Limited Impact of Soil Microorganisms on the Endophytic Bacteria of Tartary Buckwheat (Fagopyrum tataricum)
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sterilization and Source of Tartary Buckwheat
2.2. Sample Collection and Preparation
2.3. DNA Extraction and Sequencing
2.4. Sequence Processing and Statistical Analysis
2.5. Circos and Co-Occurrence Network
3. Results
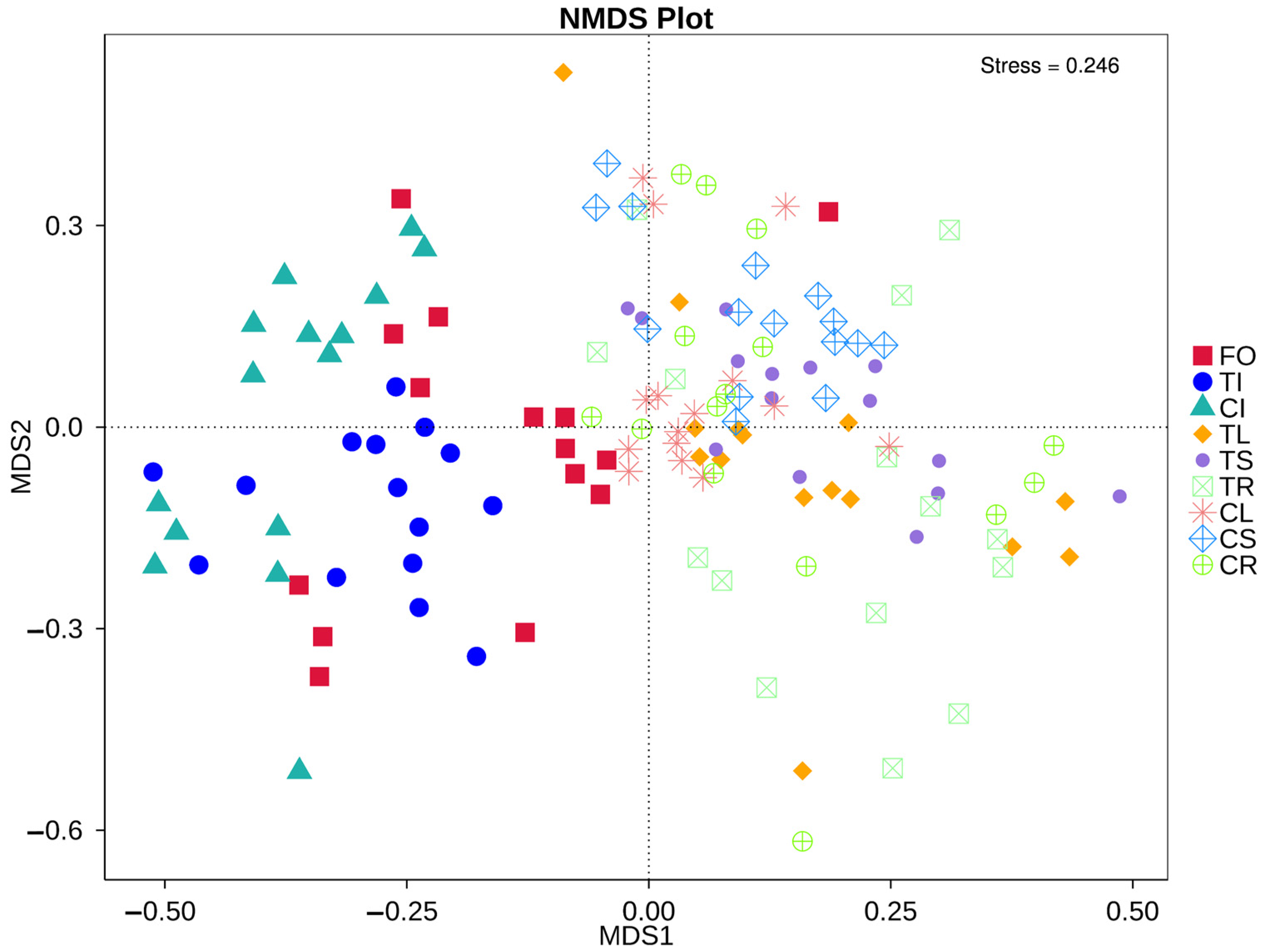
3.1. Comparison of Alpha Diversity in Different Tissues of Tartary Buckwheat
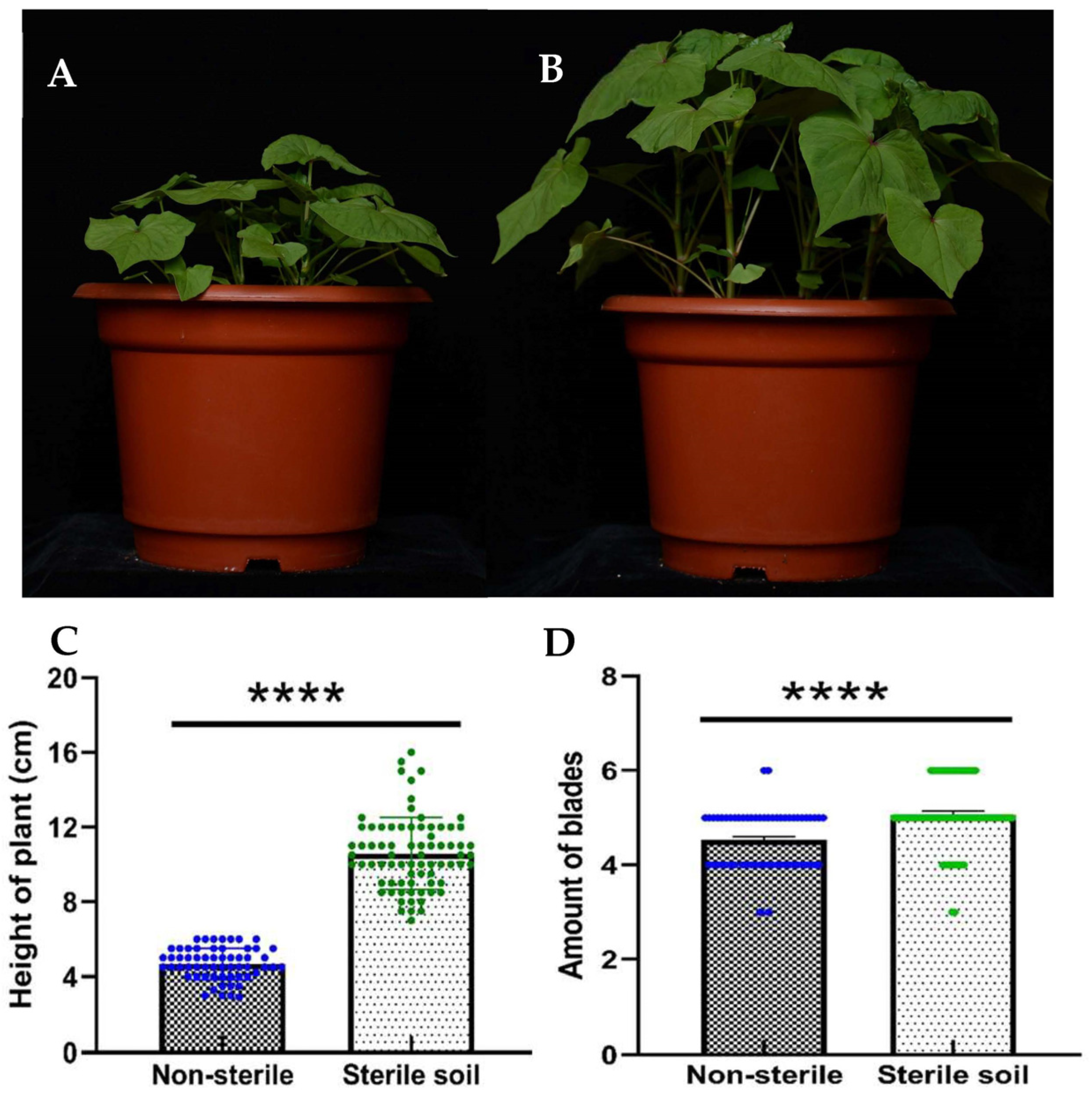
3.2. Comparison of Plant Height and Leaf Number of Tartary Buckwheat Grown in Sterilized and Non-Sterilized Soil
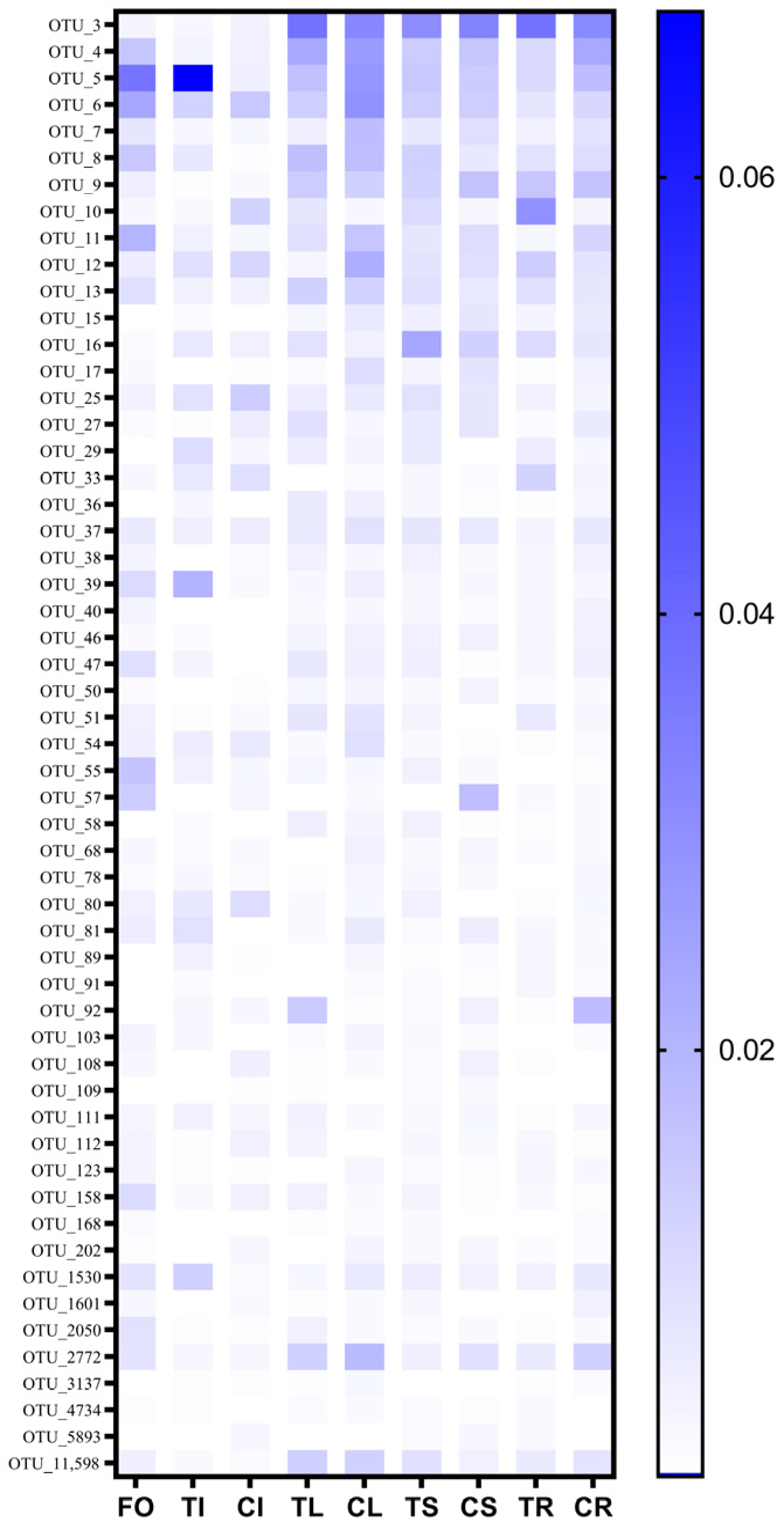
3.3. Assembly of Microbial Community in Different Plant Parts and Core Microbiome Identification
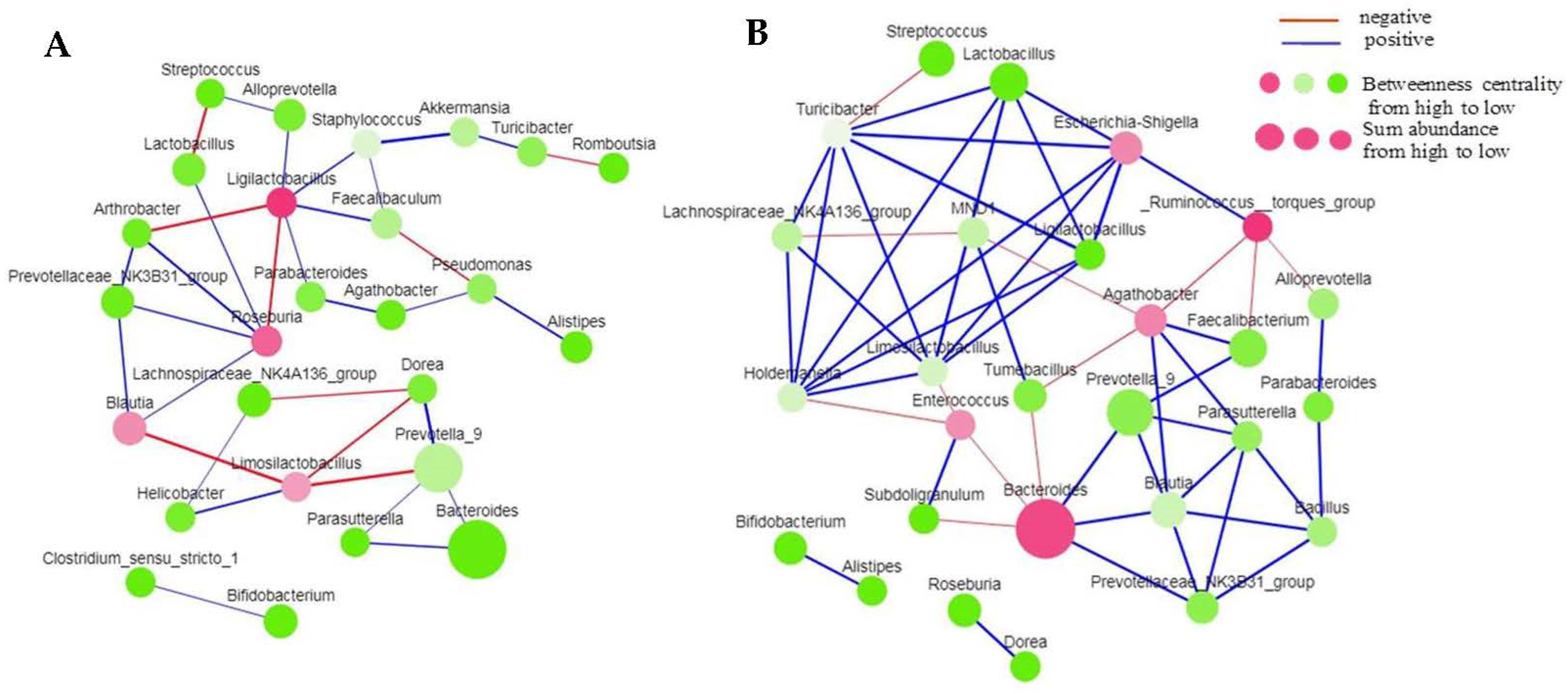
3.4. Co-Occurrence Network of the Bacterial Community in Different Tissues of Tartary Buckwheat
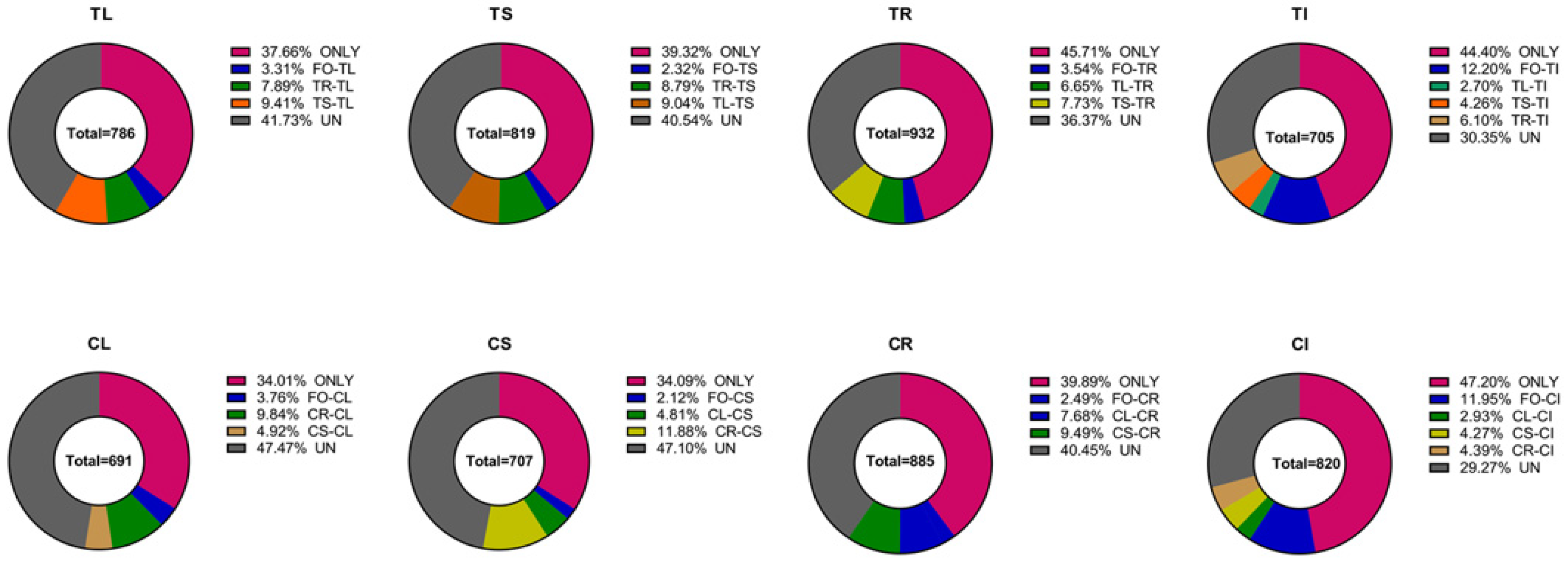
3.5. The Proportion of Endophytic Bacteria in Different Tissues of Tartary Buckwheat
4. Discussion
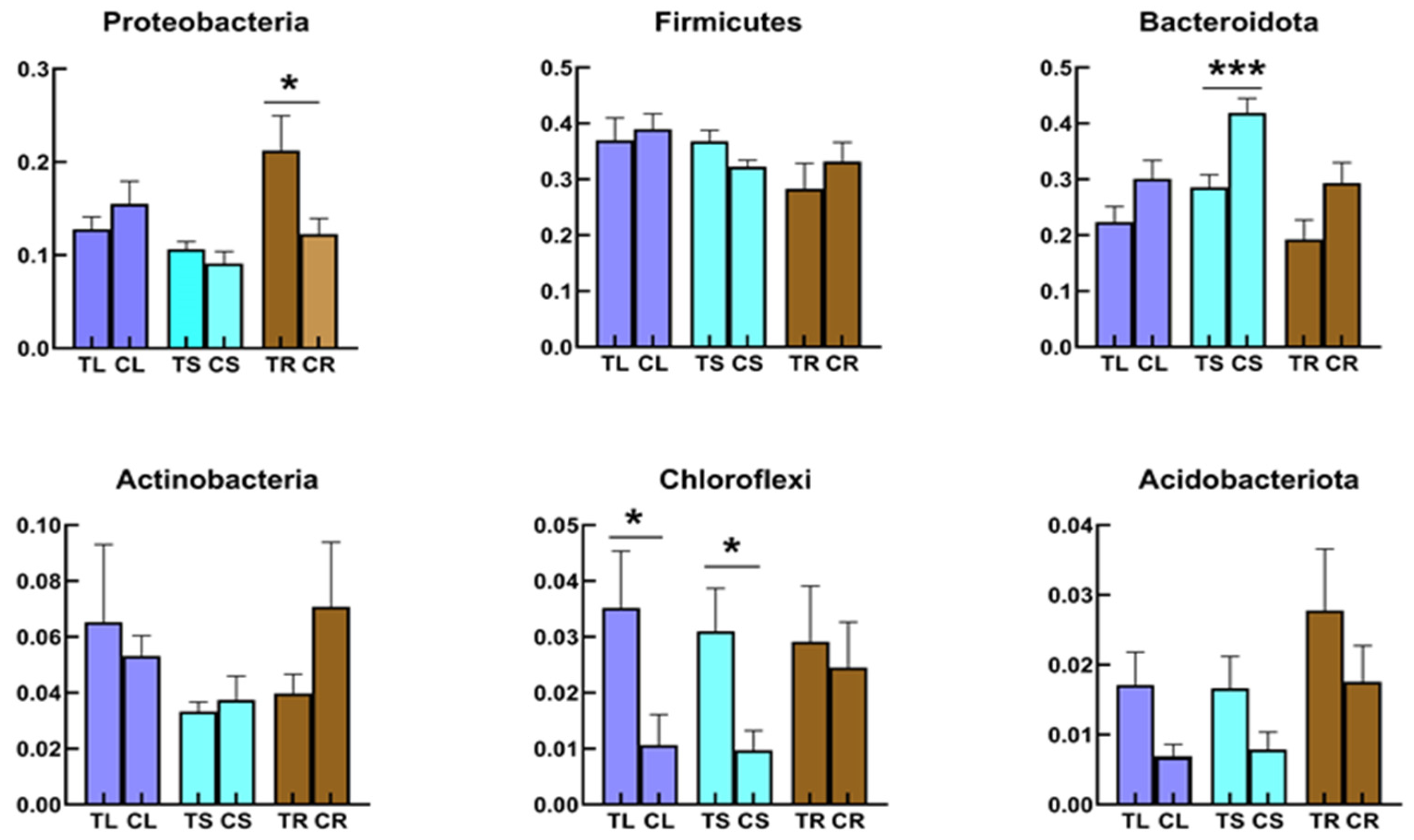
4.1. Relationship between Soil Microbiota and Tartary Buckwheat Microbiome
4.2. Seed Microbiota Associated with Endophytic Microorganisms of Tartary Buckwheat
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef]
- Wang, H.; Chen, R.F.; Iwashita, T.; Shen, R.F.; Ma, J.F. Physiological Characterization of Aluminum Tolerance and Accumulation in Tartary and Wild Buckwheat. New Phytol. 2015, 205, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Bonafaccia, G.; Marocchini, M.; Kreft, I. Composition and technological properties of the flour and bran form common and tatary buckwheat. Food Chem. 2003, 80, 9–15. [Google Scholar] [CrossRef]
- Fabjan, N.; Rode, J.; Kosǐr, I.J.; Wang, Z.; Zhang, Z.; Kreft, I. Tartary Buckwheat (Fagopyrum Tataricum Gaertn.) as a Source of Dietary Rutin and Quercitrin. J. Agric. Food Chem. 2003, 51, 6452–6455. [Google Scholar] [CrossRef]
- Gupta, N.; Naik, P.K.; Chauhan, R.S. Differential Transcript Profiling through CDNA-AFLP Showed Complexity of Rutin Biosynthesis and Accumulation in Seeds of a Nutraceutical Food Crop (Fagopyrum Spp.). BMC Genom. 2012, 13, 231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; He, M.; Fan, Y.; Zhao, H.; Gao, B.; Yang, K.; Li, F.; Tang, Y.; Gao, Q.; Lin, T.; et al. Resequencing of Global Tartary Buckwheat Accessions Reveals Multiple Domestication Events and Key Loci Associated with Agronomic Traits. Genome Biol. 2021, 22, 23. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; De Beeck, M.O.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural Variability and Niche Differentiation in the Rhizosphere and Endosphere Bacterial Microbiome of Field-Grown Poplar Trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef]
- Bergna, A.; Cernava, T.; Rändler, M.; Grosch, R.; Zachow, C.; Berg, G. Tomato Seeds Preferably Transmit Plant Beneficial Endophytes. Phytobiomes J. 2018, 2, 183–193. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moënne-Loccoz, Y. Let the Core Microbiota Be Functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef]
- Karasov, T.L.; Kniskern, J.M.; Gao, L.; Deyoung, B.J.; Ding, J.; Dubiella, U.; Lastra, R.O.; Nallu, S.; Roux, F.; Innes, R.W.; et al. The Long-Term Maintenance of a Resistance Polymorphism through Diffuse Interactions. Nature 2014, 512, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Finkel, O.M.; Salas-González, I.; Castrillo, G.; Conway, J.M.; Law, T.F.; Teixeira, P.J.P.L.; Wilson, E.D.; Fitzpatrick, C.R.; Jones, C.D.; Dangl, J.L. A Single Bacterial Genus Maintains Root Growth in a Complex Microbiome. Nature 2020, 587, 103–108. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Nelson, E.B. The Seed Microbiome: Origins, Interactions, and Impacts. Plant Soil 2018, 422, 7–34. [Google Scholar] [CrossRef]
- Blaney, C.S.; Kotanen, P.M. Persistence in the Seed Bank: The Effects of Fungi and Invertebrates on Seeds of Native and Exotic Plants. Ecoscience 2002, 9, 509–517. [Google Scholar] [CrossRef]
- Luo, J.; Tao, Q.; Jupa, R.; Liu, Y.; Wu, K.; Song, Y.; Li, J.; Huang, Y.; Zou, L.; Liang, Y.; et al. Role of Vertical Transmission of Shoot Endophytes in Root-Associated Microbiome Assembly and Heavy Metal Hyperaccumulation in Sedum Alfredii. Environ. Sci. Technol. 2019, 53, 6954–6963. [Google Scholar] [CrossRef] [PubMed]
- Chi, F.; Shen, S.H.; Cheng, H.P.; Jing, Y.X.; Yanni, Y.G.; Dazzo, F.B. Ascending Migration of Endophytic Rhizobia, from Roots to Leaves, inside Rice Plants and Assessment of Benefits to Rice Growth Physiology. Appl. Environ. Microbiol. 2005, 71, 7271–7278. [Google Scholar] [CrossRef] [PubMed]
- Moroenyane, I.; Tremblay, J.; Yergeau, É. Soybean Microbiome Recovery after Disruption Is Modulated by the Seed and Not the Soil Microbiome. Phytobiomes J. 2021, 5, 418–431. [Google Scholar] [CrossRef]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the Root Microbiome: Bacterial Root Endophytes and Plant Growth Promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef]
- Links, M.G.; Demeke, T.; Gräfenhan, T.; Hill, J.E.; Hemmingsen, S.M.; Dumonceaux, T.J. Simultaneous Profiling of Seed-Associated Bacteria and Fungi Reveals Antagonistic Interactions between Microorganisms within a Shared Epiphytic Microbiome on Triticum and Brassica Seeds. New Phytol. 2014, 202, 542–553. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and Diversity of Seed Associated Endophytes in Zea across Boundaries of Evolution, Ethnography and Ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Jiang, T.; Liu, Y.X.; Bai, Y.C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.W.; Bai, Y.; Osbourn, A. A Specialized Metabolic Network Selectively Modulates Arabidopsis Root Microbiota. Science 2019, 364, 80–88. [Google Scholar] [CrossRef]
- Witkowicz, R.; Biel, W.; Skrzypek, E.; Chłopicka, J.; Gleń-Karolczyk, K.; Krupa, M.; Prochownik, E.; Galanty, A. Microorganisms and Biostimulants Impact on the Antioxidant Activity of Buckwheat (Fagopyrum Esculentum Moench) Sprouts. Antioxidants 2020, 9, 584. [Google Scholar] [CrossRef]
- Likar, M.; Bukovnik, U.; Kreft, I.; Chrungoo, N.K.; Regvar, M. Mycorrhizal Status and Diversity of Fungal Endophytes in Roots of Common Buckwheat (Fagopyrum esculentum) and Tartary Buckwheat (F. tataricum). Mycorrhiza 2008, 18, 309–315. [Google Scholar] [CrossRef]
- Ren, Q.; Sun, L.; Wu, H.; Wang, Y.; Wang, Z.; Zheng, F.; Lu, X.; Xu, J. The Changes of Microbial Community and Flavor Compound in the Fermentation Process of Chinese Rice Wine Using Fagopyrum tataricum Grain as Feedstock. Sci. Rep. 2019, 9, 3365. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 1, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILV A ribosomal RNA gene database project, improved data processingand web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; van Elsas, J.D. Properties of Bacterial Endophytes and Their Proposed Role in Plant Growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Newman, L.; Vangronsveld, J. Exploiting Plant-Microbe Partnerships to Improve Biomass Production and Remediation. Trends Biotechnol. 2009, 27, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, C.V.; Kjøller, R.; Raaijmakers, J.M.; Riber, L.; Christensen, S.; Rasmussen, S.; Christensen, J.H.; Dahl, A.B.; Westergaard, J.C.; Nielsen, M.; et al. Extension of Plant Phenotypes by the Foliar Microbiome. Annu. Rev. Plant Biol. 2021, 72, 823–846. [Google Scholar] [CrossRef] [PubMed]
- Brader, G.; Compant, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic Potential of Endophytic Bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef]
- Lee, S.M.; Kong, H.G.; Song, G.C.; Ryu, C.M. Disruption of Firmicutes and Actinobacteria Abundance in Tomato Rhizosphere Causes the Incidence of Bacterial Wilt Disease. ISME J. 2021, 15, 330–347. [Google Scholar] [CrossRef]
- Weller, D.M.; Raaijmakers, J.M.; McSpadden Gardener, B.B.; Thomashow, L.S. Microbial Populations Responsible for Specific Soil Suppressiveness to Plant Pathogens. Annu. Rev. Phytopathol. 2002, 40, 309–348. [Google Scholar] [CrossRef] [PubMed]
- Bakker, P.A.H.M.; Pieterse, C.M.J.; de Jonge, R.; Berendsen, R.L. The Soil-Borne Legacy. Cell 2018, 172, 1178–1180. [Google Scholar] [CrossRef]
- Grady, K.L.; Sorensen, J.W.; Stopnisek, N.; Guittar, J.; Shade, A. Assembly and Seasonality of Core Phyllosphere Microbiota on Perennial Biofuel Crops. Nat. Commun. 2019, 10, 4135. [Google Scholar] [CrossRef] [PubMed]
- Maignien, L.; DeForce, E.A.; Chafee, M.E.; Murat Eren, A.; Simmons, S.L. Ecological Succession and Stochastic Variation in the Assembly of Arabidopsis thaliana Phyllosphere Communities. mBio 2014, 5, e00682-13. [Google Scholar] [CrossRef]
- Ottesen, A.R.; Gorham, S.; Reed, E.; Newell, M.J.; Ramachandran, P.; Canida, T.; Allard, M.; Evans, P.; Brown, E.; White, J.R. Using a Control to Better Understand Phyllosphere Microbiota. PLoS ONE 2016, 11, e0163482. [Google Scholar] [CrossRef]
- Dos-Santos, C.M.; de Souza, D.G.; Balsanelli, E.; Cruz, L.M.; de Souza, E.M.; Baldani, J.I.; Schwab, S. A Culture-Independent Approach to Enrich Endophytic Bacterial Cells from Sugarcane Stems for Community Characterization. Microb. Ecol. 2017, 74, 453–465. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.S.C.; Okura, V.K.; Armanhi, J.S.L.; Jorrín, B.; Lozano, N.; Da Silva, M.J.; González-Guerrero, M.; De Araújo, L.M.; Verza, N.C.; Bagheri, H.C.; et al. Unlocking the Bacterial and Fungal Communities Assemblages of Sugarcane Microbiome. Sci. Rep. 2016, 6, 28774. [Google Scholar] [CrossRef] [PubMed]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional Overlap of the Arabidopsis Leaf and Root Microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Barret, M.; Briand, M.; Bonneau, S.; Préveaux, A.; Valière, S.; Bouchez, O.; Hunault, G.; Simoneau, P.; Jacquesa, M.A. Emergence Shapes the Structure of the Seed Microbiota. Appl. Environ. Microbiol. 2015, 81, 1257–1266. [Google Scholar] [CrossRef]
- Cottyn, B.; Debode, J.; Regalado, E.; Mew, T.W.; Swings, J. Phenotypic and Genetic Diversity of Rice Seed-Associated Bacteria and Their Role in Pathogenicity and Biological Control. J. Appl. Microbiol. 2009, 107, 885–897. [Google Scholar] [CrossRef]
- Guo, X.; Van Iersel, M.W.; Chen, J.; Brackett, R.E.; Beuchat, L.R. Evidence of Association of Salmonellae with Tomato Plants Grown Hydroponically in Inoculated Nutrient Solution. Appl. Environ. Microbiol. 2002, 68, 3639–3643. [Google Scholar] [CrossRef]
- Van Overbeek, L.S.; Franke, A.C.; Nijhuis, E.H.M.; Groeneveld, R.M.W.; da Rocha, U.N.; Lotz, L.A.P. Bacterial Communities Associated with Chenopodium Album and Stellaria Media Seeds from Arable Soils. Microb. Ecol. 2011, 62, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, K.K.; Jeon, J.; Harris, W.A.; Lee, Y.H. Domestication of Oryza Species Eco-Evolutionarily Shapes Bacterial and Fungal Communities in Rice Seed. Microbiome 2020, 8, 20. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and Heritability of the Maize Rhizosphere Microbiome under Field Conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and Representative Bacterial Community of Maize Roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef]
- Liu, J.; Abdelfattah, A.; Norelli, J.; Burchard, E.; Schena, L.; Droby, S.; Wisniewski, M. Apple Endophytic Microbiota of Different Rootstock/Scion Combinations Suggests a Genotype-Specific Influence. Microbiome 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Nomura, K.; Wang, X.; Sohrabi, R.; Xu, J.; Yao, L.; Paasch, B.C.; Ma, L.; Kremer, J.; Cheng, Y.; et al. A Plant Genetic Network for Preventing Dysbiosis in the Phyllosphere. Nature 2020, 580, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Fernández-González, A.J.; Cardoni, M.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Villadas, P.J.; Fernández-López, M.; Mercado-Blanco, J. Linking Belowground Microbial Network Changes to Different Tolerance Level towards Verticillium Wilt of Olive. Microbiome 2020, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Cram, J.A.; Xia, L.C.; Needham, D.M.; Sachdeva, R.; Sun, F.; Fuhrman, J.A. Cross-Depth Analysis of Marine Bacterial Networks Suggests Downward Propagation of Temporal Changes. ISME J. 2015, 9, 2573–2586. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Sun, B.; Li, H.; Liu, M.; Chen, L.; Zhou, S. Aggregate-Related Changes in Network Patterns of Nematodes and Ammonia Oxidizers in an Acidic Soil. Soil Biol. Biochem. 2015, 88, 101–109. [Google Scholar] [CrossRef]
- Ma, B.; Wang, Y.; Ye, S.; Liu, S.; Stirling, E.; Gilbert, J.A.; Faust, K.; Knight, R.; Jansson, J.K.; Cardona, C.; et al. Earth Microbial Co-Occurrence Network Reveals Interconnection Pattern across Microbiomes. Microbiome 2020, 8, 82. [Google Scholar] [CrossRef]

| Groups | Simpson | Shannon | chao1 | ACE | ||||
|---|---|---|---|---|---|---|---|---|
| p-Value | Sig | p-Value | Sig | p-Value | Sig | p-Value | Sig | |
| FO vs. TL | 0.027 | * | 0.014 | * | 0.023 | * | 0.018 | * |
| FO vs. TS | 3.00 × 10−4 | *** | 1.00 × 10−4 | *** | 0.084 | 0.151 | ||
| FO vs. TR | 0.023 | * | 0.003 | ** | 9.00 × 10−4 | *** | 0.004 | ** |
| FO vs. CL | 0.566 | 0.111 | 0.531 | 0.322 | ||||
| FO vs. CS | 0.006 | ** | 0.005 | ** | 0.904 | 0.968 | ||
| FO vs. CR | 0.003 | ** | 0.001 | ** | 0.023 | * | 0.012 | * |
| CL vs. TL | 0.414 | 0.371 | 0.098 | 0.165 | ||||
| CS vs. TS | 0.386 | 0.273 | 0.065 | 0.140 | ||||
| CR vs. TR | 0.477 | 0.731 | 0.278 | 0.681 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhu, X.; Dong, Y.; Chen, Y.; Li, M.; Li, C. Limited Impact of Soil Microorganisms on the Endophytic Bacteria of Tartary Buckwheat (Fagopyrum tataricum). Microorganisms 2023, 11, 2085. https://doi.org/10.3390/microorganisms11082085
Liu X, Zhu X, Dong Y, Chen Y, Li M, Li C. Limited Impact of Soil Microorganisms on the Endophytic Bacteria of Tartary Buckwheat (Fagopyrum tataricum). Microorganisms. 2023; 11(8):2085. https://doi.org/10.3390/microorganisms11082085
Chicago/Turabian StyleLiu, Xuyan, Xishen Zhu, Yumei Dong, Yan Chen, Meifang Li, and Chengyun Li. 2023. "Limited Impact of Soil Microorganisms on the Endophytic Bacteria of Tartary Buckwheat (Fagopyrum tataricum)" Microorganisms 11, no. 8: 2085. https://doi.org/10.3390/microorganisms11082085
APA StyleLiu, X., Zhu, X., Dong, Y., Chen, Y., Li, M., & Li, C. (2023). Limited Impact of Soil Microorganisms on the Endophytic Bacteria of Tartary Buckwheat (Fagopyrum tataricum). Microorganisms, 11(8), 2085. https://doi.org/10.3390/microorganisms11082085






