Arabia and increasing trends in the antimicrobial aesistance of Enterobacteriaceae. Cureus 2023, 15, e35050. [Google Scholar] [CrossRef] [PubMed]
- Alraddadi, B.M.; Heaphy, E.L.G.; Aljishi, Y.; Ahmed, W.; Eljaaly, K.; Al-Turkistani, H.H.; Alshukairi, A.N.; Qutub, M.O.; Alodini, K.; Alosaimi, R.; et al. Molecular epidemiology and outcome of carbapenem-resistant Enterobacterales in Saudi Arabia. BMC Infect. Dis. 2022, 22, 542. [Google Scholar] [CrossRef] [PubMed]
- Al-Zahrani, I.A.; Alsiri, B.A. The emergence of carbapenem-resistant Klebsiella pneumoniae isolates producing OXA-48 and NDM in the Southern (Asir) province, Saudi Arabia. Saudi Med. J. 2018, 39, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Strateva, T.; Yordanov, D. Pseudomonas aeruginosa—A phenomenon of bacterial resistance. J. Med. Microbiol. 2009, 58, 1133–1148. [Google Scholar] [CrossRef] [PubMed]
- Alhussain, F.A.; Yenugadhati, N.; Al Eidan, F.A.; Al Johani, S.; Badri, M. Risk factors, antimicrobial susceptibility pattern and patient outcomes of Pseudomonas aeruginosa infection: A matched case-control study. J. Infect. Public Health 2021, 14, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Alshammari, H.O.; Somily, A.; Yahia Qattan, M.; Alsubki, R.A.; Moussa, I.M. Susceptibility pattern of multi-drug resistance Pseudomonas aeruginosa isolates from tertiary care hospital in Riyadh, KSA. J. King Saud. Univ. Sci. 2023, 35, 102702. [Google Scholar] [CrossRef]
- Hafiz, T.A.; Bin Essa, E.A.; Alharbi, S.R.; Alyami, A.S.; Alkudmani, Z.S.; Mubaraki, M.A.; Alturki, N.A.; Alotaibi, F. Epidemiological, microbiological, and clinical characteristics of multi-resistant Pseudomonas aeruginosa isolates in King Fahad Medical City, Riyadh, Saudi Arabia. Trop. Med. Infect. Dis. 2023, 8, 205. [Google Scholar] [CrossRef]
- Khan, M.A.; Faiz, A. Antimicrobial resistance patterns of Pseudomonas aeruginosa in tertiary care hospitals of Makkah and Jeddah. Ann. Saudi Med. 2016, 36, 23–28. [Google Scholar] [CrossRef]
- Towner, K.J. Acinetobacter: An old friend, but a new enemy. J. Hosp. Infect. 2009, 73, 355–363. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Karageorgopoulos, D.E. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: Need for international harmonization in terminology. Clin. Infect. Dis. 2008, 46, 1121–1122. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Husain, S.; Potoski, B.A.; McCurry, K.R.; Paterson, D.L. Extensively drug-resistant Acinetobacter baumannii. Emerg. Infect. Dis. 2009, 15, 980–982. [Google Scholar] [CrossRef] [PubMed]
- Marie, M.A.; Krishnappa, L.G.; Alzahrani, A.J.; Mubaraki, M.A.; Alyousef, A.A. A prospective evaluation of synergistic effect of sulbactam and tazobactam combination with meropenem or colistin against multidrug resistant Acinetobacter baumannii. Bosn. J. Basic Med. Sci. 2015, 15, 24–29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shah, M.W.; Yasir, M.; Farman, M.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Alawi, M.; El-Hossary, D.; Azhar, E.I. Antimicrobial susceptibility and molecular characterization of clinical strains of Acinetobacter baumannii in Western Saudi Arabia. Microb. Drug Resist. 2019, 25, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Syed Khaja, A.S.; Hossain, A.; Alenazi, F.; Said, K.B.; Moursi, S.A.; Almalaq, H.A.; Mohamed, H.; Rakha, E. Molecular characterization and antibiogram of Acinetobacter baumannii clinical isolates recovered from the patients with ventilator-associated pneumonia. Healthcare 2022, 10, 2210. [Google Scholar] [CrossRef] [PubMed]
- Hafiz, T.A.; Alghamdi, S.S.; Mubaraki, M.A.; Alghamdi, S.S.M.; Alothaybi, A.; Aldawood, E.; Alotaibi, F. A two-year retrospective study of multidrug-resistant Acinetobacter baumannii respiratory infections in critically Ill patients: Clinical and microbiological findings. J. Infect. Public Health 2023, 16, 313–319. [Google Scholar] [CrossRef]
- Aloraifi, R.I.; Alharthi, A.F.; Almefleh, A.A.; Alamri, A.H.; Alobud, A.S.; Bawazeer, R.A.; Alswaji, A.A.; Alalwan, B.; Aldriwesh, M.G.; Al Johani, S.M.; et al. Prevalence of carbapenem non-susceptible Gram-negative bacteria at tertiary care hospitals in Saudi Arabia. Cureus 2023, 15, e33767. [Google Scholar] [CrossRef]
- AlAmri, A.M.; AlQurayan, A.M.; Sebastian, T.; AlNimr, A.M. Molecular surveillance of multidrug-resistant Acinetobacter baumannii. Curr. Microbiol. 2020, 77, 335–342. [Google Scholar] [CrossRef]
- Gill, C.M.; Aktathorn, E.; Alfouzan, W.; Bourassa, L.; Brink, A.; Burnham, C.D.; Canton, R.; Carmeli, Y.; Falcone, M.; Kiffer, C.; et al. Multicenter, prospective validation of a phenotypic algorithm to guide carbapenemase testing in carbapenem-resistant Pseudomonas aeruginosa using the ERACE-PA global surveillance program. Open Forum Infect. Dis. 2022, 9, ofab617. [Google Scholar] [CrossRef] [PubMed]
- Saudi Vision 2030. Available online: http://vision2030.gov.sa/en (accessed on 8 May 2017).
- Alsowaida, Y.S.; Thabit, A.K.; Almangour, T.A.; Bin Saleh, K.; Mahrous, A.; Saeed Almutairi, M.; Alshehail, B.; Aljefri, D.; Mohzari, Y.; Alfahad, W.; et al. Infectious diseases pharmacy practice, education, and research in Saudi Arabia: A review and future perspectives by the Infectious Diseases Pharmacy Specialty Network at the Saudi Society of Clinical Pharmacy. Saudi Pharm. J. 2022, 30, 1836–1843. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Antimicrobial Resistance Surveillance System: Manual for Early Implementation; World Health Organization: Geneva, Switzerland, 2015.
- The Center for Disease Dynamics, E.a.P. ResistanceMap. Available online: https://resistancemap.onehealthtrust.org/ (accessed on 28 May 2017).


| Antibiotic | Staphylococcus aureus (n = 5650) | MRSA (n = 3097) | VRE (n = 24) | Enterococcus Species * (n = 2266) | Enterococcus faecalis (n = 149) |
|---|---|---|---|---|---|
| Amoxicillin/clavulanic acid | 72.1 | 16.9 | - | - | - |
| Amikacin | - | 23 | - | - | - |
| Ampicillin | 47.4 | 5.2 | 100 | 83 | 95 |
| Azithromycin | 50 | - | - | - | - |
| Cefazolin | 100 | 100 | - | - | - |
| Ceftriaxone | 66.7 | - | - | - | - |
| Cefotaxime | - | 14.4 | - | - | - |
| Cefoxitin | 56.9 | 11.5 ** | 100 | 29.7 | - |
| Chloramphenicol | 54.6 | - | - | - | - |
| Ciprofloxacin | 74 | 58.6 | - | 35.9 | 67 |
| Clindamycin | 88.9 | 62.4 | 28.6 | 27.9 | 5 |
| Cloxacillin | 82.9 | 25.6 | - | 82.6 | - |
| Daptomycin | 100 | 97.9 | - | - | - |
| Doxycycline | 53.4 | 51 | - | 17.7 | - |
| Erythromycin | 73.9 | 52.6 | - | 28.8 | 9 |
| Fosfomycin | 90.5 | 100 | - | - | - |
| Fusidic Acid | 94.1 | 58.4 | - | - | - |
| Gentamicin | 86.2 | 59.9 | - | 37.4 | 50 |
| Imipenem | 36.8 | 18.2 | - | - | - |
| Levofloxacin | 76.5 | 49.3 | 71.4 | 64 | 55 |
| Linezolid | 80.5 | 86.8 | 87.9 | 97.3 | 97 |
| Moxifloxacin | 83 | 44.9 | 85.7 | - | 55 |
| Mupirocin | 80.9 | 96.6 | - | - | - |
| Nitrofurantoin | 84.7 | 68.1 | 43 | 80.9 | 95 |
| Oxacillin | 78.3 | 4.3 ** | - | - | - |
| Penicillin | 50.1 | 0.3 | 85.7 | - | 85 |
| Quinupristin | 100 | - | - | - | 5 |
| Rifampin | 94.1 | 84 | - | - | - |
| Streptomycin | - | - | - | - | 70 |
| Quinupristin/dalfopristin | 94.1 | - | - | - | - |
| Teicoplanin | 97.1 | 91 | 38.5 | - | - |
| Tigecycline | 86.2 | 62.5 | 100 | 100 | |
| Tetracycline | 75.2 | 69.2 | - | 35.3 | 85 |
| Tobramycin | 54.5 | - | - | - | |
| Trimethoprim/sulfamethoxazole | 82.6 | 55.6 | 28.6 | 67.4 | - |
| Vancomycin | 87.7 | 86.9 | 0 | 93 | 97 |
| Antibiotic | Escherichia coli (n = 50,589) | Klebsiella pneumoniae (n = 18,539) | Klebsiella aerogenes (n = 858) | Klebsiella oxytoca (n = 739) | Proteus mirabilis (n = 5604) | Providencia stuartii (n = 1733) | Morganella morganii (n = 1754) | Enterobacter cloacae (n = 1363) | Citrobacter freundii (n = 596) |
|---|---|---|---|---|---|---|---|---|---|
| Amikacin | 82.5 | 81.3 | 30 | 94.7 | 58 | 76.6 | 73 | 72.3 | 88.1 |
| Amoxicillin/ clavulanic acid | 58.8 | 52.5 | 12.2 | 33.3 | 49.9 | 19.6 | 24.4 | 40 | 9.1 |
| Ampicillin | 33.4 | 17.3 | 6.5 | 5.9 | 25.3 | 16.1 | 4.9 | 27 | 0 |
| Cefepime | 62.3 | 59.4 | 9.8 | 60.7 | 38.5 | 38.4 | 14.5 | 47.9 | 45.7 |
| Cefoxitin | 77.2 | 69.4 | 53.5 | 66.7 | 71.4 | 73.1 | 73.1 | 28 | 41.5 |
| Ceftazidime | 65.2 | 57.9 | 12.5 | 26 | 36.5 | 37.3 | 12.8 | 57.3 | 37.6 |
| Ceftriaxone | 50.9 | 44.1 | - | - | 20.2 | 54.5 | - | - | - |
| Cefuroxime | 46.4 | 43.8 | - | - | 27.4 | 27.3 | 0 | 45.5 | - |
| Ciprofloxacin | 58.5 | 59.9 | 11.1 | 11.9 | 35.6 | 18.7 | 38.4 | 70.2 | 54.2 |
| Colistin | 79.3 | 83.7 | 53.8 | 71.4 | 1.6 | 1.5 | 0 | 60.9 | 93.3 |
| Ertapenem | 82.4 | 65.7 | 66.7 | 100 | 72.8 | 77.2 | - | 87.5 | 85.7 |
| Gentamicin | 74.2 | 69.4 | 28 | 58.2 | 39.1 | 21.7 | 31.3 | 70.3 | 58.6 |
| Imipenem | 88.5 | 75.6 | 94.2 | 98.1 | 67.2 | 54.9 | 95.6 | 95.9 | 71 |
| Levofloxacin | 51.9 | 60.3 | 20.5 | 28.9 | 20 | 19.5 | 13.1 | 55.6 | 37.9 |
| Meropenem | 89.1 | 73.8 | 100 | 100 | 82.2 | 73.1 | 94.1 | 50 | 42.9 |
| Nitrofurantoin | 77.5 | 44.2 | 33.3 | 38.7 | 19.7 | 7 | 3 | 61 | 63.1 |
| Piperacillin/ tazobactam | 77.7 | 64.4 | 58.5 | 70.8 | 70.8 | 72 | 53.3 | 75.2 | 65.2 |
| Tigecycline | 85.4 | 76.4 | 91.7 | 100 | 0 | 48.4 | 50 | 82.6 | 91.7 |
| Trimethoprim/ sulfamethoxazole | 51.5 | 54.7 | 40 | 23.1 | 25.9 | 33 | 38.7 | 52.6 | 29.3 |
| Antibiotic | Escherichia coli (Suspected ESBL) (n = 152) | Escherichia coli (Confirmed ESBL) (n = 7277) | Klebsiella pneumoniae (Confirmed ESBL) (n = 1382) |
|---|---|---|---|
| Amikacin | 87.2 | 84.6 | 83.2 |
| Amoxicillin | 0 | - | - |
| Amoxicillin/clavulanic acid | 38.7 | 39.7 | 41.6 |
| Ampicillin | 6 | 8.8 | 16.1 |
| Aztreonam | 17.3 | 5.2 | 0 |
| Cefepime | 30.2 | 28.7 | 33.3 |
| Cefoxitin | 61.5 | 78.2 | 87.7 |
| Ceftazidime | 26.3 | 14.5 | 9.6 |
| Ceftriaxone | 25.7 | 22.7 | - |
| Cefuroxime | 3.7 | - | - |
| Ciprofloxacin | 23.6 | 32.5 | 49.1 |
| Gentamicin | 51.3 | 60.1 | 46.2 |
| Imipenem | 91.7 | 90.4 | 97.9 |
| Levofloxacin | 17.4 | - | - |
| Meropenem | 98 | 88 | 99.1 |
| Nitrofurantoin | 66 | 79.1 | 38.4 |
| Piperacillin | 0 | - | - |
| Piperacillin/tazobactam | 70.9 | 68.2 | 71.3 |
| Tigecycline | 100 | 75.3 | 72 |
| Trimethoprim/sulfamethoxazole | 22.6 | 39.9 | 36.1 |
| Antibiotic | Escherichia coli (Suspected CRE) (n = 41) | Escherichia coli (Confirmed CRE) (n = 106) | Klebsiella pneumoniae (Suspected CRE) (n = 42) | Klebsiella pneumoniae (Confirmed CRE) (n = 337) | Klebsiella pneumoniae (Suspected CRE/ESBL) (n = 78) |
|---|---|---|---|---|---|
| Amikacin | 100 | 62.1 | 71.4 | 52.4 | 42.3 |
| Amoxicillin/clavulanic acid | - | 0 | - | 0 | 9 |
| Cefepime | 60.9 | 2 | 26.2 | 0.33 | 24.4 |
| Cefotaxime | 48.8 | 25 | 21.4 | 0 | - |
| Ceftazidime | 68.3 | 1.6 | 19 | 3.8 | 8.7 |
| Ceftriaxone | 48.8 | 4 | 21.4 | 0.5 | 26 |
| Ciprofloxacin | - | 29.5 | - | 3.9 | 23 |
| Colistin | 100 | 97.5 | 100 | 92 | - |
| Gentamicin | 53.7 | 44.3 | 52.3 | 25.8 | 55 |
| Imipenem | 95.1 | 21.4 | 50 | 11.1 | 75.6 |
| Meropenem | 95.1 | 28.3 | 50 | 11.8 | 70.5 |
| Piperacillin/tazobactam | - | 0 | - | 0 | 24.4 |
| Tigecycline | - | 92.2 | - | 89.8 | - |
| Trimethoprim/sulfamethoxazole | - | 52.1 | - | 7 | 29 |
| Antibiotic | Pseudomonas aeruginosa (n = 8995) | Acinetobacter baumannii * (n = 1082) |
|---|---|---|
| Amikacin | 63.85 | 36.3 |
| Aztreonam | 48.45 | - |
| Cefepime | 52.1 | - |
| Ceftazidime | 50.4 | - |
| Ciprofloxacin | 54.3 | 1.25 |
| Colistin | 79.25 | 97.5 |
| Gentamicin | 51.79 | 16 |
| Imipenem | 50.7 | 17.6 |
| Levofloxacin | 40.2 | 2.8 |
| Meropenem | 32.84 | 14.4 |
| Piperacillin/tazobactam | 60.3 | - |
| Tigecycline | - | 21.4 |
| Tobramycin | 80 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thabit, A.K.; Alabbasi, A.Y.; Alnezary, F.S.; Almasoudi, I.A. An Overview of Antimicrobial Resistance in Saudi Arabia (2013–2023) and the Need for National Surveillance. Microorganisms 2023, 11, 2086. https://doi.org/10.3390/microorganisms11082086
Thabit AK, Alabbasi AY, Alnezary FS, Almasoudi IA. An Overview of Antimicrobial Resistance in Saudi Arabia (2013–2023) and the Need for National Surveillance. Microorganisms. 2023; 11(8):2086. https://doi.org/10.3390/microorganisms11082086
Chicago/Turabian StyleThabit, Abrar K., Afaq Y. Alabbasi, Faris S. Alnezary, and Imtinan A. Almasoudi. 2023. "An Overview of Antimicrobial Resistance in Saudi Arabia (2013–2023) and the Need for National Surveillance" Microorganisms 11, no. 8: 2086. https://doi.org/10.3390/microorganisms11082086
APA StyleThabit, A. K., Alabbasi, A. Y., Alnezary, F. S., & Almasoudi, I. A. (2023). An Overview of Antimicrobial Resistance in Saudi Arabia (2013–2023) and the Need for National Surveillance. Microorganisms, 11(8), 2086. https://doi.org/10.3390/microorganisms11082086






