Altered Urine Microbiome in Male Children and Adolescents with Attention-Deficit Hyperactivity Disorder
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Sample Collection
2.2. Bacterial and EV Isolation and DNA Extraction from Clinical Samples
2.3. PCR Amplification, Library Construction, and Sequencing of 16S rRNA Gene Variable Regions
2.4. Analysis of Bacterial Composition in the Microbiome
2.5. Statistical Analysis
3. Results
3.1. Demographic Characteristics
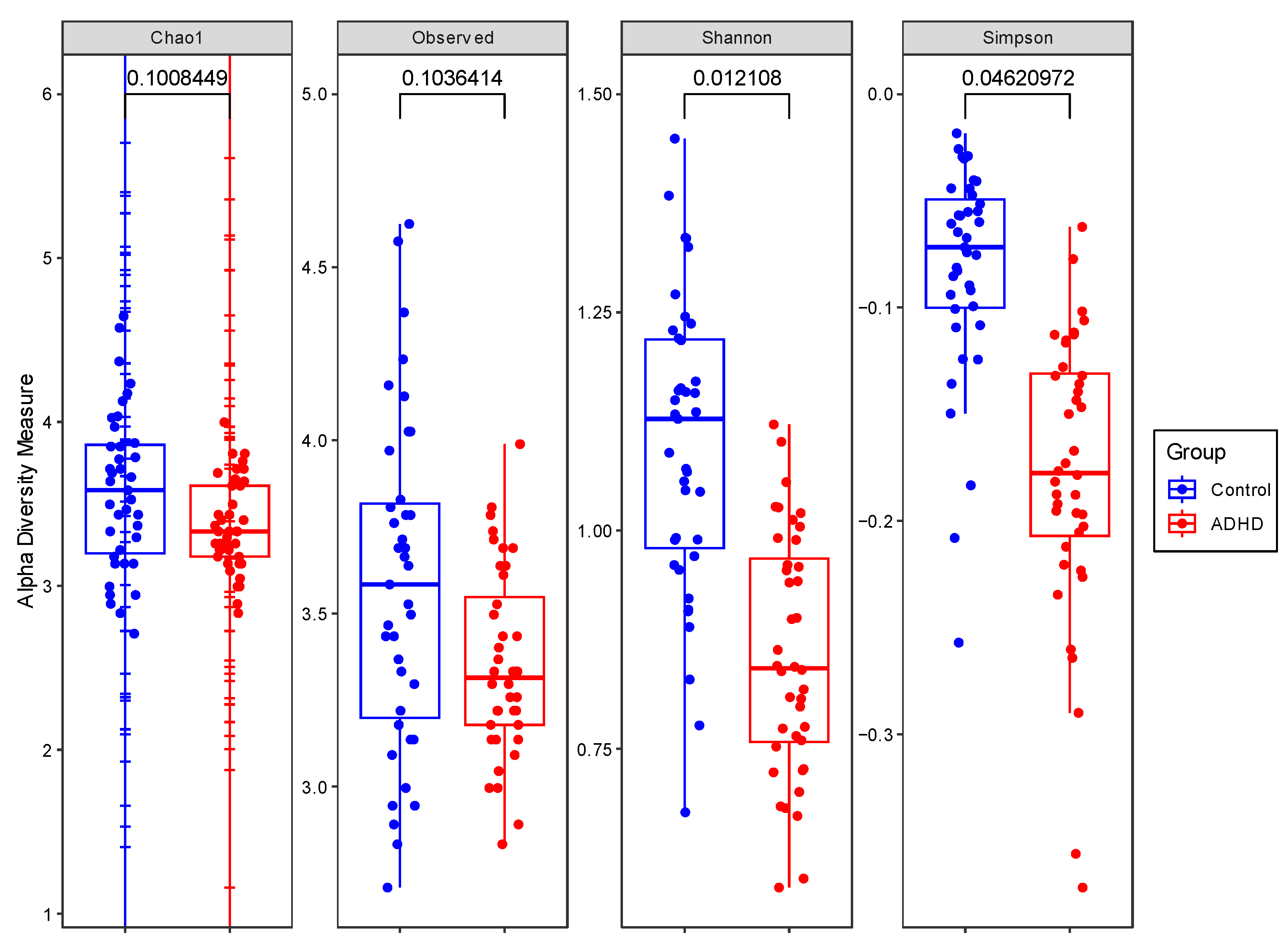
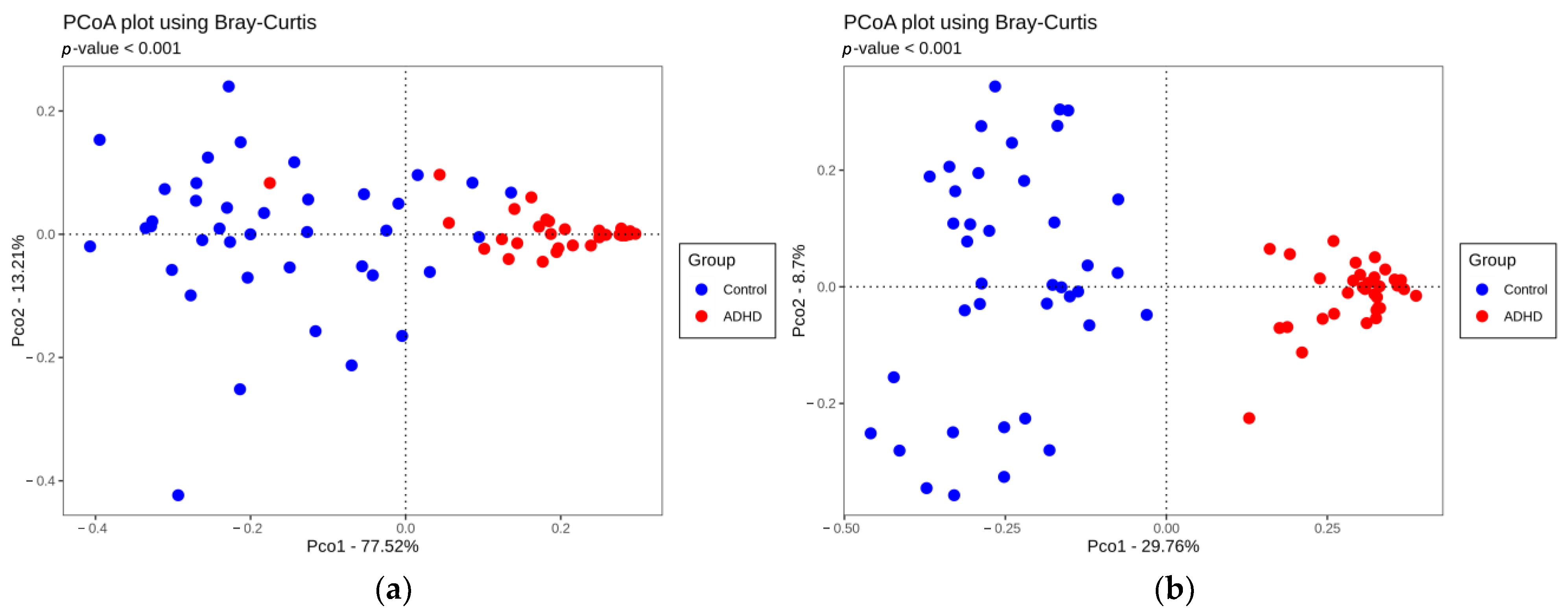
3.2. Comparison of Alpha and Beta Diversity between ADHD and Controls
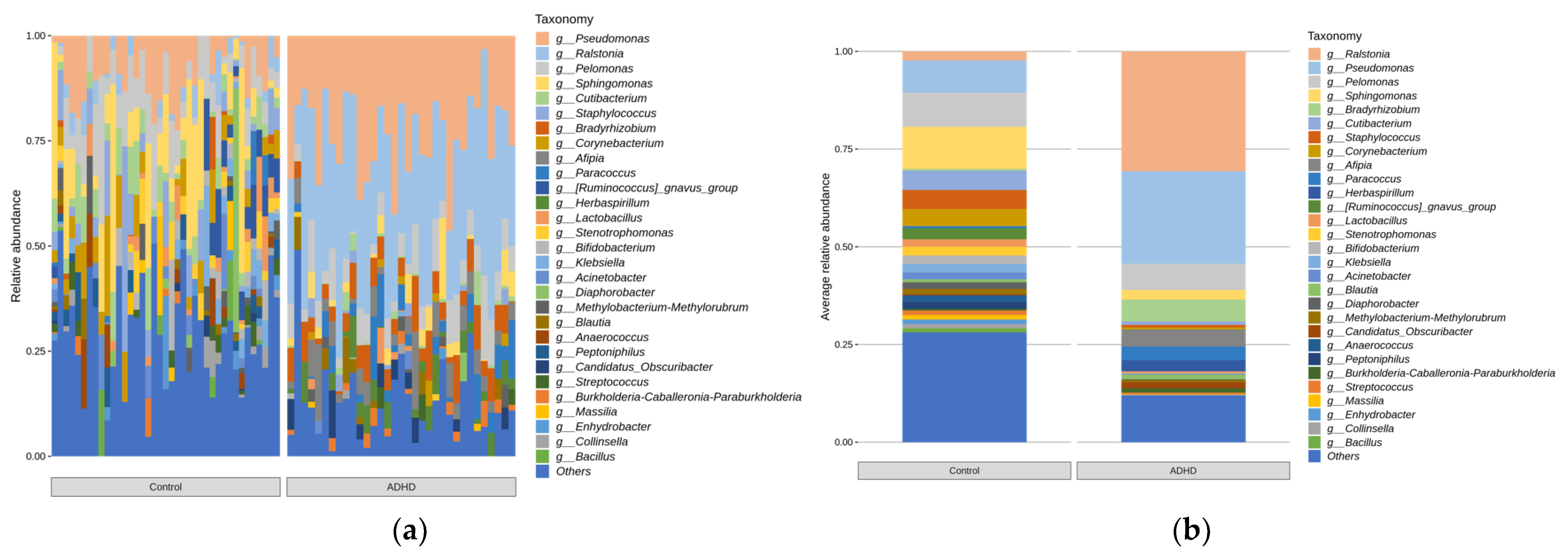
3.3. Relative Microbiota Abundance Differences between ADHD and Controls
3.4. Correlation between Baseline ADHD Symptom Scores and ADHD-Specific Taxa
4. Discussion
4.1. Comparison of Alpha and Beta Diversity between ADHD and Controls
4.2. Relative Microbiota Abundance Differences between ADHD and Controls
4.3. Correlation between Baseline Evaluation Scores and ADHD-Specific Taxa
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association (APA): Washington, DC, USA, 2013; p. 280. [Google Scholar]
- Biederman, J.; Newcorn, J.; Sprich, S. Comorbidity of Attention Deficit Hyperactivity Disorder with Conduct, Depressive, Anxiety, and Other Disorders. Am. J. Psychiatry 1991, 148, 564–577. [Google Scholar] [CrossRef]
- Spencer, T.J.; Biederman, J.; Mick, E. Attention-Deficit/Hyperactivity Disorder: Diagnosis, Lifespan, Comorbidities, and Neurobiology. Ambul. Pediatr. 2007, 7, 73–81. [Google Scholar] [CrossRef]
- Biederman, J.; Monuteaux, M.C.; Doyle, A.E.; Seidman, L.J.; Wilens, T.E.; Ferrero, F.; Morgan, C.L.; Faraone, S.V. Impact of Executive Function Deficits and Attention-Deficit/Hyperactivity Disorder (ADHD) on Academic Outcomes in Children. J. Consult. Clin. Psychol. 2004, 72, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, Y.; Odisho, D.; Wu, S.; Yi, C.; Oliver, B.G. Can Biomarkers Be Used to Diagnose Attention Deficit Hyperactivity Disorder? Front. Psychiatry 2023, 14, 1026616. [Google Scholar] [CrossRef] [PubMed]
- Hebebrand, J.; Antel, J. Squaring the Circle? On the Search for Circulating Biomarkers in Polygenic Psychiatric Disorders. Eur. Child Adolesc. Psychiatry 2014, 23, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Sherwin, E.; Dinan, T.G.; Cryan, J.F. Recent Developments in Understanding the Role of the Gut Microbiota in Brain Health and Disease. Ann. N. Y. Acad. Sci. 2018, 1420, 5–25. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.C.; Olson, C.A.; Hsiao, E.Y. Interactions between the Microbiota, Immune and Nervous Systems in Health and Disease. Nat. Neurosci. 2017, 20, 145–155. [Google Scholar] [CrossRef]
- Wang, Y.; Kasper, L.H. The Role of Microbiome in Central Nervous System Disorders. Brain Behav. Immun. 2014, 38, 1–12. [Google Scholar] [CrossRef]
- Aarts, E.; Ederveen, T.H.A.; Naaijen, J.; Zwiers, M.P.; Boekhorst, J.; Timmerman, H.M.; Smeekens, S.P.; Netea, M.G.; Buitelaar, J.K.; Franke, B.; et al. Gut Microbiome in ADHD and Its Relation to Neural Reward Anticipation. PLoS ONE 2017, 12, e0183509. [Google Scholar] [CrossRef]
- Prehn-Kristensen, A.; Zimmermann, A.; Tittmann, L.; Lieb, W.; Schreiber, S.; Baving, L.; Fischer, A. Reduced Microbiome Alpha Diversity in Young Patients with ADHD. PLoS ONE 2018, 13, e0200728. [Google Scholar] [CrossRef]
- Jiang, H.-Y.; Zhou, Y.-Y.; Zhou, G.-L.; Li, Y.-C.; Yuan, J.; Li, X.-H.; Ruan, B. Gut Microbiota Profiles in Treatment-Naïve Children with Attention Deficit Hyperactivity Disorder. Behav. Brain Res. 2018, 347, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Szopinska-Tokov, J.; Dam, S.; Naaijen, J.; Konstanti, P.; Rommelse, N.; Belzer, C.; Buitelaar, J.; Franke, B.; Bloemendaal, M.; Aarts, E.; et al. Investigating the Gut Microbiota Composition of Individuals with Attention-Deficit/Hyperactivity Disorder and Association with Symptoms. Microorganisms 2020, 8, 406. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Ge, W.-R.; Zhang, S.; Sun, Y.-L.; Wang, B.; Yang, G. Case-Control Study of the Effects of Gut Microbiota Composition on Neurotransmitter Metabolic Pathways in Children With Attention Deficit Hyperactivity Disorder. Front. Neurosci. 2020, 14, 127. [Google Scholar] [CrossRef] [PubMed]
- Casas, L.; Karvonen, A.M.; Kirjavainen, P.V.; Täubel, M.; Hyytiäinen, H.; Jayaprakash, B.; Lehmann, I.; Standl, M.; Pekkanen, J.; Heinrich, J. Early Life Home Microbiome and Hyperactivity/Inattention in School-Age Children. Sci. Rep. 2019, 9, 17355. [Google Scholar] [CrossRef]
- Cheng, S.; Han, B.; Ding, M.; Wen, Y.; Ma, M.; Zhang, L.; Qi, X.; Cheng, B.; Li, P.; Kafle, O.P.; et al. Identifying Psychiatric Disorder-Associated Gut Microbiota Using Microbiota-Related Gene Set Enrichment Analysis. Brief. Bioinform. 2020, 21, 1016–1022. [Google Scholar] [CrossRef]
- Wang, L.J.; Yang, C.Y.; Chou, W.J.; Lee, M.J.; Chou, M.C.; Kuo, H.C.; Yeh, Y.M.; Lee, S.Y.; Huang, L.H.; Li, S.C. Gut Microbiota and Dietary Patterns in Children with Attention-Deficit/Hyperactivity Disorder. Eur. Child Adolesc. Psychiatry 2020, 29, 287–297. [Google Scholar] [CrossRef]
- Pärtty, A.; Kalliomäki, M.; Wacklin, P.; Salminen, S.; Isolauri, E. A Possible Link between Early Probiotic Intervention and the Risk of Neuropsychiatric Disorders Later in Childhood: A Randomized Trial. Pediatr. Res. 2015, 77, 823–828. [Google Scholar] [CrossRef]
- Stevens, A.J.; Purcell, R.V.; Darling, K.A.; Eggleston, M.J.F.; Kennedy, M.A.; Rucklidge, J.J. Human Gut Microbiome Changes during a 10 Week Randomised Control Trial for Micronutrient Supplementation in Children with Attention Deficit Hyperactivity Disorder. Sci. Rep. 2019, 9, 10128. [Google Scholar] [CrossRef] [PubMed]
- Wojciuk, B.; Salabura, A.; Grygorcewicz, B.; Kędzierska, K.; Ciechanowski, K.; Dołęgowska, B. Urobiome: In Sickness and in Health. Microorganisms 2019, 7, 548. [Google Scholar] [CrossRef]
- Peyronnet, B.; Mironska, E.; Chapple, C.; Cardozo, L.; Oelke, M.; Dmochowski, R.; Amarenco, G.; Gamé, X.; Kirby, R.; van der Aa, F.; et al. A Comprehensive Review of Overactive Bladder Pathophysiology: On the Way to Tailored Treatment. Eur. Urol. 2019, 75, 988–1000. [Google Scholar] [CrossRef]
- Tang, J. Microbiome in the Urinary System—A Review. AIMS Microbiol. 2017, 3, 143. [Google Scholar] [CrossRef] [PubMed]
- Gerber, D.; Forster, C.S.; Hsieh, M. The Role of the Genitourinary Microbiome in Pediatric Urology: A Review. Curr. Urol. Rep. 2018, 19, 13. [Google Scholar] [CrossRef] [PubMed]
- Kawalec, A.; Zwolińska, D. Emerging Role of Microbiome in the Prevention of Urinary Tract Infections in Children. Int. J. Mol. Sci. 2022, 23, 870. [Google Scholar] [CrossRef] [PubMed]
- Perez-Carrasco, V.; Soriano-Lerma, A.; Soriano, M.; Gutiérrez-Fernández, J.; Garcia-Salcedo, J.A. Urinary Microbiome: Yin and Yang of the Urinary Tract. Front. Cell. Infect. Microbiol. 2021, 11, 617002. [Google Scholar] [CrossRef] [PubMed]
- Leue, C.; Kruimel, J.; Vrijens, D.; Masclee, A.; van Os, J.; van Koeveringe, G. Functional Urological Disorders: A Sensitized Defence Response in the Bladder-Gut-Brain Axis. Nat. Rev. Urol. 2017, 14, 153–163. [Google Scholar] [CrossRef]
- de Almeida Vasconcelos, M.M.; Netto, J.M.B.; Arana, I.E.; Teixeira, I.B.; Lima, E.M.; Carvalho, T.A.; de Bessa Junior, J.; de Carvalho Mrad, F.C. Association between Attention Deficit Hyperactivity Disorder and Lower Urinary Tract Symptoms in Children and Adolescents in a Community Setting. Int. Braz. J. Urol. 2021, 47, 969–978. [Google Scholar] [CrossRef]
- Mahjani, B.; Koskela, L.R.; Mahjani, C.G.; Janecka, M.; Batuure, A.; Hultman, C.M.; Reichenberg, A.; Buxbaum, J.D.; Akre, O.; Grice, D.E. Systematic Review and Meta-Analysis: Relationships between Attention-Deficit/Hyperactivity Disorder and Urinary Symptoms in Children. Eur. Child Adolesc. Psychiatry 2022, 31, 663–670. [Google Scholar] [CrossRef]
- Richarte, V.; Sánchez-Mora, C.; Corrales, M.; Fadeuilhe, C.; Vilar-Ribó, L.; Arribas, L.; Garcia, E.; Rosales-Ortiz, S.K.; Arias-Vasquez, A.; Soler-Artigas, M.; et al. Gut Microbiota Signature in Treatment-Naïve Attention-Deficit/Hyperactivity Disorder. Transl. Psychiatry 2021, 11, 382. [Google Scholar] [CrossRef]
- Srikantha, P.; Mohajeri, M.H. The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef]
- Daneberga, Z.; Nakazawa-Miklasevica, M.; Berga-Svitina, E.; Murmane, D.; Isarova, D.; Cupane, L.; Masinska, M.; Nartisa, I.; Lazdane, A.; Miklasevics, E. Urinary Organic Acids Spectra in Children with Altered Gut Microbiota Composition and Autistic Spectrum Disorder. Nord. J. Psychiatry 2021, 76, 523–529. [Google Scholar] [CrossRef]
- Gevi, F.; Belardo, A.; Zolla, L. A Metabolomics Approach to Investigate Urine Levels of Neurotransmitters and Related Metabolites in Autistic Children. Biochim. Biophys. Acta 2020, 1866, 165859. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Liu, D.; Wang, Y.; Zeng, T.; Peng, Y. Urinary 3-(3-Hydroxyphenyl)-3-Hydroxypropionic Acid, 3-Hydroxyphenylacetic Acid, and 3-Hydroxyhippuric Acid Are Elevated in Children with Autism Spectrum Disorders. Biomed. Res. Int. 2016, 2016, 9485412. [Google Scholar] [CrossRef]
- Wu, P.; Chen, Y.; Zhao, J.; Zhang, G.; Chen, J.; Wang, J.; Zhang, H. Urinary Microbiome and Psychological Factors in Women with Overactive Bladder. Front. Cell. Infect. Microbiol. 2017, 7, 488. [Google Scholar] [CrossRef]
- Kang, C.-S.; Ban, M.; Choi, E.-J.; Moon, H.-G.; Jeon, J.-S.; Kim, D.-K.; Park, S.-K.; Jeon, S.G.; Roh, T.-Y.; Myung, S.-J.; et al. Extracellular Vesicles Derived from Gut Microbiota, Especially Akkermansia Muciniphila, Protect the Progression of Dextran Sulfate Sodium-Induced Colitis. PLoS ONE 2013, 8, e76520. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.Y.; Rho, M.; You, Y.A.; Kwon, E.J.; Kim, M.H.; Kym, S.; Jee, Y.K.; Kim, Y.K.; Kim, Y.J. 16S RRNA Gene-Based Metagenomic Analysis Reveals Differences in Bacteria-Derived Extracellular Vesicles in the Urine of Pregnant and Non-Pregnant Women. Exp. Mol. Med. 2016, 48, e208. [Google Scholar] [CrossRef]
- Jang, S.C.; Kim, S.R.; Yoon, Y.J.; Park, K.S.; Kim, J.H.; Lee, J.; Kim, O.Y.; Choi, E.J.; Kim, D.K.; Choi, D.S.; et al. In Vivo Kinetic Biodistribution of Nano-Sized Outer Membrane Vesicles Derived from Bacteria. Small 2015, 11, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Park, J.Y.; Lee, E.H.; Yang, J.; Jeong, B.R.; Kim, Y.K.; Seoh, J.Y.; Lee, S.H.; Han, P.L.; Kim, E.J. Rapid Assessment of Microbiota Changes in Individuals with Autism Spectrum Disorder Using Bacteria-Derived Membrane Vesicles in Urine. Exp. Neurobiol. 2017, 26, 307. [Google Scholar] [CrossRef]
- Sheehan, D.V.; Sheehan, K.H.; Shytle, R.D.; Janavs, J.; Bannon, Y.; Rogers, J.E.; Milo, K.M.; Stock, S.L.; Wilkinson, B. Reliability and Validity of the Mini International Neuropsychiatric Interview for Children and Adolescents (MINI-KID). J. Clin. Psychiatry 2010, 71, 313–326. [Google Scholar] [CrossRef]
- Kwak, K.J.; Park, H.W.; Kim, C.T. A Study for the Standardization of Korean WISC-3 (1). Korean J. Dev. Psychol. 2002, 15, 19–33. [Google Scholar]
- Storm, D.W.; Copp, H.L.; Halverson, T.M.; Du, J.; Juhr, D.; Wolfe, A.J. A Child’s Urine Is Not Sterile: A Pilot Study Evaluating the Pediatric Urinary Microbiome. J. Pediatr. Urol. 2022, 18, 383–392. [Google Scholar] [CrossRef]
- Kassiri, B.; Shrestha, E.; Kasprenski, M.; Antonescu, C.; Florea, L.D.; Sfanos, K.S.; Wang, M.H. A Prospective Study of the Urinary and Gastrointestinal Microbiome in Prepubertal Males. Urology 2019, 131, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Clarridge, J.E. Impact of 16S RRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S.; Jordan, G.; Chamberlain, S. Phyloseq: Handling and Analysis of High-Throughput Microbiome Census Data. 2023. Available online: https://rdrr.io/bioc/phyloseq/ (accessed on 26 July 2023).
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human Gut Microbiome Viewed across Age and Geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Checa-Ros, A.; Jeréz-Calero, A.; Molina-Carballo, A.; Campoy, C.; Muñoz-Hoyos, A. Current Evidence on the Role of the Gut Microbiome in ADHD Pathophysiology and Therapeutic Implications. Nutrients 2021, 13, 249. [Google Scholar] [CrossRef]
- Ha, S.; Oh, D.; Lee, S.; Park, J.; Ahn, J.; Choi, S.; Cheon, K.-A. Altered Gut Microbiota in Korean Children with Autism Spectrum Disorders. Nutrients 2021, 13, 3300. [Google Scholar] [CrossRef]
- de Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; de Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal Microbiota and Metabolome of Children with Autism and Pervasive Developmental Disorder Not Otherwise Specified. PLoS ONE 2013, 8, 76993. [Google Scholar] [CrossRef]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal Flora and Gastrointestinal Status in Children with Autism -- Comparisons to Typical Children and Correlation with Autism Severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low Relative Abundances of the Mucolytic Bacterium Akkermansia Muciniphila and Bifidobacterium Spp. in Feces of Children with Autism. Appl. Environ. Microbiol. 2011, 77, 6718. [Google Scholar] [CrossRef] [PubMed]
- Strati, F.; Cavalieri, D.; Albanese, D.; de Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New Evidences on the Altered Gut Microbiota in Autism Spectrum Disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing Study of Fecal Microflora of Autistic and Control Children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Chomel, B.B.; Kasten, R.W.; Stuckey, M.J.; Breitschwerdt, E.B.; Maggi, R.G.; Henn, J.B.; Koehler, J.E.; Chang, C. Experimental Infection of Cats with Afipia Felis and Various Bartonella Species or Subspecies. Vet. Microbiol. 2014, 172, 505–510. [Google Scholar] [CrossRef]
- Swann, J.R.; Diaz Heijtz, R.; Mayneris-Perxachs, J.; Arora, A.; Isaksson, J.; Bölte, S.; Tammimies, K. Characterizing the Metabolomic Signature of Attention-Deficit Hyperactivity Disorder in Twins. Neuropharmacology 2023, 234, 109562. [Google Scholar] [CrossRef]
- Jašarević, E.; Morrison, K.E.; Bale, T.L. Sex Differences in the Gut Microbiome–Brain Axis across the Lifespan. Philos. Trans. R. Soc. B 2016, 371, 20150122. [Google Scholar] [CrossRef]
- Mowlem, F.D.; Rosenqvist, M.A.; Martin, J.; Lichtenstein, P.; Asherson, P.; Larsson, H. Sex Differences in Predicting ADHD Clinical Diagnosis and Pharmacological Treatment. Eur. Child Adolesc. Psychiatry 2019, 28, 481–489. [Google Scholar] [CrossRef]


| Scales | Mean ± SD |
|---|---|
| CBCL | |
| Attention problems subscale a | 61.30 ± 7.16 |
| DSM-oriented ADHD subscale a | 64.30 ± 11.91 |
| ADHD-RS | |
| Inattention | 11.03 ± 5.35 |
| Hyperactivity/impulsivity | 10.55 ± 5.18 |
| Total | 21.58 ± 9.92 |
| Phylum Level | Firmicutes b | Actinobacteriota b |
|---|---|---|
| CBCL | ||
| Attention | 0.154 (0.392) | 0.138 (0.445) |
| Problems | ||
| Subscale a | ||
| DSM oriented | 0.167 (0.352) | 0.157 (0.383) |
| ADHD | ||
| Subscale a | ||
| ADHD-RS | ||
| Inattention | −0.057 (0.154) | 0.003 (0.987) |
| Hyperactivity | 0.025 (0.889) | 0.114 (0.528) |
| Total | −0.016 (0.930) | 0.043 (0.813) |
| Genus level | Ralstonia b | Afipia b |
| CBCL | ||
| Attention | −0.289 (0.103) | 0.427 × (0.013) |
| Problems | ||
| Subscale a | ||
| DSM | −0.339 (0.054) | 0.375 × (0.032) |
| oriented | ||
| ADHD | ||
| subscale a | ||
| ADHD-RS | ||
| Inattention | −0.012 (0.947) | −0.076 (0.676) |
| H/I | 0.049 (0.786) | −0.149 (0.409) |
| Total | 0.015 (0.936) | −0.093 (0.607) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, Y.J.; Shin, B.; Lee, S.-H.; Park, S.; Kim, Y.-K.; Kim, J.-J.; Kim, E. Altered Urine Microbiome in Male Children and Adolescents with Attention-Deficit Hyperactivity Disorder. Microorganisms 2023, 11, 2063. https://doi.org/10.3390/microorganisms11082063
Cho YJ, Shin B, Lee S-H, Park S, Kim Y-K, Kim J-J, Kim E. Altered Urine Microbiome in Male Children and Adolescents with Attention-Deficit Hyperactivity Disorder. Microorganisms. 2023; 11(8):2063. https://doi.org/10.3390/microorganisms11082063
Chicago/Turabian StyleCho, Yoon Jae, Bokyoung Shin, Sung-Ha Lee, Sangmin Park, Yoon-Keun Kim, Jae-Jin Kim, and Eunjoo Kim. 2023. "Altered Urine Microbiome in Male Children and Adolescents with Attention-Deficit Hyperactivity Disorder" Microorganisms 11, no. 8: 2063. https://doi.org/10.3390/microorganisms11082063
APA StyleCho, Y. J., Shin, B., Lee, S.-H., Park, S., Kim, Y.-K., Kim, J.-J., & Kim, E. (2023). Altered Urine Microbiome in Male Children and Adolescents with Attention-Deficit Hyperactivity Disorder. Microorganisms, 11(8), 2063. https://doi.org/10.3390/microorganisms11082063


_Di_Marco.png)



