Influenza A Virus in Pigs in Senegal and Risk Assessment of Avian Influenza Virus (AIV) Emergence and Transmission to Human
Abstract
1. Introduction
2. Materials and Methods
2.1. Surveillance and Sample Collection
2.2. Detection of Swine Influenza A Viruses (swIAVs) by RT-qPCR
2.2.1. RNA Extraction from Pigs’ Nasal Swabs
2.2.2. Screening of Influenza Viruses
2.3. Complete IAV Genome Amplification and NGS Sequencing
2.3.1. cDNA Synthesis
2.3.2. Multi-Segment PCR of Swine Influenza A Virus (swIAV)
2.3.3. Next Generation Sequencing
2.4. Phylogenetic Analysis
2.5. Hemagglutination Inhibition (HI) Assays of Serum Samples
2.6. Statistical Analysis
2.7. Ethical Statement and Permission
3. Results
3.1. Sample Collection
3.2. Detection of IAVs
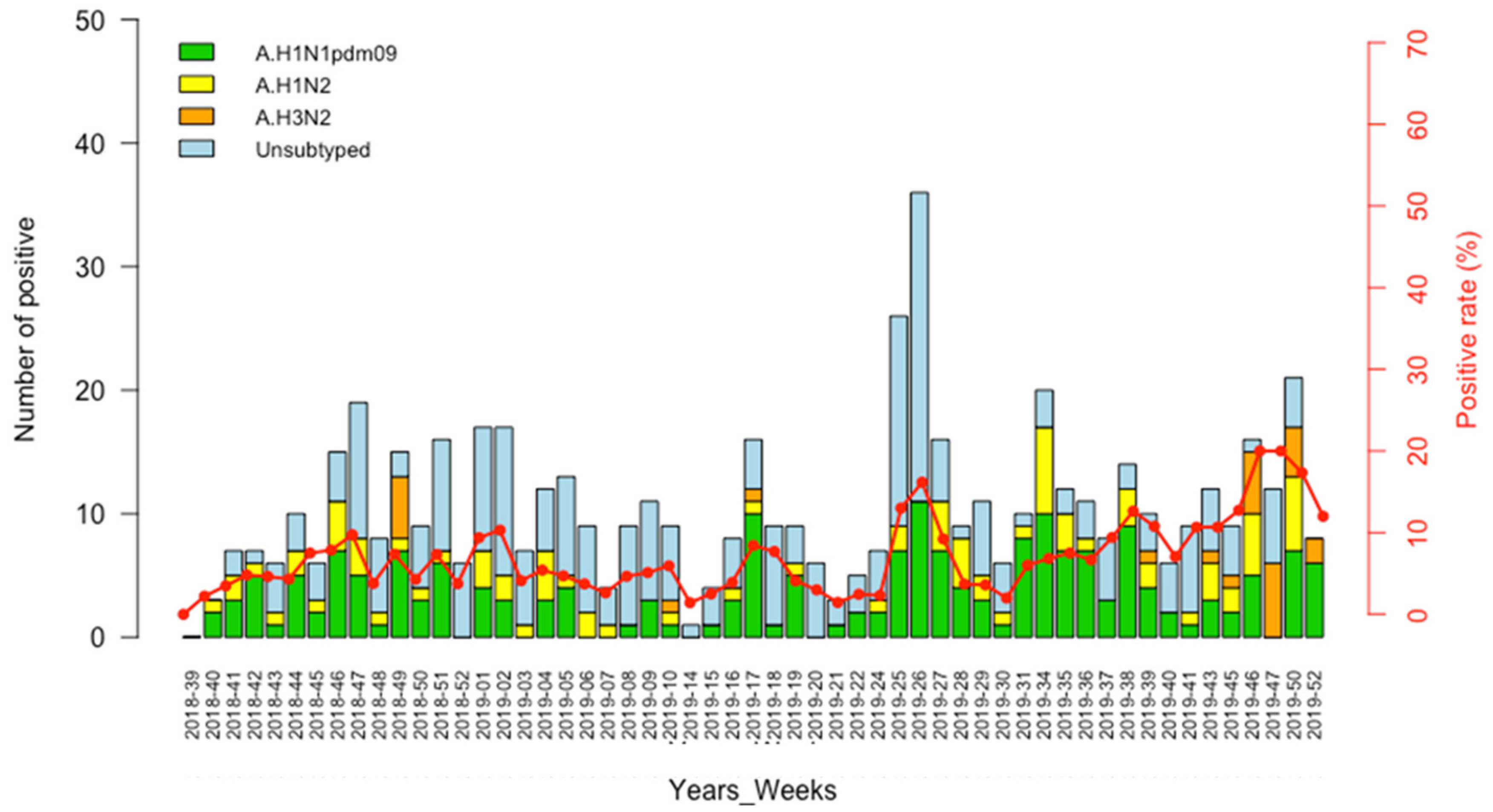
3.3. Circulation Pattern of Swine Influenza A Viruses (swIAVs)
3.4. Phylogenetic Analysis of IAV Strains Circulating in Senegalese Pigs, 2018–2019
3.5. Genetic Analysis of Amino Acid Residues of Senegalese Strains
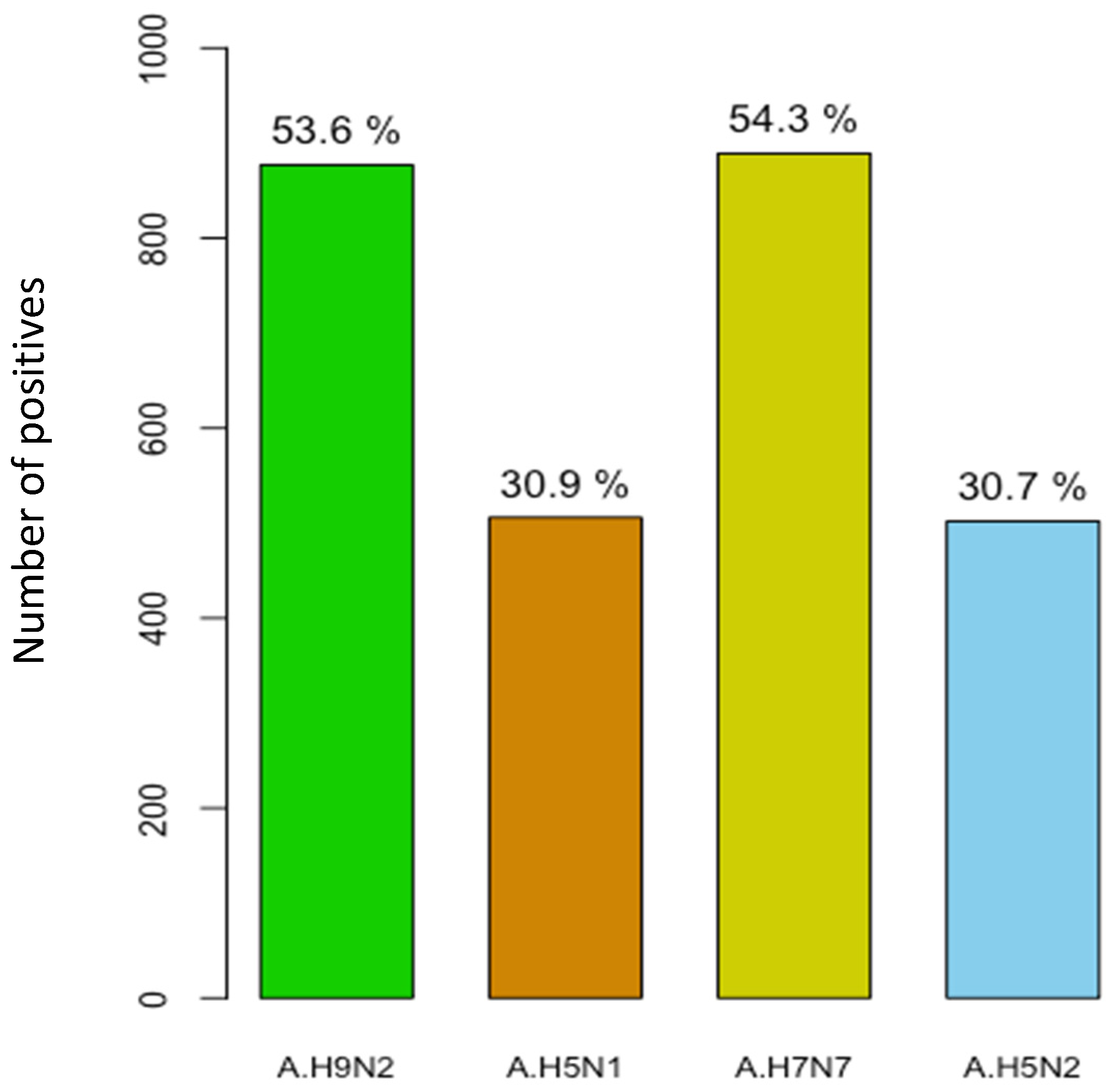
3.6. Serological Evidence of AIV Infection in Pigs
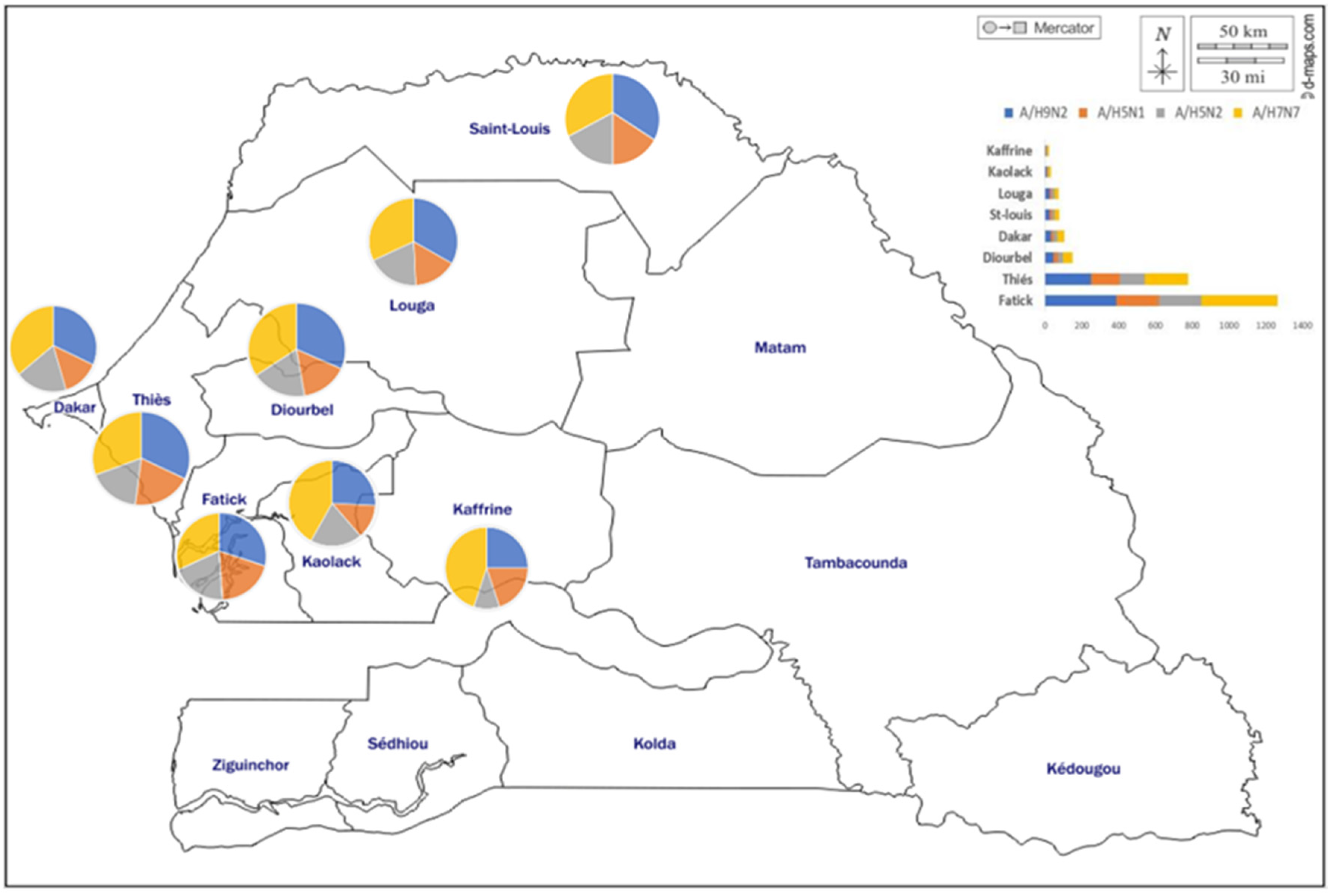
3.7. Geographical Distribution of AIV Seropositive Cases
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Touré, C.T.; Fall, A.; Andriamandimby, S.F.; Jallow, M.M.; Goudiaby, D.; Kiori, D.; Sy, S.; Diaw, Y.; Ndiaye, K.N.; Mbaye, F.; et al. Epidemiology and Molecular Analyses of Influenza B Viruses in Senegal from 2010 to 2019. Viruses 2022, 14, 1063. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Asha, K.; Khanna, M.; Ronsard, L.; Meseko, C.A.; Sanicas, M. The emerging influenza virus threat: Status and new prospects for its therapy and control. Arch. Virol. 2018, 163, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Dawson, W.K.; Lazniewski, M.; Plewczynski, D. RNA structure interactions and ribonucleoprotein processes of the influenza A virus. Brief. Funct. Genom. 2018, 17, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Vemula, S.V.; Zhao, J.; Liu, J.; Wang, X.; Biswas, S.; Hewlett, I. Current approaches for diagnosis of influenza virus infections in humans. Viruses 2016, 8, 96. [Google Scholar] [CrossRef]
- Kalonda, A.; Phonera, M.; Saasa, N.; Kajihara, M.; Sutcliffe, C.G.; Sawa, H.; Takada, A.; Simulundu, E. Influenza A and D viruses in non-human mammalian hosts in africa: A systematic review and meta-analysis. Viruses 2021, 13, 2411. [Google Scholar] [CrossRef]
- Fodor, E.; Te Velthuis, A.J. Structure and function of the influenza virus transcription and replication machinery. Cold Spring Harb. Perspect. Med. 2020, 10, a038398. [Google Scholar] [CrossRef]
- Jester, B.; Uyeki, T.; Jernigan, D. Readiness for responding to a severe pandemic 100 years after 1918. Am. J. Epidemiol. 2018, 187, 2596. [Google Scholar] [CrossRef]
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006, 12, 9. [Google Scholar] [CrossRef]
- O’donovan, T.; Donohoe, L.; Ducatez, M.F.; Meyer, G.; Ryan, E. Seroprevalence of influenza D virus in selected sample groups of Irish cattle, sheep and pigs. Ir. Vet. J. 2019, 72, 11. [Google Scholar] [CrossRef]
- Ducatez, M.F.; Pelletier, C.; Meyer, G. Influenza D virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368. [Google Scholar] [CrossRef]
- Henritzi, D.; Petric, P.P.; Lewis, N.S.; Graaf, A.; Pessia, A.; Starick, E.; Breithaupt, A.; Strebelow, G.; Luttermann, C.; Parker, L.M.K.; et al. Surveillance of European domestic pig populations identifies an emerging reservoir of potentially zoonotic swine influenza A viruses. Cell Host Microbe 2020, 28, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Kotey, E.N.; Asante, I.A.; Adusei-Poku, M.; Arjarquah, A.; Ampadu, R.; Rodgers, D.; Nyarko, E.O.; Asiedu, W.; Dafeamekpor, C.; Wiley, M.R.; et al. Phylogenetic and genetic characterization of influenza A H9N2 viruses isolated from backyard poultry in selected farms in Ghana. Vet. Med. Sci. 2022, 8, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Osoro, E.M.; Lidechi, S.; Marwanga, D.; Nyaundi, J.; Mwatondo, A.; Muturi, M.; Ng’ang’a, Z.; Njenga, K. Seroprevalence of influenza A virus in pigs and low risk of acute respiratory illness among pig workers in Kenya. Environ. Health Prev. Med. 2019, 24, 53. [Google Scholar] [CrossRef]
- Webby, R.J.; Webster, R.G. Emergence of influenza A viruses. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2001, 356, 1817–1828. [Google Scholar] [CrossRef]
- Takemae, N.; Harada, M.; Nguyen, P.T.; Nguyen, T.; Nguyen, T.N.; To, T.L.; Nguyen, T.D.; Pham, V.P.; Le, V.T.; Do, H.T.; et al. Influenza A viruses of swine (IAV-S) in Vietnam from 2010 to 2015: Multiple introductions of A (H1N1) pdm09 viruses into the pig population and diversifying genetic constellations of enzootic IAV-S. J. Virol. 2017, 91, e01490-16. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Qi, W.; Chen, J.; Zhu, W.; Huang, Z.; Xie, J.; Zhang, G. Seroepidemiological evidence of avian influenza A virus transmission to pigs in southern China. J. Clin. Microbiol. 2013, 51, 601–602. [Google Scholar] [CrossRef][Green Version]
- Song, Y.; Zhang, Y.; Zhang, B.; Chen, L.; Zhang, M.; Wang, J.; Jiang, Y.; Yang, C.; Jiang, T. Identification, genetic analysis, and pathogenicity of classical swine H1N1 and human-swine reassortant H1N1 influenza viruses from pigs in China. Viruses 2020, 12, 55. [Google Scholar] [CrossRef]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef]
- Mena, I.; Nelson, M.I.; Quezada-Monroy, F.; Dutta, J.; Cortes-Fernández, R.; Lara-Puente, J.H.; Castro-Peralta, F.; Cunha, L.F.; Trovão, N.S.; Lozano-Dubernard, B.; et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 2016, 5, e16777. [Google Scholar] [CrossRef]
- Watson, S.J.; Langat, P.; Reid, S.M.; Lam TT, Y.; Cotten, M.; Kelly, M.; Van Reeth, K.; Qiu, Y.; Simon, G.; Bonin, E.; et al. Molecular epidemiology and evolution of influenza viruses circulating within European swine between 2009 and 2013. J. Virol. 2015, 89, 9920–9931. [Google Scholar] [CrossRef]
- Kim, J.I.; Lee, I.; Park, S.; Lee, S.; Hwang, M.W.; Bae, J.Y.; Heo, J.; Kim, D.; Jang, S.-I.; Kim, K.; et al. Phylogenetic analysis of a swine influenza A (H3N2) virus isolated in Korea in 2012. PLoS ONE 2014, 9, e88782. [Google Scholar] [CrossRef] [PubMed]
- Biggerstaff, M.; Reed, C.; Epperson, S.; Jhung, M.A.; Gambhir, M.; Bresee, J.S.; Jernigan, D.B.; Swerdlow, D.L.; Finelli, L. Estimates of the number of human infections with influenza A (H3N2) variant virus, United States, August 2011–April 2012. Clin. Infect. Dis. 2013, 57 (Suppl. S1), S12–S15. [Google Scholar] [CrossRef] [PubMed]
- Meseko, C.A.; Odaibo, G.N.; Olaleye, D.O. Detection and isolation of 2009 pandemic influenza A/H1N1 virus in commercial piggery, Lagos Nigeria. Vet. Microbiol. 2014, 168, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.; Larsen, L.E.; Dürrwald, R.; Foni, E.; Harder, T.; Van Reeth, K.; Markowska-Daniel, I.; Reid, S.M.; Dan, A.; Maldonado, J.; et al. European surveillance network for influenza in pigs: Surveillance programs, diagnostic tools and Swine influenza virus subtypes identified in 14 European countries from 2010 to 2013. PLoS ONE 2014, 9, e115815. [Google Scholar] [CrossRef]
- Baudon, E.; Peyre, M.; Tung, D.D.; Thi Nga, P.; Khong, N.V.; Cowling, B.J.; Peiris, M. Surveillance of swine influenza viruses in sentinel familial farms in Hung Yen province in Northern Vietnam in 2013–2014. Zoonoses Public Health 2020, 67, 213–221. [Google Scholar] [CrossRef]
- Lo, F.T.; Zecchin, B.; Diallo, A.A.; Racky, O.; Tassoni, L.; Diop, A.; Diouf, M.; Diouf, M.; Samb, Y.N.; Pastori, A.; et al. Intercontinental spread of Eurasian highly pathogenic avian influenza A (H5N1) to Senegal. Emerg. Infect. Dis. 2022, 28, 234. [Google Scholar] [CrossRef]
- Jallow, M.M.; Fall, A.; Barry, M.A.; Diop, B.; Sy, S.; Goudiaby, D.; Fall, M.; Enouf, V.; Niang, M.N.; Dia, N. Genetic characterization of the first detected human case of low pathogenic avian influenza A/H9N2 in sub-Saharan Africa, Senegal. Emerg. Microbes Infect. 2020, 9, 1092–1095. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. CDC Protocol of Realtime RTPCR for Influenza A (H1N1); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2009.
- Zhou, B.; Donnelly, M.E.; Scholes, D.T.; St George, K.; Hatta, M.; Kawaoka, Y.; Wentworth, D.E. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J. Virol. 2009, 83, 10309–10313. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hill, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Oxford University: Oxford, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- World Health Organization. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Yuan, Z.; Zhu, W.; Chen, Y.; Zhou, P.; Cao, Z.; Xie, J.; Zhang, C.; Ke, C.; Qi, W.; Su, S.; et al. Serological surveillance of H5 and H9 avian influenza A viral infections among pigs in Southern China. Microb. Pathog. 2013, 64, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Rith, S.; Netrabukkana, P.; Sorn, S.; Mumford, E.; Mey, C.; Holl, D.; Goutard, F.; Y, B.; Fenwick, S.; Robertson, I.; et al. Serologic evidence of human influenza virus infections in swine populations, Cambodia. Influenza Other Respir. Viruses 2013, 7, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.W.; Barr, I.G.; Loh, R.; Levy, A.; Tempone, S.; O’Dea, M.; Watson, J.; Wong, F.Y.K.; Effler, P.V. Respiratory illness in a piggery associated with the first identified outbreak of swine influenza in Australia: Assessing the risk to human health and zoonotic potential. Trop. Med. Infect. Dis. 2019, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Harder, T.C.; grosse Beilage, E.; Lange, E.; Meiners, C.; Döhring, S.; Pesch, S.; Noé, T.; Grund, C.; Beer, M.; Starick, E. Expanded cocirculation of stable subtypes, emerging lineages, and new sporadic reassortants of porcine influenza viruses in swine populations in Northwest Germany. J. Virol. 2013, 87, 10460–10476. [Google Scholar] [CrossRef]
- Dia, N.; Ndiaye, M.N.; de Lourdes Monteiro, M.; Koivogui, L.; Bara, M.O.; Diop, O.M. A subregional analysis of epidemiologic and genetic characteristics of influenza A (H1N1) pdm09 in Africa: Senegal, Cape Verde, Mauritania, and Guinea, 2009–2010. Am. J. Trop. Med. Hyg. 2013, 88, 946. [Google Scholar] [CrossRef]
- Niang, M.N.; Barry, M.A.; Talla, C.; Mbengue, A.; Sarr, F.D.; Ba, I.O.; Hedible, B.G.; Ndoye, B.; Vray, M.; Dia, N.; et al. Estimation of the burden of flu-association influenza-like illness visits on total clinic visits through the sentinel influenza monitoring system in Senegal during the 2013–2015 influenza seasons. Epidemiol. Infect. 2018, 146, 2049–2055. [Google Scholar] [CrossRef]
- Niang, M.N.; Dosseh, A.; Ndiaye, K.; Sagna, M.; Gregory, V.; Goudiaby, D.; Hay, A.; Diop, O.M. Sentinel surveillance for influenza in Senegal, 1996–2009. J. Infect. Dis. 2012, 206 (Suppl. S1), S129–S135. [Google Scholar] [CrossRef]
- Li, H.; Leng, H.; Tang, S.; Su, C.; Xu, Y.; Wang, Y.; Lv, J.; Zhang, S.; Feng, Y.; Song, S.; et al. Prevalence, genetics and evolutionary properties of Eurasian avian-like H1N1 swine influenza viruses in Liaoning. Viruses 2022, 14, 643. [Google Scholar] [CrossRef]
- Osbjer, K.; Berg, M.; Sokerya, S.; Chheng, K.; San, S.; Davun, H.; Magnusson, U.; Olsen, B.; Zohari, S. Influenza A virus in backyard pigs and poultry in rural Cambodia. Transbound. Emerg. Dis. 2017, 64, 1557–1568. [Google Scholar] [CrossRef]
- Karlsson, E.A.; Ciuoderis, K.; Freiden, P.J.; Seufzer, B.; Jones, J.C.; Johnson, J.; Parra, R.; Gongora, A.; Cardenas, D.; Barajas, D.; et al. Prevalence and characterization of influenza viruses in diverse species in Los Llanos, Colombia: Prevalence of influenza viruses in Colombia. Emerg. Microbes Infect. 2013, 2, 1–10. [Google Scholar] [CrossRef]
- Baudon, E.; Chu, D.K.; Tung, D.D.; Thi Nga, P.; Vu Mai Phuong, H.; Le Khanh Hang, N.; Thanh, L.T.; Thuy, N.T.; Khanh, N.C.; Mai, L.Q.; et al. Swine influenza viruses in Northern Vietnam in 2013–2014. Emerg. Microbes Infect. 2018, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, M.R.; Kandeil, A.; El-Shesheny, R.; Shehata, M.M.; McKenzie, P.P.; Webby, R.J.; Ali, M.A.; Kayali, G. Evidence of infection with avian, human, and swine influenza viruses in pigs in Cairo, Egypt. Arch. Virol. 2018, 163, 359–364. [Google Scholar] [CrossRef]
- Kyriakis, C.S.; Brown, I.H.; Foni, E.; Kuntz-Simon, G.; Maldonado, J.; Madec, F.; Essen, S.C.; Chiapponi, C.; Van Reeth, K. Virological surveillance and preliminary antigenic characterization of influenza viruses in pigs in five European countries from 2006 to 2008. Zoonoses Public Health 2011, 58, 93–101. [Google Scholar] [CrossRef]
- Corzo, C.A.; Culhane, M.; Juleen, K.; Stigger-Rosser, E.; Ducatez, M.F.; Webby, R.J.; Lowe, J.F. Active surveillance for influenza A virus among swine, midwestern United States, 2009–2011. Emerg. Infect. Dis. 2013, 19, 954. [Google Scholar] [CrossRef] [PubMed]
- Osoro, E.M.; Lidechi, S.; Nyaundi, J.; Marwanga, D.; Mwatondo, A.; Muturi, M.; Ng’ang’a, Z.; Njenga, K. Detection of pandemic influenza A/H1N1/pdm09 virus among pigs but not in humans in slaughterhouses in Kenya, 2013–2014. BMC Res. Notes 2019, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Song, D.S.; Kang, B.K.; Oh, J.S.; Park, B.K. Serologic surveillance of swine H1 and H3 and avian H5 and H9 influenza A virus infections in swine population in Korea. Prev. Vet. Med. 2007, 79, 294–303. [Google Scholar] [CrossRef]
- Ali, A.; Khatri, M.; Wang, L.; Saif, Y.M.; Lee, C.W. Identification of swine H1N2/pandemic H1N1 reassortant influenza virus in pigs, United States. Vet. Microbiol. 2012, 158, 60–68. [Google Scholar] [CrossRef]
- Ciacci-Zanella, J.R.; Schaefer, R.; Gava, D.; Haach, V.; Cantão, M.E.; Coldebella, A. Influenza A virus infection in Brazilian swine herds following the introduction of pandemic 2009 H1N1. Vet. Microbiol. 2015, 180, 118–122. [Google Scholar] [CrossRef]
- Abe, H.; Mine, J.; Parchariyanon, S.; Takemae, N.; Boonpornprasert, P.; Ubonyaem, N.; Patcharasinghawut, P.; Nuansrichay, B.; Tanikawa, T.; Tsunekuni, R.; et al. Co-infection of influenza A viruses of swine contributes to effective shuffling of gene segments in a naturally reared pig. Virology 2015, 484, 203–212. [Google Scholar] [CrossRef]
- Maya-Badillo, B.A.; Ojeda-Flores, R.; Chaves, A.; Reveles-Félix, S.; Orta-Pineda, G.; Martínez-Mercado, M.J.; Saavedra-Montañez, M.; Segura-Velázquez, R.; Sanvicente, M.; Sánchez-Betancourt, J.I. Eco-epidemiological evidence of the transmission of avian and human influenza A viruses in wild pigs in Campeche, Mexico. Viruses 2020, 12, 528. [Google Scholar] [CrossRef]
- Chiapponi, C.; Prosperi, A.; Moreno, A.; Baioni, L.; Faccini, S.; Manfredi, R.; Zanni, I.; Gabbi, V.; Calanchi, I.; Fusaro, A.; et al. Genetic variability among swine Influenza viruses in Italy: Data analysis of the period 2017–2020. Viruses 2021, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Markin, A.; Ciacci Zanella, G.; Arendsee, Z.W.; Zhang, J.; Krueger, K.M.; Gauger, P.C.; Baker, A.L.V.; Anderson, T.K. Reverse-zoonoses of 2009 H1N1 pandemic influenza A viruses and evolution in United States swine results in viruses with zoonotic potential. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Kirisawa, R.; Ogasawara, Y.; Yoshitake, H.; Koda, A.; Furuya, T. Genomic reassortants of pandemic A (H1N1) 2009 virus and endemic porcine H1 and H3 viruses in swine in Japan. J. Vet. Med. Sci. 2014, 76, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Brockwell-Staats, C.; Webster, R.G.; Webby, R.J. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influenza Other Respir. Viruses 2009, 3, 207–213. [Google Scholar] [CrossRef]
- Ngo, L.T.; Hiromoto, Y.; Pham, V.P.; Le HT, H.; Nguyen, H.T.; Le, V.T.; Takemae, N.; Saito, T. Isolation of novel triple-reassortant swine H3N2 influenza viruses possessing the hemagglutinin and neuraminidase genes of a seasonal influenza virus in Vietnam in 2010. Influenza Other Respir. Viruses 2012, 6, 6–10. [Google Scholar] [CrossRef]
- Fan, X.; Zhu, H.; Zhou, B.; Smith, D.K.; Chen, X.; Lam, T.T.Y.; Poon, L.L.M.; Peiris, M.; Guan, Y. Emergence and dissemination of a swine H3N2 reassortant influenza virus with 2009 pandemic H1N1 genes in pigs in China. J. Virol. 2012, 86, 2375–2378. [Google Scholar] [CrossRef][Green Version]
- Franck, N.; Queguiner, S.; Gorin, S.; Eveno, E.; Fablet, C.; Madec, F.; Kuntz-Simon, G. Molecular epidemiology of swine influenza virus in France: Identification of novel H1N1 reassortants. In Proceedings of the 5th International Symposium on Emerging and Re-Emerging Pig Diseases, Krakow, Poland, 24–27 June 2007; Volume 250, pp. 24–27. [Google Scholar]
- Olsen, C.W.; Karasin, A.I.; Carman, S.; Li, Y.; Bastien, N.; Ojkic, D.; Alves, D.; Charbonneau, G.; Henning, B.M.; Low, D.E.; et al. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg. Infect. Dis. 2006, 12, 1132. [Google Scholar] [CrossRef]
- Sharma, V.; Sharma, M.; Dhull, D.; Kaushik, S.; Kaushik, S. Phylogenetic analysis of the hemagglutinin gene of influenza A (H1N1) pdm09 and A (H3N2) virus isolates from Haryana, India. VirusDisease 2019, 30, 336–343. [Google Scholar] [CrossRef]
- Liu, S.; Zhu, W.; Feng, Z.; Gao, R.; Guo, J.; Li, X.; Liu, J.; Wang, D.; Shu, Y. Substitution of D701N in the PB2 protein could enhance the viral replication and pathogenicity of Eurasian avian-like H1N1 swine influenza viruses. Emerg. Microbes Infect. 2018, 7, 1–10. [Google Scholar] [CrossRef]
- Jiao, P.; Tian, G.; Li, Y.; Deng, G.; Jiang, Y.; Liu, C.; Liu, W.; Bu, Z.; Kawaoka, Y.; Chen, H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J. Virol. 2008, 82, 1146–1154. [Google Scholar] [CrossRef]
- Cao, Z.; Zeng, W.; Hao, X.; Huang, J.; Cai, M.; Zhou, P.; Zhang, G. Continuous evolution of influenza A viruses of swine from 2013 to 2015 in Guangdong, China. PLoS ONE 2019, 14, e0217607. [Google Scholar] [CrossRef] [PubMed]
- Mon, P.P.; Thurain, K.; Janetanakit, T.; Nasamran, C.; Bunpapong, N.; Aye, A.M.; San, Y.Y.; Tun, T.N.; Amonsin, A. Swine influenza viruses and pandemic H1N1-2009 infection in pigs, Myanmar. Transbound. Emerg. Dis. 2020, 67, 2653–2666. [Google Scholar] [CrossRef] [PubMed]
- Krumbholz, A.; Schmidtke, M.; Bergmann, S.; Motzke, S.; Bauer, K.; Stech, J.; Dürrwald, R.; Wutzler, P.; Zell, R. High prevalence of amantadine resistance among circulating European porcine influenza A viruses. J. Gen. Virol. 2009, 90, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Rolling, T.; Koerner, I.; Zimmermann, P.; Holz, K.; Haller, O.; Staeheli, P.; Kochs, G. Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J. Virol. 2009, 83, 6673–6680. [Google Scholar] [CrossRef]
- Iwatsuki-Horimoto, K.; Horimoto, T.; Fujii, Y.; Kawaoka, Y. Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J. Virol. 2004, 78, 10149–10155. [Google Scholar] [CrossRef]
- Kuo, R.L.; Krug, R.M. Influenza a virus polymerase is an integral component of the CPSF30-NS1A protein complex in infected cells. J. Virol. 2009, 83, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Meseko, C.; Globig, A.; Ijomanta, J.; Joannis, T.; Nwosuh, C.; Shamaki, D.; Harder, T.; Hoffman, D.; Pohlmann, A.; Beer, M.; et al. Evidence of exposure of domestic pigs to Highly Pathogenic Avian Influenza H5N1 in Nigeria. Sci. Rep. 2018, 8, 5900. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, T.; Li, X.; Jiang, Z.; Jiang, Q.; Chen, Q.; Tu, X.; Chen, Z.; Chang, J.; Li, L.; et al. Serological evidence of H7, H5 and H9 avian influenza virus co-infection among herons in a city park in Jiangxi, China. Sci. Rep. 2014, 4, 6345. [Google Scholar] [CrossRef]
- Song, X.H.; Xiao, H.; Huang, Y.; Fu, G.; Jiang, B.; Kitamura, Y.; Liu, W.; Liu, D.; Gao, G.F.; Liu, W. Serological surveillance of influenza A virus infection in swine populations in Fujian province, China: No evidence of naturally occurring H5N1 infection in pigs. Zoonoses Public Health 2010, 57, 291–298. [Google Scholar] [CrossRef]


| Tested | Positive Inf A | H and N Types | Detection Rates of Influenza A Subtypes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subtypes, n (%) | N (%) | N (%) | H1 | N1 | H3 | N2 | A/H1N1pdm09 | A/H3N2 | A/H1N2 | Unsubtyped |
| Region, n (%) | ||||||||||
| Dakar | 69 (4.08) | 19 (3.6) | 6 (2.8) | 6 (3.0) | 2 (7.4) | 4 (4.0) | 6 (3.03) | 2 (7.4) | 2 (2.3) | 11 (4.07) |
| Diourbel | 109 (6.4) | 41 (7.9) | 21 (9.8) | 18 (8.8) | 0 (0.0) | 10 (9.8) | 18 (9.1) | 0 (0) | 10 (11.7) | 20 (7.4) |
| Fatick | 778 (46) | 232 (44.6) | 95 (44.4) | 90 (44.1) | 13 (48.1) | 44 (43.1) | 86 (43.4) | 13 (48.1) | 37 (43.5) | 120 (44.44) |
| Kafrine | 15 (0.9) | 3 (0.57) | 2 (0.9) | 2 (1) | 0 (0.0) | 0 (0.0) | 2 (1.01) | 0 (0) | 0 (0) | 1 (0.37) |
| Kaolack | 20 (1.2) | 2 (0.38) | 1 (0.5) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0) | 0 (0) | 1 (0.37) |
| Louga | 43 (2.5) | 11 (2.1) | 2 (0.9) | 1 (0.5) | 2 (7.4) | 4 (4.0) | 1 (0.5) | 2 (7.4) | 2 (2.3) | 6 (2.2) |
| Saint louis | 44 (2.6) | 9 (1.7) | 3 (1.4) | 3 (1.5) | 0 (0.0) | 0 (0.0) | 3 (1.5) | 0 (0) | 0 (0) | 6 (2.2) |
| Thiés | 449 (26.5) | 153 (29.4) | 62 (29.0) | 58 (28.4) | 7 (25.9) | 30 (29.4) | 57 (28.8) | 7 (25.9) | 24 (28.2) | 83 (30.7) |
| Missing | 164 (9.7) | 50 (9.6) | 22 (10.3) | 25 (12.2) | 3 (11.1) | 10 (9.8) | 24 (12.1) | 3 (11.1) | 10 (11.7) | 22 (8.1) |
| Years, n (%) | ||||||||||
| 2018 | 410 (24.2) | 113 (21.7) | 49 (22.9) | 47 (23.0) | 5 (18.5) | 24 (23.5) | 47 (23.7) | 5 (18.5) | 19 (22.3) | 56 (20.4) |
| 2019 | 1281 (75.7) | 407 (78.3) | 165 (77.1) | 157 (77.0) | 22 (81.5) | 78 (76.5) | 151 (76.3) | 22 (81.5) | 66 (77.6) | 219 (79.6) |
| Total | 1691 | 520 (30.7) | 214 (100) | 204 (100) | 27 (100) | 102 (100) | 198 (38.07) | 27 (5.2) | 85 (16.3) | 275 (51.9) |
| Virus | Subtype | Cleavage Site | HA | NA | M2 | PA | NS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 204 | 206 | 240 | 153 | 154 | 241 | 274 | 294 | 26 | 27 | 30 | 31 | 34 | 560 | 19 | 106 | |||
| A/GuangdongMaonan/SWL1536/2019 a | H1N1 | PSIQSR/GLF | V | E | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/0096/2018 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/0196/2018 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | S | L | M | |||
| A/swine/SEN/0410/2018 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/0023/2019 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/0500/2019 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/0773/2019 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/0783/2019 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/0872/2019 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/1024/2019 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/1174/2019 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | V | |||
| A/swine/SEN/1176/2019 b | H1N1 | PSIQSR/GLF | D | Q | Q | H | N | L | V | A | N | G | P | M | M | |||
| A/Dakar/31/2018 c | H1N1 | PSIQSR/GLF | D | Q | H | N | L | V | A | N | G | |||||||
| A/Dakar/26/2018 c | H1N1 | PSIQSR/GLF | Q | H | N | L | V | A | N | G | ||||||||
| A/HongKong/2671/2019 a | H3N2 | PEKQTR/GI | F | S | N | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/1174/2019 b | H3N2 | PEKQTR/GI | S | A | D | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/1241/2019 b | H3N2 | PEKQTR/GI | S | A | D | H | N | L | V | A | N | G | P | M | M | |||
| A/swine/SEN/1249/2019 b | H3N2 | PEKQTR/GI | S | A | D | H | N | L | V | A | N | G | P | M | M | |||
| A/Senegal/0024/2019 c | H3N2 | PEKQTR/GI | S | A | D | H | N | L | V | A | N | G | ||||||
| A/Senegal/0010/2019 c | H3N2 | PEKQTR/GI | D | H | N | L | I | A | N | G | ||||||||
| HI Positivity Rates to Different AIV Antigens | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Regions | No. of Sera (%) | H9N2 (%) | 95%CI | H5N1 (%) | 95%CI | H7N7 (%) | 95%CI | H5N2 (%) | 95%CI |
| Dakar | 67 (4.1) | 33 (3.8) | 2.0–5.0 | 14 (2.8) | 1.0–5.0 | 37 (4.2) | 3.0–6.0 | 20 (4.0) | 2.0–6.0 |
| Diourbel | 106 (6.5) | 47 (5.3) | 4.0–7.0 | 23 (4.5) | 3.0–7.0 | 51 (5.7) | 4.0–7.0 | 27 (5.4) | 3.6–7.8 |
| Fatick | 750 (45.8) | 388 (44.2) | 40.0–47.0 | 232 (45.8) | 41.0–50.0 | 414 (46.6) | 43.0–50.0 | 229 (45.6) | 41.0–50.0 |
| Kaffrine | 15 (0.9) | 5 (0.6) | 0.2–1.0 | 4 (0.8) | 0.2–2.0 | 9 (1.0) | 0.5–2.0 | 2 (0.4) | 0.07–1.6 |
| Kaolack | 19 (1.2) | 8 (0.9) | 0.4–2.0 | 4 (0.8) | 0.2–2.0 | 13 (1.5) | 0.8–2.5 | 6 (1.2) | 0.48–2.7 |
| Louga | 42 (2.6) | 25 (2.8) | 2.0–4.0 | 12 (2.4) | 1.2–4.2 | 24 (2.7) | 1.7–4.0 | 14 (2.8) | 1.5–4.7 |
| Saint-louis | 42 (2.6) | 26 (3.0) | 2.0–4.0 | 13 (2.6) | 1.4–4.4 | 25 (2.8) | 1.8–4.1 | 14 (2.8) | 1.5–4.7 |
| Thiés | 440 (26.9) | 249 (28.4) | 25.0–31.0 | 157 (31.0) | 27.0–35.0 | 238 (26.8) | 23.9–29.8 | 135 (26.9) | 23.0–31.0 |
| Missing | 155 (9.5) | 96 (11.0) | 9.0–13.0 | 47 (9.3) | 7.0–12.0 | 78 (8.8) | 7.0–10.8 | 55 (11.0) | 8.0–14.0 |
| Years | |||||||||
| 2018 | 394 (24.1) | 268 (30.5) | 27.0–34.0 | 210 (41.5) | 37.0–45.0 | 285 (32.0) | 29.0–35.0 | 215 (42.8) | 38.0–47.0 |
| 2019 | 1242 (75.9) | 609 (69.4) | 66.0–72.0 | 296 (58.5) | 54.0–63.0 | 604 (68.0) | 64.7–70.9 | 287 (57.2) | 53.0–61.0 |
| Total | 1636 (100) | 877 (53.6) | 51.0–56.0 | 506 (31.0) | 29.0–33.0 | 889 (54.3) | 51.8–56.7 | 502 (30.7) | 28.0–33.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jallow, M.M.; Barry, M.A.; Fall, A.; Ndiaye, N.K.; Kiori, D.; Sy, S.; Goudiaby, D.; Niang, M.N.; Fall, G.; Fall, M.; et al. Influenza A Virus in Pigs in Senegal and Risk Assessment of Avian Influenza Virus (AIV) Emergence and Transmission to Human. Microorganisms 2023, 11, 1961. https://doi.org/10.3390/microorganisms11081961
Jallow MM, Barry MA, Fall A, Ndiaye NK, Kiori D, Sy S, Goudiaby D, Niang MN, Fall G, Fall M, et al. Influenza A Virus in Pigs in Senegal and Risk Assessment of Avian Influenza Virus (AIV) Emergence and Transmission to Human. Microorganisms. 2023; 11(8):1961. https://doi.org/10.3390/microorganisms11081961
Chicago/Turabian StyleJallow, Mamadou Malado, Mamadou Aliou Barry, Amary Fall, Ndiendé Koba Ndiaye, Davy Kiori, Sara Sy, Déborah Goudiaby, Mbayame Ndiaye Niang, Gamou Fall, Malick Fall, and et al. 2023. "Influenza A Virus in Pigs in Senegal and Risk Assessment of Avian Influenza Virus (AIV) Emergence and Transmission to Human" Microorganisms 11, no. 8: 1961. https://doi.org/10.3390/microorganisms11081961
APA StyleJallow, M. M., Barry, M. A., Fall, A., Ndiaye, N. K., Kiori, D., Sy, S., Goudiaby, D., Niang, M. N., Fall, G., Fall, M., & Dia, N. (2023). Influenza A Virus in Pigs in Senegal and Risk Assessment of Avian Influenza Virus (AIV) Emergence and Transmission to Human. Microorganisms, 11(8), 1961. https://doi.org/10.3390/microorganisms11081961







