Sphingomonas sediminicola Dae20 Is a Highly Promising Beneficial Bacteria for Crop Biostimulation Due to Its Positive Effects on Plant Growth and Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Cultures
2.2. In Vitro Arabidopsis Assay
2.3. Plant Growth Promoting Traits of S. sediminicola Dae20
2.4. Brassicaceae Assays
2.5. Statistical Analysis
3. Results
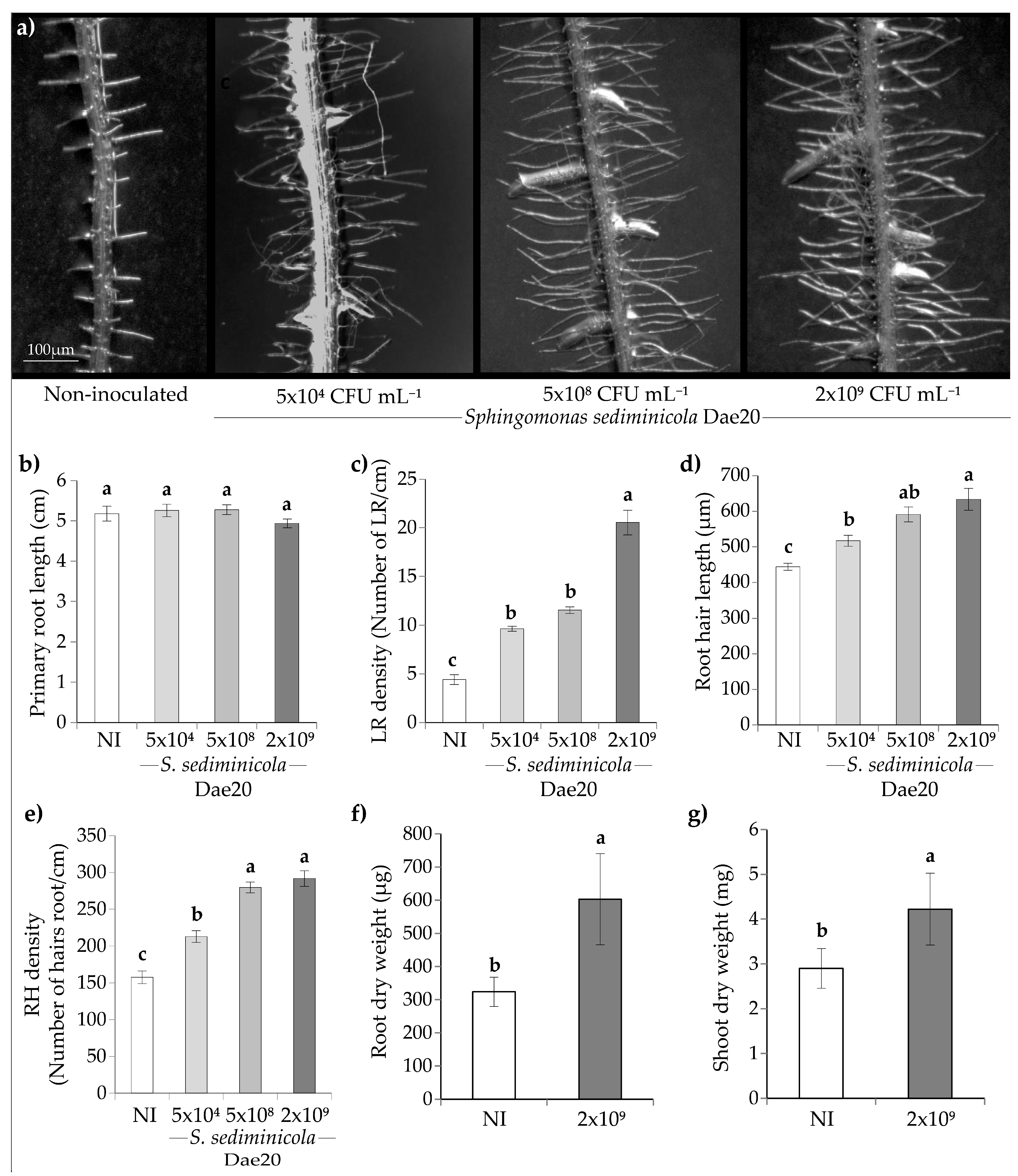
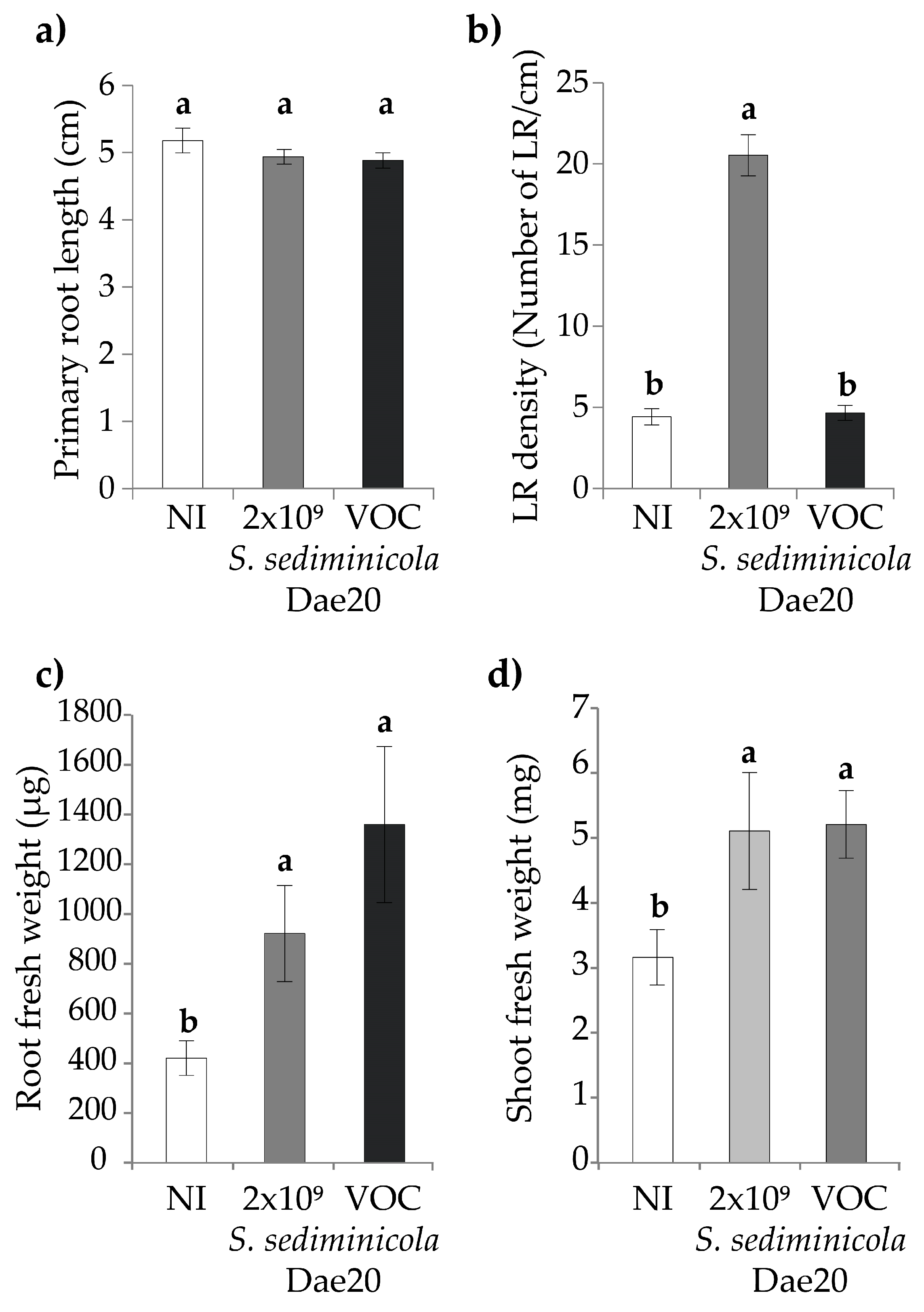
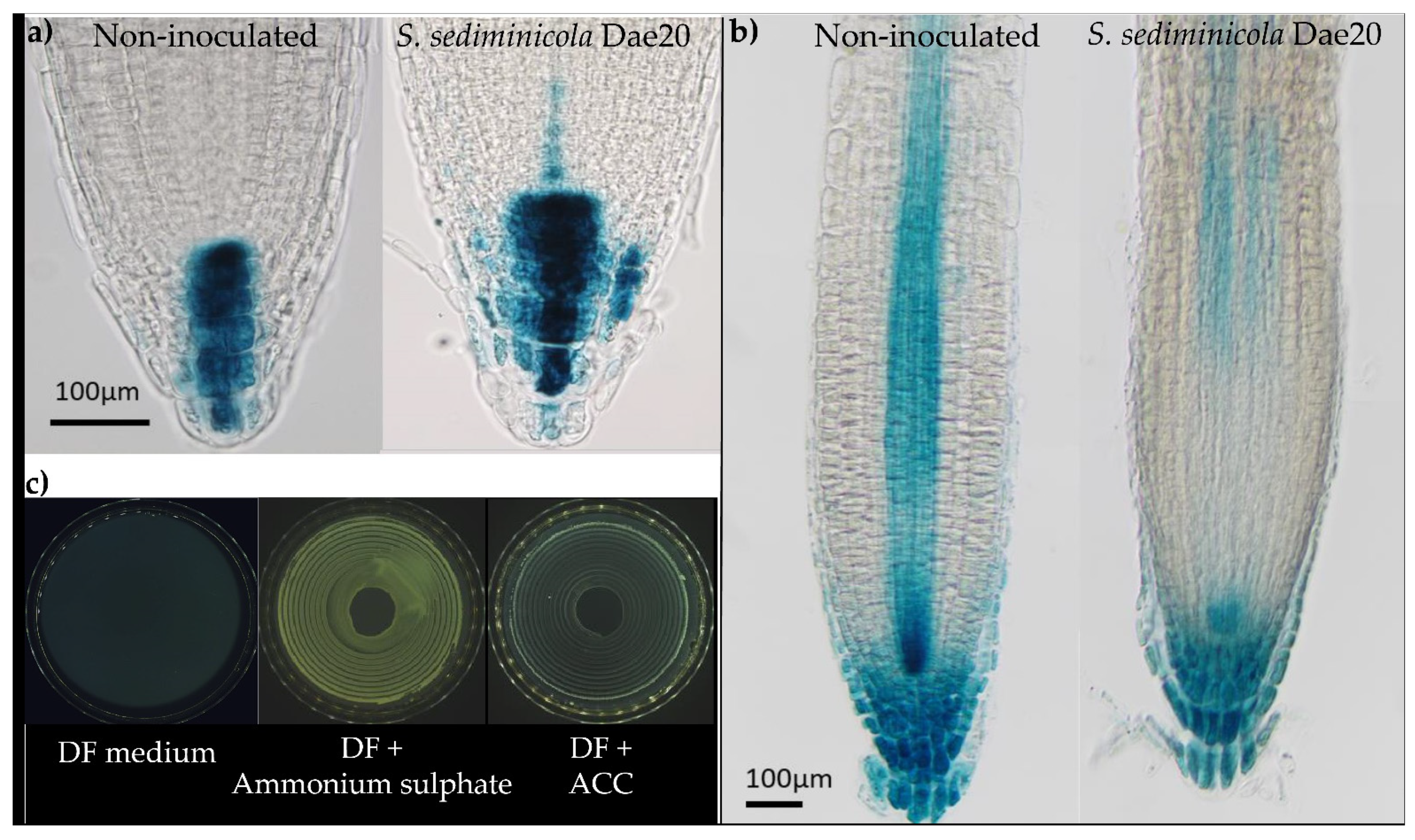
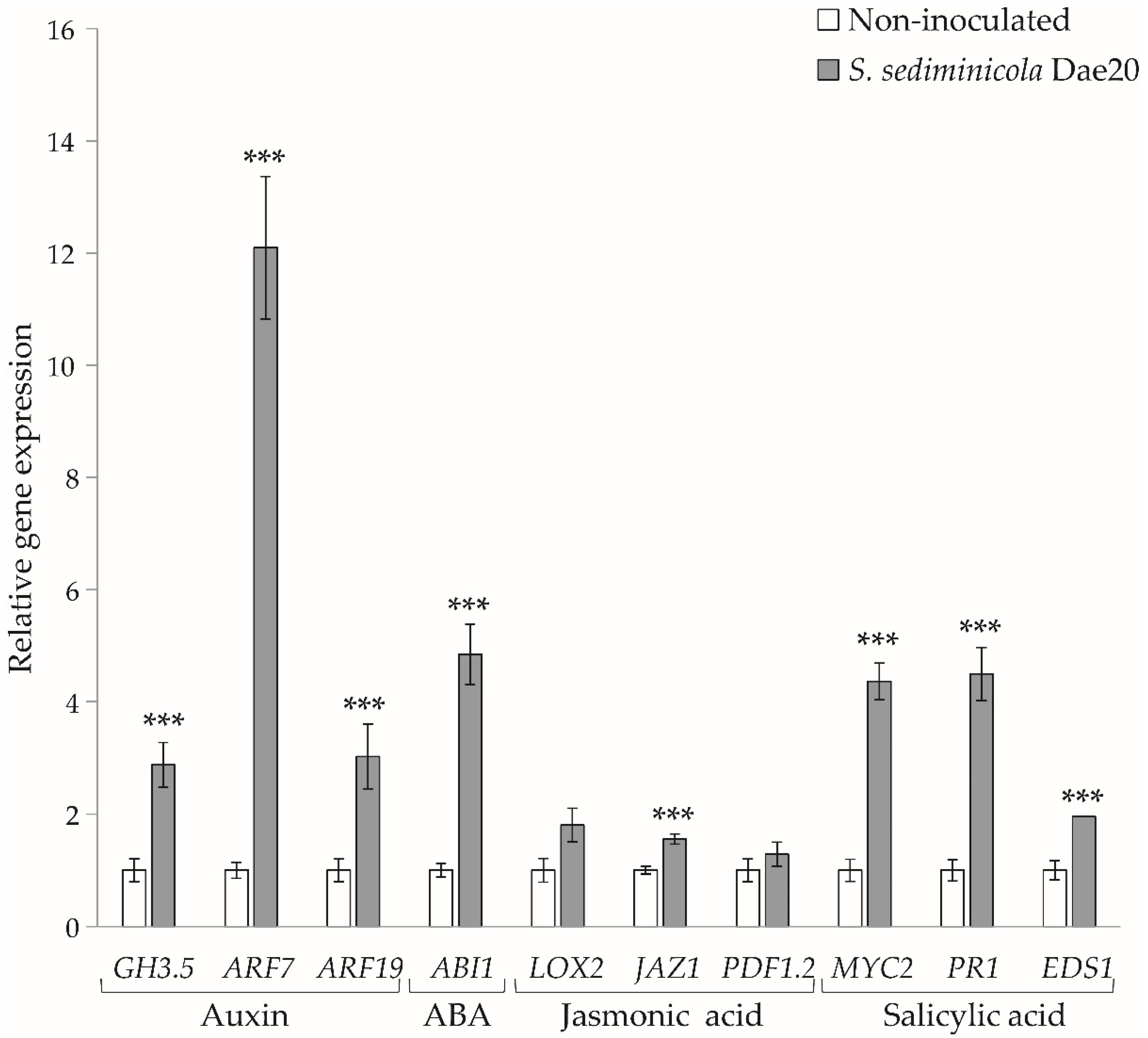
3.1. S. sediminicola Dae20 Stimulated the Root System of A. thaliana
3.2. S. sediminicola Dae20 Has Several PGPR Characteristics
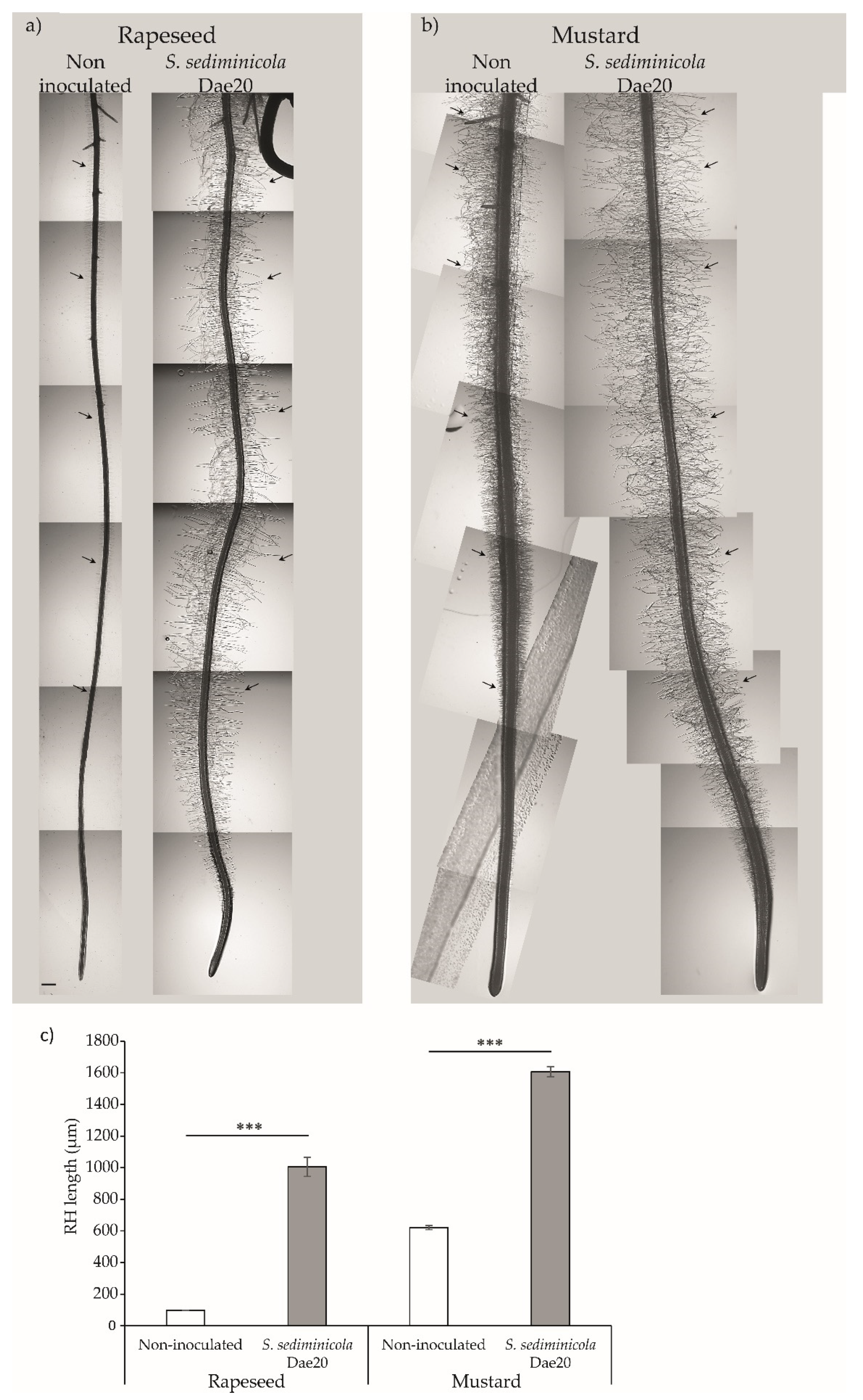
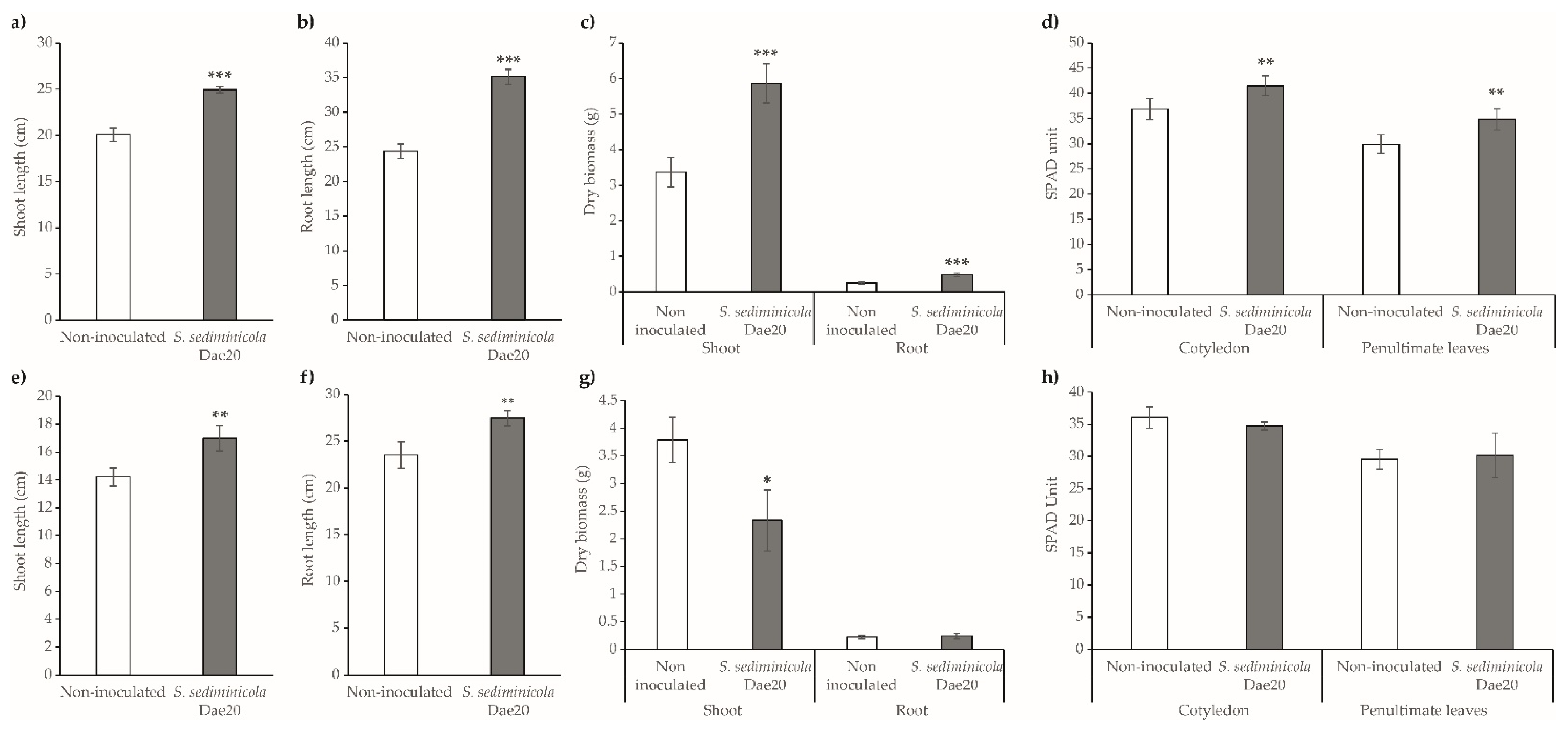
3.3. S. sediminicola Dae20 Enhances the Growth and Root System of Brassicaceae Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baweja, P.; Kumar, S.; Kumar, G. Fertilizers and Pesticides: Their Impact on Soil Health and Environment. Soil Health 2020, 265–285. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Nazari, M.; Antar, M.; Msimbira, L.A.; Naamala, J.; Lyu, D.; Rabileh, M.; Zajonc, J.; Smith, D.L. PGPR in Agriculture: A Sustainable Approach to Increasing Climate Change Resilience. Front. Sustain. Food Syst. 2021, 5, 667546. [Google Scholar] [CrossRef]
- Haskett, T.L.; Tkacz, A.; Poole, P.S. Engineering Rhizobacteria for Sustainable Agriculture. ISME J. 2021, 15, 949–964. [Google Scholar] [CrossRef]
- Hassan, M.K.; McInroy, J.A.; Kloepper, J.W. The Interactions of Rhizodeposits with Plant Growth-Promoting Rhizobacteria in the Rhizosphere: A Review. Agriculture 2019, 9, 142. [Google Scholar] [CrossRef]
- Venturi, V.; Bez, C. A Call to Arms for Cell–Cell Interactions between Bacteria in the Plant Microbiome. Trends Plant Sci. 2021, 26, 1126–1132. [Google Scholar] [CrossRef]
- Koprivova, A.; Kopriva, S. Plant Secondary Metabolites Altering Root Microbiome Composition and Function. Curr. Opin. Plant Biol. 2022, 67, 102227. [Google Scholar] [CrossRef]
- Geddes, B.A.; Kearsley, J.; Morton, R.; diCenzo, G.C.; Finan, T.M. Chapter Eight—The Genomes of Rhizobia. Adv. Bot. Res. 2020, 94, 213–249. [Google Scholar] [CrossRef]
- Kumari, B.; Mallick, M.; Solanki, M.; Solanki, A.; Hora, A.; Guo, W. Plant Growth Promoting Rhizobacteria (PGPR): Modern Prospects for Sustainable Agriculture. Plant Health Under Biot. Stress 2019, 2, 109–127. [Google Scholar] [CrossRef]
- Grover, M.; Bodhankar, S.; Sharma, A.; Sharma, P.; Singh, J.; Nain, L. PGPR Mediated Alterations in Root Traits: Way Toward Sustainable Crop Production. Front. Sustain. Food Syst. 2021, 4, 618230. [Google Scholar] [CrossRef]
- Bhattacharjee, R.B.; Jourand, P.; Chaintreuil, C.; Dreyfus, B.; Singh, A.; Mukhopadhyay, S.N. Indole Acetic Acid and ACC Deaminase-Producing Rhizobium leguminosarum Bv. Trifolii SN10 Promote Rice Growth, and in the Process Undergo Colonization and Chemotaxis. Biol. Fertil. Soils 2012, 48, 173–182. [Google Scholar] [CrossRef]
- van Wees, S.C.; de Swart, E.A.; van Pelt, J.A.; van Loon, L.C.; Pieterse, C.M. Enhancement of Induced Disease Resistance by Simultaneous Activation of Salicylate- and Jasmonate-Dependent Defense Pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 8711–8716. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Gui, Y.; Li, Z.; Jiang, C.-H.; Guo, J.; Niu, D. Induced Systemic Resistance for Improving Plant Immunity by Beneficial Microbes. Plants 2022, 11, 386. [Google Scholar] [CrossRef] [PubMed]
- Hazra, D.; Purkait, A. Role of Biostimulant Formulations in Crop Production: An Overview. Int. J. Agric. Sci. Vet. Med. 2020, 8, 38–48. [Google Scholar]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.-S.; Patra, J.K. Revitalization of Plant Growth Promoting Rhizobacteria for Sustainable Development in Agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef]
- Xie, C.-H.; Yokota, A. Sphingomonas azotifigens Sp. Nov., a Nitrogen-Fixing Bacterium Isolated from the Roots of Oryza Sativa. Int. J. Syst. Evol. Microbiol. 2006, 56, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.J.; Jo, E.J.; Yoon, H.S.; Song, G.C.; Jeon, C.O.; Chung, Y.R. 2011 Sphingomonas oryziterrae Sp. Nov. and Sphingomonas jinjuensis Sp. Nov. Isolated from Rhizosphere Soil of Rice (Oryza sativa L.). Int. J. Syst. Evol. Microbiol. 2011, 61, 2389–2394. [Google Scholar] [CrossRef]
- Khan, A.L.; Waqas, M.; Kang, S.-M.; Al-Harrasi, A.; Hussain, J.; Al-Rawahi, A.; Al-Khiziri, S.; Ullah, I.; Ali, L.; Jung, H.-Y.; et al. Bacterial Endophyte Sphingomonas Sp. LK11 Produces Gibberellins and IAA and Promotes Tomato Plant Growth. J. Microbiol. 2014, 52, 689–695. [Google Scholar] [CrossRef]
- Luo, Y.; Zhou, M.; Zhao, Q.; Wang, F.; Gao, J.; Sheng, H.; An, L. Complete Genome Sequence of Sphingomonas Sp. Cra20, a Drought Resistant and Plant Growth Promoting Rhizobacteria. Genomics 2020, 112, 3648–3657. [Google Scholar] [CrossRef]
- Mazoyon, C.; Hirel, B.; Pecourt, A.; Catterou, M.; Gutierrez, L.; Sarazin, V.; Dubois, F.; Duclercq, J. Sphingomonas sediminicola Is an Endosymbiotic Bacterium Able to Induce the Formation of Root Nodules in Pea (Pisum sativum L.) and to Enhance Plant Biomass Production. Microorganisms 2023, 11, 199. [Google Scholar] [CrossRef]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, Efflux-Dependent Auxin Gradients as a Common Module for Plant Organ Formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.E.; Benfey, P.N. Organization and Cell Differentiation in Lateral Roots of Arabidopsis thaliana. Development 1997, 124, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Arvidsson, S.; Kwasniewski, M.; Riaño-Pachón, D.M.; Mueller-Roeber, B. QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Goswami, D.; Pithwa, S.; Dhandhukia, P.; Thakker, J.N. Delineating Kocuria turfanensis 2M4 as a Credible PGPR: A Novel IAA-Producing Bacteria Isolated from Saline Desert. J. Plant Interact. 2014, 9, 566–576. [Google Scholar] [CrossRef]
- Bryan, N.S.; Grisham, M.B. Methods to Detect Nitric Oxide and Its Metabolites in Biological Samples. Free Radic. Biol. Med. 2007, 43, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Arora, N.K.; Verma, M. Modified Microplate Method for Rapid and Efficient Estimation of Siderophore Produced by Bacteria. 3 Biotech 2017, 7, 381. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for Isolating and Characterizing ACC Deaminase-Containing Plant Growth-Promoting Rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef]
- Dworkin, M.; Foster, J.W. Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [CrossRef]
- Li, T.; Li, Y.; Shi, Z.; Wang, S.; Wang, Z.; Liu, Y.; Wen, X.; Mo, F.; Han, J.; Liao, Y. Crop Development Has More Influence on Shaping Rhizobacteria of Wheat than Tillage Practice and Crop Rotation Pattern in an Arid Agroecosystem. Appl. Soil Ecol. 2021, 165, 104016. [Google Scholar] [CrossRef]
- Pikovskaya, R. Mobilization of Phosphorus in Soil in Connection with Vital Activity of Some Microbial Species. Mikrobiologiya 1948, 17, 362–370. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Mazoyon, C.; Firmin, S.; Bensaddek, L.; Pecourt, A.; Chabot, A.; Faucon, M.-P.; Sarazin, V.; Dubois, F.; Duclercq, J. Optimizing Crop Production with Bacterial Inputs: Insights into Chemical Dialogue between Sphingomonas sediminicola and Pisum sativum. Microorganisms 2023, 11, 1847. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhao, C.; Wang, E.; Raza, A.; Yin, C. Bacillus Amyloliquefaciens as an Excellent Agent for Biofertilizer and Biocontrol in Agriculture: An Overview for Its Mechanisms. Microbiol. Res. 2022, 259, 127016. [Google Scholar] [CrossRef] [PubMed]
- Hamid, B.; Zaman, M.; Farooq, S.; Fatima, S.; Sayyed, R.; Baba, Z.; Sheikh, T.; Reddy, M.; El Enshasy, H.; Gafur, A.; et al. Bacterial Plant Biostimulants: A Sustainable Way towards Improving Growth, Productivity, and Health of Crops. Sustainability 2021, 13, 2856. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.d.C.; Flores, A.; Rojas-Sánchez, B.; Urtis-Flores, C.A.; Morales-Cedeño, L.R.; Valencia-Marin, M.F.; Chávez-Avila, S.; Rojas-Solis, D.; Santoyo, G. Plant Growth-Promoting Bacteria as Bioinoculants: Attributes and Challenges for Sustainable Crop Improvement. Agronomy 2021, 11, 1167. [Google Scholar] [CrossRef]
- Ambrosini, A.; de Souza, R.; Passaglia, L.M.P. Ecological Role of Bacterial Inoculants and Their Potential Impact on Soil Microbial Diversity. Plant Soil 2016, 400, 193–207. [Google Scholar] [CrossRef]
- Peng, Q.; Shaaban, M.; Wu, Y.; Hu, R.; Wang, B.; Wang, J. The Diversity of Iron Reducing Bacteria Communities in Subtropical Paddy Soils of China. Appl. Soil Ecol. 2016, 101, 20–27. [Google Scholar] [CrossRef]
- Dong, W.; Liu, E.; Yan, C.; Tian, J.; Zhang, H.; Zhang, Y. Impact of No Tillage vs. Conventional Tillage on the Soil Bacterial Community Structure in a Winter Wheat Cropping Succession in Northern China. Eur. J. Soil Biol. 2017, 80, 35–42. [Google Scholar] [CrossRef]
- Alahmad, A.; Decocq, G.; Spicher, F.; Kheirbeik, L.; Kobaissi, A.; Tetu, T.; Dubois, F.; Duclercq, J. Cover Crops in Arable Lands Increase Functional Complementarity and Redundancy of Bacterial Communities. J. Appl. Ecol. 2019, 56, 651–664. [Google Scholar] [CrossRef]
- Lv, X.; Wang, Q.; Zhang, X.; Hao, J.; Li, L.; Chen, W.; Li, H.; Wang, Y.; Ma, C.; Wang, J.; et al. Community Structure and Associated Networks of Endophytic Bacteria in Pea Roots throughout Plant Life Cycle. Plant Soil 2021, 468, 225–238. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, F.; Huang, Y.; Zhou, M.; Gao, J.; Yan, T.; Sheng, H.; An, L. Sphingomonas Sp. Cra20 Increases Plant Growth Rate and Alters Rhizosphere Microbial Community Structure of Arabidopsis thaliana Under Drought Stress. Front. Microbiol. 2019, 10, 1221. [Google Scholar] [CrossRef]
- Chen, B.; Shen, J.; Zhang, X.; Pan, F.; Yang, X.; Feng, Y. The Endophytic Bacterium, Sphingomonas SaMR12, Improves the Potential for Zinc Phytoremediation by Its Host, Sedum alfredii. PLoS ONE 2014, 9, e106826. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ge, C.; Xu, S.; Wu, Y.; Sahito, Z.; Ma, L.; Pan, F.; Zhou, Q.; Huang, L.; Feng, Y.; et al. The Endophytic Bacterium Sphingomonas SaMR12 Alleviates Cd Stress in Oilseed Rape through Regulation of the GSH-AsA Cycle and Antioxidative Enzymes. BMC Plant Biol. 2020, 20, 63. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Modulation of Plant Ethylene Levels by the Bacterial Enzyme ACC Deaminase. FEMS Microbiol. Lett. 2005, 251, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Q.; Tian, X.; Mu, L.; Ji, M.; Wang, X.; Li, N.; Liu, F.; Shu, J.; Crawford, N.M.; et al. PHB3 Regulates Lateral Root Primordia Formation via NO-Mediated Degradation of AUXIN/INDOLE-3-ACETIC ACID Proteins. J. Exp. Bot. 2022, 73, 4034–4045. [Google Scholar] [CrossRef] [PubMed]
- Altamura, M.M.; Piacentini, D.; Della Rovere, F.; Fattorini, L.; Falasca, G.; Betti, C. New Paradigms in Brassinosteroids, Strigolactones, Sphingolipids, and Nitric Oxide Interaction in the Control of Lateral and Adventitious Root Formation. Plants 2023, 12, 413. [Google Scholar] [CrossRef]
- Tangaromsuk, J.; Pokethitiyook, P.; Kruatrachue, M.; Upatham, E.S. Cadmium Biosorption by Sphingomonas paucimobilis Biomass. Bioresour. Technol. 2002, 85, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.L.; Waqas, M.; Asaf, S.; Kamran, M.; Shahzad, R.; Bilal, S.; Khan, M.A.; Kang, S.-M.; Kim, Y.-H.; Yun, B.-W.; et al. Plant Growth-Promoting Endophyte Sphingomonas Sp. LK11 Alleviates Salinity Stress in Solanum pimpinellifolium. Environ. Exp. Bot. 2017, 133, 58–69. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, X.; Cao, Z.; Zhao, K.; Wang, S.; Chen, M.; Hu, X. Growth-Promoting Sphingomonas paucimobilis ZJSH1 Associated with Dendrobium officinale through Phytohormone Production and Nitrogen Fixation. Microb. Biotechnol. 2014, 7, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Halo, B.A.; Khan, A.L.; Waqas, M.; Al-Harrasi, A.; Hussain, J.; Ali, L.; Adnan, M.; Lee, I.-J. Endophytic Bacteria (Sphingomonas Sp. LK11) and Gibberellin Can Improve Solanum lycopersicum Growth and Oxidative Stress under Salinity. J. Plant Interact. 2015, 10, 117–125. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, M.A.; Khan, A.L.; Waqas, M.; Shahzad, R.; Kim, A.-Y.; Kang, S.-M.; Lee, I.-J. Bacterial Endophytes from Arid Land Plants Regulate Endogenous Hormone Content and Promote Growth in Crop Plants: An Example of Sphingomonas Sp. and Serratia marcescens. J. Plant Interact. 2017, 12, 31–38. [Google Scholar] [CrossRef]
- Menon, R.R.; Kumari, S.; Kumar, P.; Verma, A.; Krishnamurthi, S.; Rameshkumar, N. Sphingomonas pokkalii Sp. Nov., a Novel Plant Associated Rhizobacterium Isolated from a Saline Tolerant Pokkali Rice and Its Draft Genome Analysis. Syst. Appl. Microbiol. 2019, 42, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against Leaf-Pathogenic Pseudomonas syringae by Sphingomonas Strains in a Controlled Model System. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.L.; Collavino, M.M.; Sansberro, P.A.; Mroginski, L.A.; Galdeano, E. Diversity of Endophytic Fungal and Bacterial Communities in Ilex paraguariensis Grown under Field Conditions. World J. Microbiol. Biotechnol. 2016, 32, 61. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Joseph, C.M.; Yang, G.; Phillips, D.A.; Nelson, L.M. Evaluation of Bacteria Isolated from Rice for Plant Growth Promotion and Biological Control of Seedling Disease of Rice. Can. J. Microbiol. 2001, 47, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Saharan, B.; Nehra, V. Plant Growth Promoting Rhizobacteria: A Critical Review. Life Sci. Med. Res. 2011, 21, 30. [Google Scholar]
- Enebe, M.C.; Babalola, O.O. The Influence of Plant Growth-Promoting Rhizobacteria in Plant Tolerance to Abiotic Stress: A Survival Strategy. Appl. Microbiol. Biotechnol. 2018, 102, 7821–7835. [Google Scholar] [CrossRef]
- Shaffique, S.; Khan, M.A.; Wani, S.H.; Pande, A.; Imran, M.; Kang, S.-M.; Rahim, W.; Khan, S.A.; Bhatta, D.; Kwon, E.-H.; et al. A Review on the Role of Endophytes and Plant Growth Promoting Rhizobacteria in Mitigating Heat Stress in Plants. Microorganisms 2022, 10, 1286. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Forero, S.K.; Rojas, E.; Beggah, S.; van der Meer, J.R. Comparison of Differential Gene Expression to Water Stress among Bacteria with Relevant Pollutant-Degradation Properties. Environ. Microbiol. Rep. 2016, 8, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Seo, J.; Peng, Y.; Jeon, D.; Lee, J.H.; Kim, C.Y.; Lee, J. A Nostoxanthin-Producing Bacterium, Sphingomonas nostoxanthinifaciens Sp. Nov., Alleviates the Salt Stress of Arabidopsis Seedlings by Scavenging of Reactive Oxygen Species. Front. Microbiol. 2023, 14, 1101150. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, H.; Pu, Q.; Zhang, C.; Chen, Y.; Lin, Z.; Hu, X.; Li, O. Complete Genome of Sphingomonas paucimobilis ZJSH1, an Endophytic Bacterium from Dendrobium Officinale with Stress Resistance and Growth Promotion Potential. Arch. Microbiol. 2023, 205, 132. [Google Scholar] [CrossRef] [PubMed]
- Latz, E.; Eisenhauer, N.; Scheu, S.; Jousset, A. Plant identity drives the expression of biocontrol factors in a rhizosphere bacterium across a plant diversity gradient. Funct. Ecol. 2015, 29, 1225–1234. [Google Scholar] [CrossRef]
| PGPR Properties | Sphingomonas sediminicola Dae20 |
|---|---|
| IAA production (µg/mg dry weight of bacteria) | 1.25 ± 0.04 |
| Nitric oxide production (µg/mg dry weight/minutes) | 175.81 ± 12.26 |
| Percent siderophore unit (psu) | 78.09 ± 4.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazoyon, C.; Catterou, M.; Alahmad, A.; Mongelard, G.; Guénin, S.; Sarazin, V.; Dubois, F.; Duclercq, J. Sphingomonas sediminicola Dae20 Is a Highly Promising Beneficial Bacteria for Crop Biostimulation Due to Its Positive Effects on Plant Growth and Development. Microorganisms 2023, 11, 2061. https://doi.org/10.3390/microorganisms11082061
Mazoyon C, Catterou M, Alahmad A, Mongelard G, Guénin S, Sarazin V, Dubois F, Duclercq J. Sphingomonas sediminicola Dae20 Is a Highly Promising Beneficial Bacteria for Crop Biostimulation Due to Its Positive Effects on Plant Growth and Development. Microorganisms. 2023; 11(8):2061. https://doi.org/10.3390/microorganisms11082061
Chicago/Turabian StyleMazoyon, Candice, Manuella Catterou, Abdelrahman Alahmad, Gaëlle Mongelard, Stéphanie Guénin, Vivien Sarazin, Fréderic Dubois, and Jérôme Duclercq. 2023. "Sphingomonas sediminicola Dae20 Is a Highly Promising Beneficial Bacteria for Crop Biostimulation Due to Its Positive Effects on Plant Growth and Development" Microorganisms 11, no. 8: 2061. https://doi.org/10.3390/microorganisms11082061
APA StyleMazoyon, C., Catterou, M., Alahmad, A., Mongelard, G., Guénin, S., Sarazin, V., Dubois, F., & Duclercq, J. (2023). Sphingomonas sediminicola Dae20 Is a Highly Promising Beneficial Bacteria for Crop Biostimulation Due to Its Positive Effects on Plant Growth and Development. Microorganisms, 11(8), 2061. https://doi.org/10.3390/microorganisms11082061










