PGPB and/or AM Fungi Consortia Affect Tomato Native Rhizosphere Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Trial: Experimental Setup and Plant Growth
2.2. Microbial Community Characterization
2.3. Bioinformatic and Statistical Analyses
2.4. Root AM Colonization Assessment
3. Results
3.1. Soil Analysis
3.2. Production
3.3. Soil Microbiota Profiling
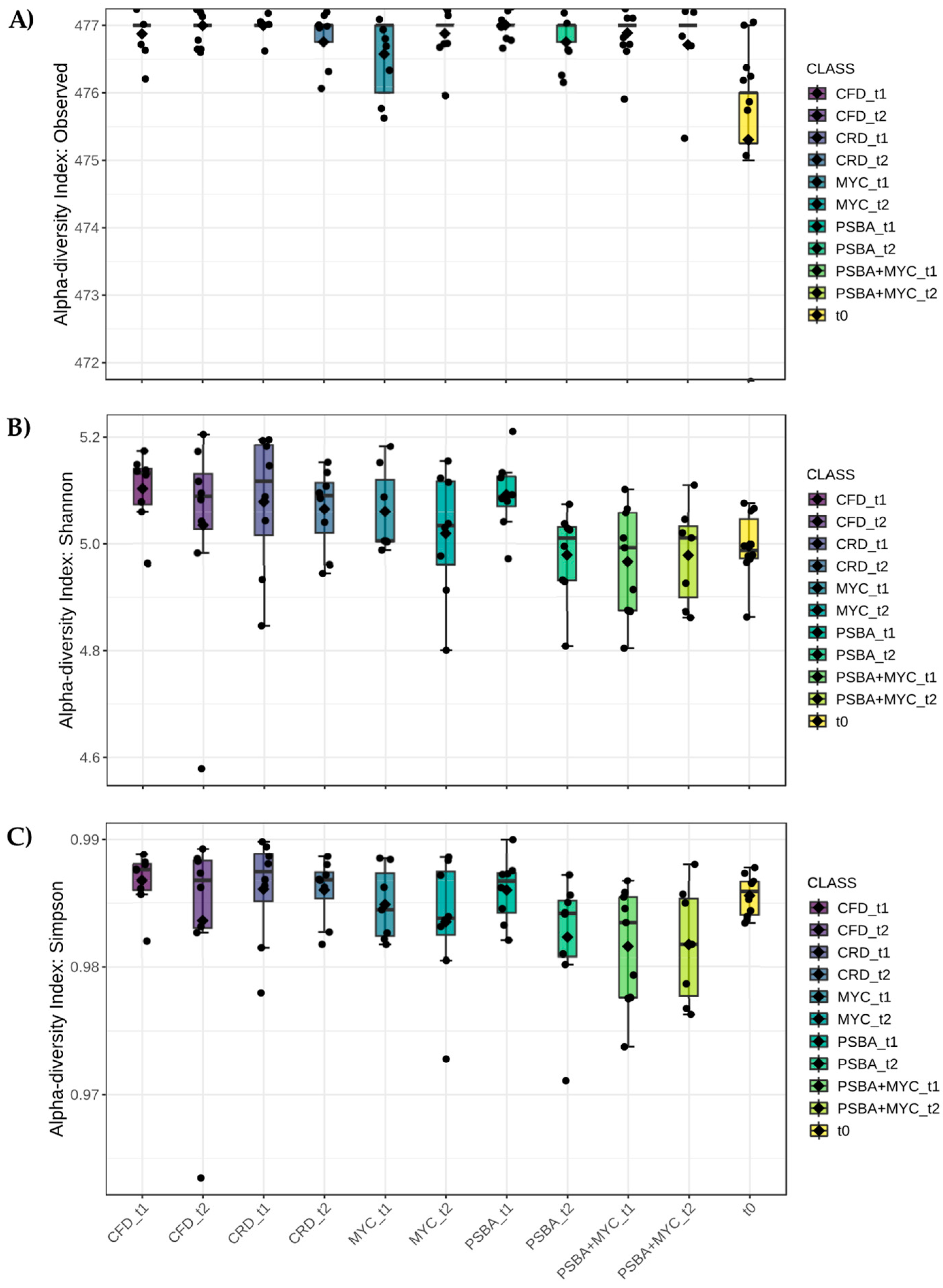
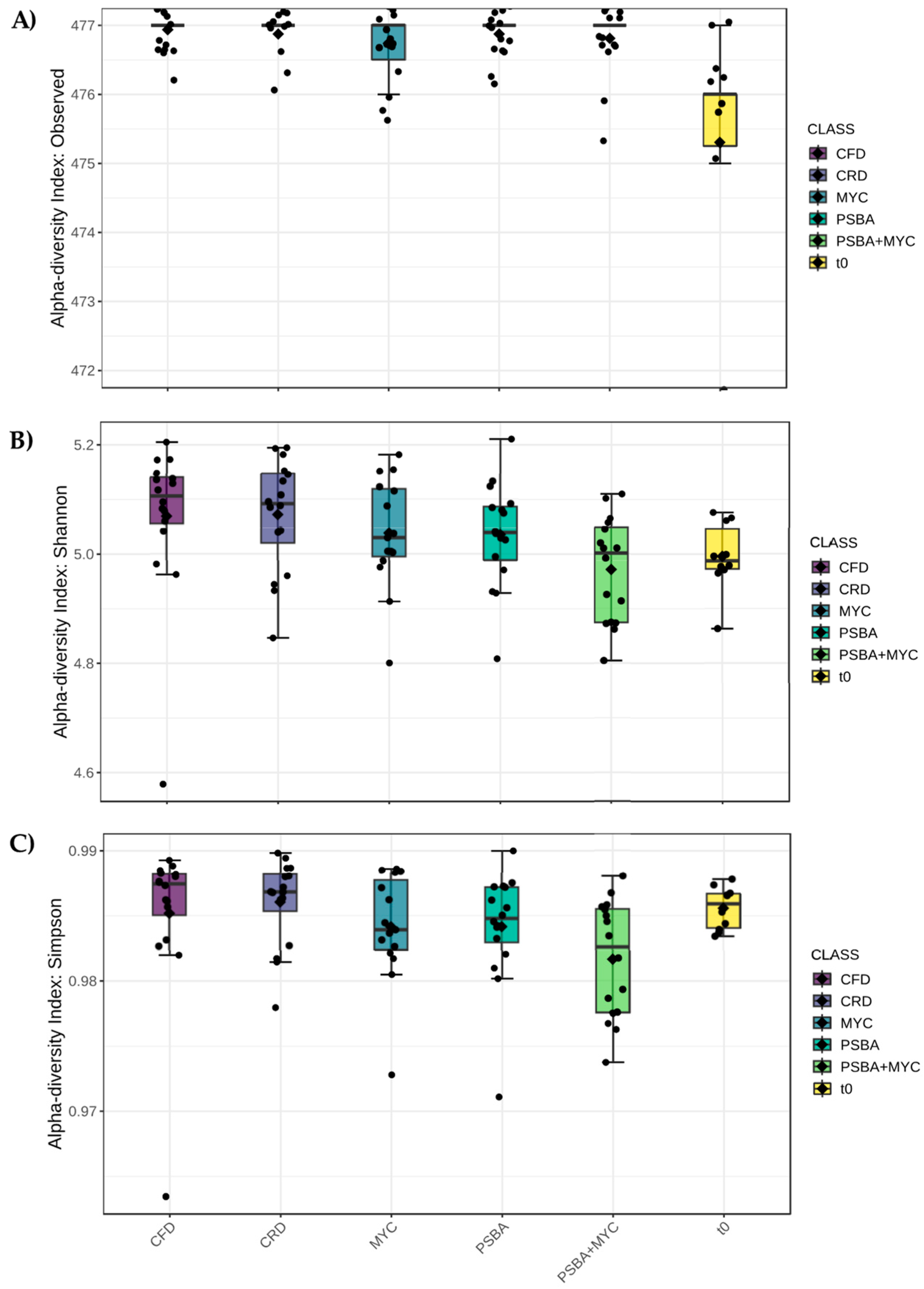
3.3.1. Alpha Diversity
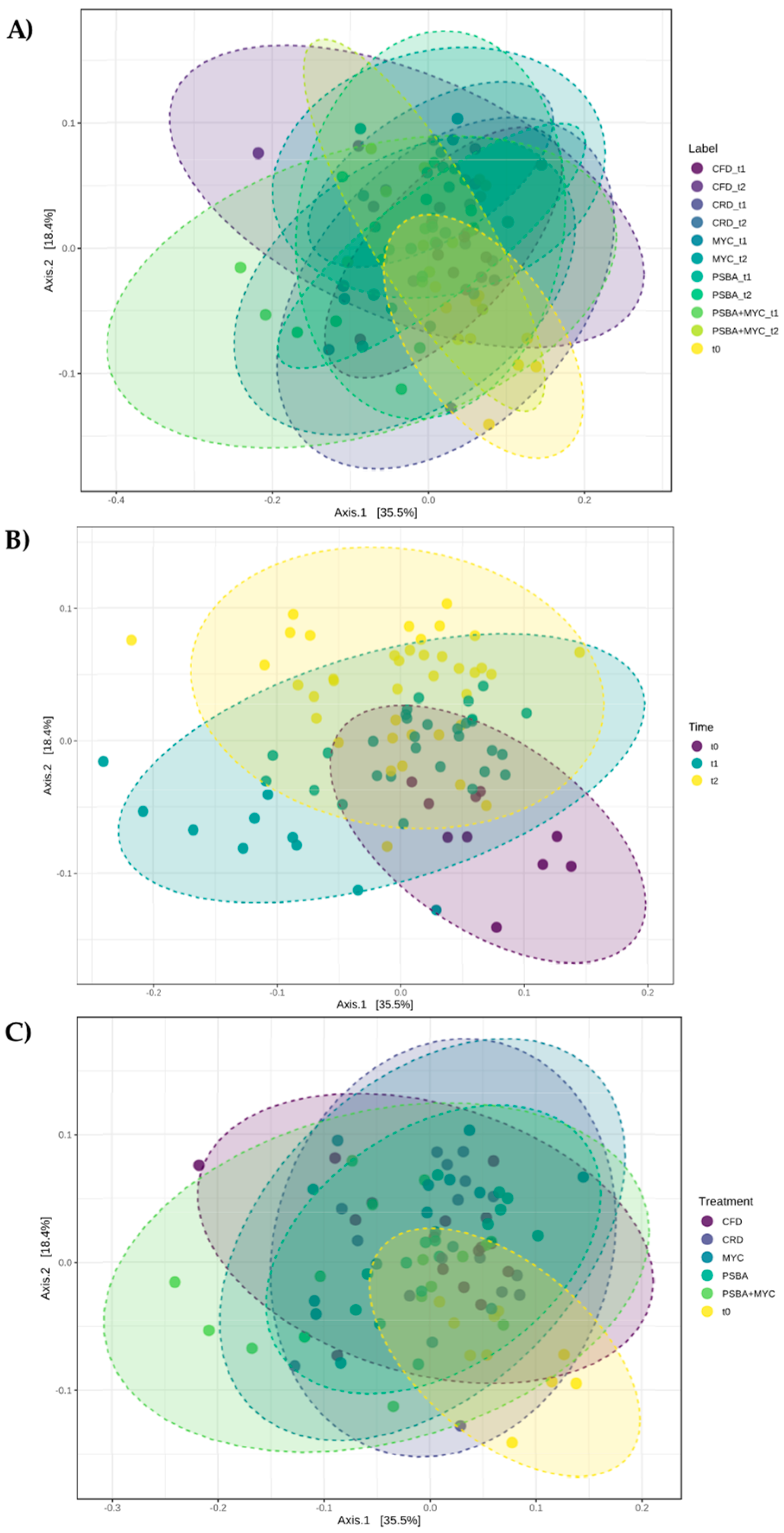
3.3.2. Beta Diversity
3.3.3. Signature
Signature Associated with the Presence of Tomato Plant
Signature Associated with the Biostimulant Treatment
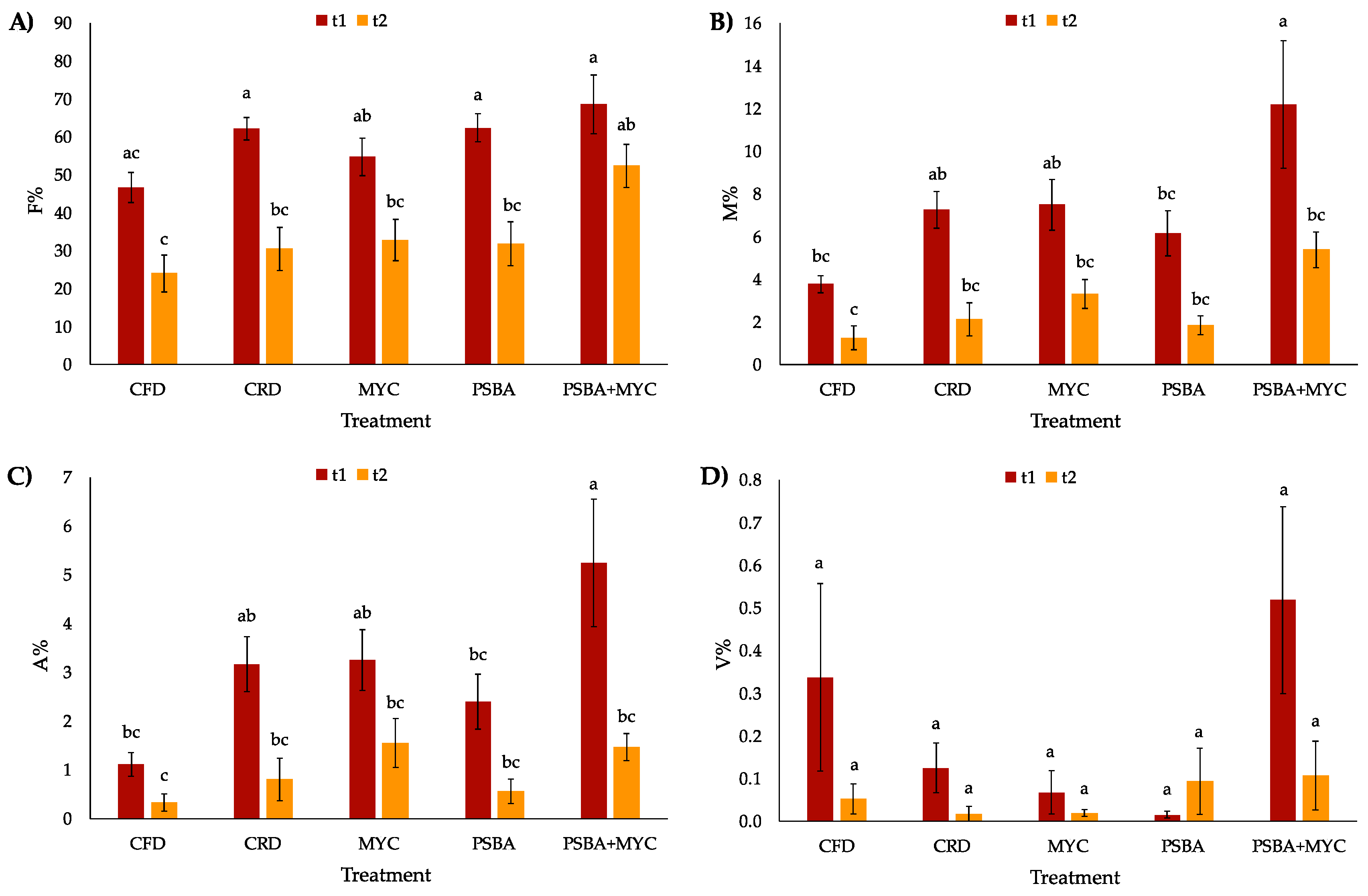
3.4. Mycorrhization Degree
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Collins, E.J.; Bowyer, C.; Tsouza, A.; Chopra, M. Tomatoes: An Extensive Review of the Associated Health Impacts of Tomatoes and Factors That Can Affect Their Cultivation. Biology 2022, 11, 239. [Google Scholar] [CrossRef]
- FAO. World Food and Agriculture—Statistical Yearbook 2021; FAO: Rome, Italy, 2021; ISBN 978-92-5-134332-6. [Google Scholar]
- Giuliani, M.M.; Gatta, G.; Cappelli, G.; Gagliardi, A.; Donatelli, M.; Fanchini, D.; De Nart, D.; Mongiano, G.; Bregaglio, S. Identifying the Most Promising Agronomic Adaptation Strategies for the Tomato Growing Systems in Southern Italy via Simulation Modeling. Eur. J. Agron. 2019, 111, 125937. [Google Scholar] [CrossRef]
- Gorrasi, S.; Pasqualetti, M.; Muñoz-Palazon, B.; Novello, G.; Mazzucato, A.; Campiglia, E.; Fenice, M. Comparison of the Peel-Associated Epiphytic Bacteria of Anthocyanin-Rich “Sun Black” and Wild-Type Tomatoes under Organic and Conventional Farming. Microorganisms 2022, 10, 2240. [Google Scholar] [CrossRef] [PubMed]
- Antoszewski, M.; Mierek-Adamska, A.; Dąbrowska, G.B. The Importance of Microorganisms for Sustainable Agriculture—A Review. Metabolites 2022, 12, 1100. [Google Scholar] [CrossRef] [PubMed]
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandić, A.; Noris, E.; Matić, S. A Review of the Most Common and Economically Important Diseases That Undermine the Cultivation of Tomato Crop in the Mediterranean Basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Cammarano, D.; Ronga, D.; Di Mola, I.; Mori, M.; Parisi, M. Impact of Climate Change on Water and Nitrogen Use Efficiencies of Processing Tomato Cultivated in Italy. Agric. Water Manag. 2020, 241, 106336. [Google Scholar] [CrossRef]
- Gamalero, E.; Bona, E.; Glick, B.R. Current Techniques to Study Beneficial Plant-Microbe Interactions. Microorganisms 2022, 10, 1380. [Google Scholar] [CrossRef]
- Kumar, S.; Diksha; Sindhu, S.S.; Kumar, R. Biofertilizers: An Ecofriendly Technology for Nutrient Recycling and Environmental Sustainability. Curr. Res. Microb. Sci. 2022, 3, 100094. [Google Scholar] [CrossRef]
- Lee, S.A.; Kim, Y.; Kim, J.M.; Chu, B.; Joa, J.-H.; Sang, M.K.; Song, J.; Weon, H.-Y. A Preliminary Examination of Bacterial, Archaeal, and Fungal Communities Inhabiting Different Rhizocompartments of Tomato Plants under Real-World Environments. Sci. Rep. 2019, 9, 9300. [Google Scholar] [CrossRef]
- Vuolo, F.; Novello, G.; Bona, E.; Gorrasi, S.; Gamalero, E. Impact of Plant-Beneficial Bacterial Inocula on the Resident Bacteriome: Current Knowledge and Future Perspectives. Microorganisms 2022, 10, 2462. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A Phylum-Level Phylogenetic Classification of Zygomycete Fungi Based on Genome-Scale Data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef]
- Giovannini, L.; Palla, M.; Agnolucci, M.; Avio, L.; Sbrana, C.; Turrini, A.; Giovannetti, M. Arbuscular Mycorrhizal Fungi and Associated Microbiota as Plant Biostimulants: Research Strategies for the Selection of the Best Performing Inocula. Agronomy 2020, 10, 106. [Google Scholar] [CrossRef]
- Lingua, G.; Massa, N.; D’Agostino, G.; Antosiano, M.; Berta, G. Mycorrhiza-Induced Differential Response to a Yellows Disease in Tomato. Mycorrhiza 2002, 12, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Lingua, G.; Franchin, C.; Todeschini, V.; Castiglione, S.; Biondi, S.; Burlando, B.; Parravicini, V.; Torrigiani, P.; Berta, G. Arbuscular Mycorrhizal Fungi Differentially Affect the Response to High Zinc Concentrations of Two Registered Poplar Clones. Environ. Pollut. 2008, 153, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Lingua, G.; Manassero, P.; Cantamessa, S.; Marsano, F.; Todeschini, V.; Copetta, A.; D’Agostino, G.; Massa, N.; Avidano, L.; et al. AM Fungi and PGP Pseudomonads Increase Flowering, Fruit Production, and Vitamin Content in Strawberry Grown at Low Nitrogen and Phosphorus Levels. Mycorrhiza 2015, 25, 181–193. [Google Scholar] [CrossRef]
- Lingua, G.; Bona, E.; Manassero, P.; Marsano, F.; Todeschini, V.; Cantamessa, S.; Copetta, A.; D’Agostino, G.; Gamalero, E.; Berta, G. Arbuscular Mycorrhizal Fungi and Plant Growth-Promoting Pseudomonads Increases Anthocyanin Concentration in Strawberry Fruits (Fragaria x Ananassa Var. Selva) in Conditions of Reduced Fertilization. Int. J. Mol. Sci. 2013, 14, 16207–16225. [Google Scholar] [CrossRef]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant Growth Promoting Rhizobacteria (PGPR) as Green Bioinoculants: Recent Developments, Constraints, and Prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.K.; Kumar, A. Disease Management of Tomato through PGPB: Current Trends and Future Perspective. 3 Biotech 2017, 7, 255. [Google Scholar] [CrossRef]
- Katsenios, N.; Andreou, V.; Sparangis, P.; Djordjevic, N.; Giannoglou, M.; Chanioti, S.; Stergiou, P.; Xanthou, M.-Z.; Kakabouki, I.; Vlachakis, D.; et al. Evaluation of Plant Growth Promoting Bacteria Strains on Growth, Yield and Quality of Industrial Tomato. Microorganisms 2021, 9, 2099. [Google Scholar] [CrossRef]
- Zuluaga, M.Y.A.; Milani, K.M.L.; Miras-Moreno, B.; Lucini, L.; Valentinuzzi, F.; Mimmo, T.; Pii, Y.; Cesco, S.; Rodrigues, E.P.; Oliveira, A.L.M. de Inoculation with Plant Growth-Promoting Bacteria Alters the Rhizosphere Functioning of Tomato Plants. Appl. Soil Ecol. 2021, 158, 103784. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.-P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial Soil Microbiome Composition and Functioning Predetermine Future Plant Health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolic Profiling of PGPR-Treated Tomato Plants Reveal Priming-Related Adaptations of Secondary Metabolites and Aromatic Amino Acids. Metabolites 2020, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, P.; Massa, N.; Cantamessa, S.; Todeschini, V.; Bona, E.; Berta, G.; Barbato, R.; Lingua, G. Tomato Responses to Funneliformis mosseae during the Early Stages of Arbuscular Mycorrhizal Symbiosis. Mycorrhiza 2020, 30, 601–610. [Google Scholar] [CrossRef]
- Zouari, I.; Salvioli, A.; Chialva, M.; Novero, M.; Miozzi, L.; Tenore, G.; Bagnaresi, P.; Bonfante, P. From Root to Fruit: RNA-Seq Analysis Shows That Arbuscular Mycorrhizal Symbiosis May Affect Tomato Fruit Metabolism. BMC Genomics 2014, 15, 221. [Google Scholar] [CrossRef] [PubMed]
- Sellitto, V.M.; Golubkina, N.A.; Pietrantonio, L.; Cozzolino, E.; Cuciniello, A.; Cenvinzo, V.; Florin, I.; Caruso, G. Tomato Yield, Quality, Mineral Composition and Antioxidants as Affected by Beneficial Microorganisms Under Soil Salinity Induced by Balanced Nutrient Solutions. Agriculture 2019, 9, 110. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.; Liu, F.; Liang, J.; Zhao, P.; Tsui, C.K.M.; Cai, L. Cross-Kingdom Synthetic Microbiota Supports Tomato Suppression of Fusarium Wilt Disease. Nat. Commun. 2022, 13, 7890. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Cantamessa, S.; Massa, N.; Manassero, P.; Marsano, F.; Copetta, A.; Lingua, G.; D’Agostino, G.; Gamalero, E.; Berta, G. Arbuscular Mycorrhizal Fungi and Plant Growth-Promoting Pseudomonads Improve Yield, Quality and Nutritional Value of Tomato: A Field Study. Mycorrhiza 2017, 27, 1–11. [Google Scholar] [CrossRef]
- Bona, E.; Todeschini, V.; Cantamessa, S.; Cesaro, P.; Copetta, A.; Lingua, G.; Gamalero, E.; Berta, G.; Massa, N. Combined Bacterial and Mycorrhizal Inocula Improve Tomato Quality at Reduced Fertilization. Sci. Hortic. 2018, 234, 160–165. [Google Scholar] [CrossRef]
- El Maaloum, S.; Elabed, A.; Alaoui-Talibi, Z.E.; Meddich, A.; Filali-Maltouf, A.; Douira, A.; Ibnsouda-Koraichi, S.; Amir, S.; El Modafar, C. Effect of Arbuscular Mycorrhizal Fungi and Phosphate-Solubilizing Bacteria Consortia Associated with Phospho-Compost on Phosphorus Solubilization and Growth of Tomato Seedlings (Solanum lycopersicum L.). Commun. Soil Sci. Plant Anal. 2020, 51, 622–634. [Google Scholar] [CrossRef]
- Bona, E.; Massa, N.; Toumatia, O.; Novello, G.; Cesaro, P.; Todeschini, V.; Boatti, L.; Mignone, F.; Titouah, H.; Zitouni, A.; et al. Climatic Zone and Soil Properties Determine the Biodiversity of the Soil Bacterial Communities Associated to Native Plants from Desert Areas of North-Central Algeria. Microorganisms 2021, 9, 1359. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Boernigen, D.; Tickle, T.L.; Morgan, X.C.; Garrett, W.S.; Huttenhower, C. Computational Meta’omics for Microbial Community Studies. Mol. Syst. Biol. 2013, 9, 666. [Google Scholar] [CrossRef] [PubMed]
- Trouvelot, A.; Kough, J.; Gianinazzi-Pearson, V. Mesure Du Taux de Mycorrhization VA d’un Système Radiculaire. Recherche de Méthodes d’estimation Ayant Une Sig- Nification Functionnelle. In Physiological and Genetical Aspects of Mycorrhizae; INRA: Paris, France, 1986; pp. 217–221. [Google Scholar]
- Ortiz-Burgos, S. Shannon-Weaver Diversity Index. In Encyclopedia of Estuaries; Encyclopedia of Earth Sciences Series; Kennish, M.J., Ed.; Springer: Dordrecht, The Netherlands, 2016; pp. 572–573. [Google Scholar]
- Hill, T.C.J.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using Ecological Diversity Measures with Bacterial Communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Emmanuel, O.C.; Babalola, O.O. Productivity and Quality of Horticultural Crops through Co-Inoculation of Arbuscular Mycorrhizal Fungi and Plant Growth Promoting Bacteria. Microbiol. Res. 2020, 239, 126569. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Lehvävirta, S.; Timonen, S.; Kasurinen, J.; Niemikapee, J.; Valkonen, J.P.T. Species-Specific Synergistic Effects of Two Plant Growth—Promoting Microbes on Green Roof Plant Biomass and Photosynthetic Efficiency. PLoS ONE 2018, 13, e0209432. [Google Scholar] [CrossRef]
- Khalid, M.; Hassani, D.; Bilal, M.; Asad, F.; Huang, D. Influence of Bio-Fertilizer Containing Beneficial Fungi and Rhizospheric Bacteria on Health Promoting Compounds and Antioxidant Activity of Spinacia oleracea, L. Bot. Stud. 2017, 58, 35. [Google Scholar] [CrossRef]
- Bonneau, L.; Huguet, S.; Wipf, D.; Pauly, N.; Truong, H.-N. Combined Phosphate and Nitrogen Limitation Generates a Nutrient Stress Transcriptome Favorable for Arbuscular Mycorrhizal Symbiosis in Medicago Truncatula. New Phytol. 2013, 199, 188–202. [Google Scholar] [CrossRef]
- Grunert, O.; Robles-Aguilar, A.A.; Hernandez-Sanabria, E.; Schrey, S.D.; Reheul, D.; Van Labeke, M.-C.; Vlaeminck, S.E.; Vandekerckhove, T.G.L.; Mysara, M.; Monsieurs, P.; et al. Tomato Plants Rather than Fertilizers Drive Microbial Community Structure in Horticultural Growing Media. Sci. Rep. 2019, 9, 9561. [Google Scholar] [CrossRef]
- Verma, R.; Annapragada, H.; Katiyar, N.; Shrutika, N.; Das, K.; Murugesan, S. Rhizobium. In Beneficial Microbes in Agroecology; Amaresan, N., Senthil Kumar, M., Annapurna, K., Krishna, K., Sankaranarayanan, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 37–54. ISBN 978-0-12-823414-3. [Google Scholar]
- Finkel, O.M.; Salas-González, I.; Castrillo, G.; Conway, J.M.; Law, T.F.; Teixeira, P.J.P.L.; Wilson, E.D.; Fitzpatrick, C.R.; Jones, C.D.; Dangl, J.L. A Single Bacterial Genus Maintains Root Growth in a Complex Microbiome. Nature 2020, 587, 103–108. [Google Scholar] [CrossRef]
- Eltlbany, N.; Baklawa, M.; Ding, G.-C.; Nassal, D.; Weber, N.; Kandeler, E.; Neumann, G.; Ludewig, U.; van Overbeek, L.; Smalla, K. Enhanced Tomato Plant Growth in Soil under Reduced P Supply through Microbial Inoculants and Microbiome Shifts. FEMS Microbiol. Ecol. 2019, 95, fiz124. [Google Scholar] [CrossRef]
- Xue, L.; Sun, B.; Yang, Y.; Jin, B.; Zhuang, G.; Bai, Z.; Zhuang, X. Efficiency and Mechanism of Reducing Ammonia Volatilization in Alkaline Farmland Soil Using Bacillus amyloliquefaciens Biofertilizer. Environ. Res. 2021, 202, 111672. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, Y.; Zhang, F.; Li, R.; Hu, Y.; Yu, H.; Tuyiringire, D.; Wang, L. Effects of Bok Choy on the Dissipation of Dibutyl Phthalate (DBP) in Mollisol and Its Possible Mechanisms of Biochemistry and Microorganisms. Ecotoxicol. Environ. Saf. 2019, 181, 284–291. [Google Scholar] [CrossRef]
- Kour, D.; Rana, K.L.; Yadav, A.N.; Yadav, N.; Kumar, M.; Kumar, V.; Vyas, P.; Dhaliwal, H.S.; Saxena, A.K. Microbial Biofertilizers: Bioresources and Eco-Friendly Technologies for Agricultural and Environmental Sustainability. Biocatal. Agric. Biotechnol. 2020, 23, 101487. [Google Scholar] [CrossRef]
- Toribio, A.J.; Jurado, M.M.; Suárez-Estrella, F.; López-González, J.A.; Martínez-Gallardo, M.R.; López, M.J. Application of Sonicated Extracts of Cyanobacteria and Microalgae for the Mitigation of Bacterial Canker in Tomato Seedlings. J. Appl. Phycol. 2021, 33, 3817–3829. [Google Scholar] [CrossRef]
- Marghoob, M.U.; Rodriguez-Sanchez, A.; Imran, A.; Mubeen, F.; Hoagland, L. Diversity and Functional Traits of Indigenous Soil Microbial Flora Associated with Salinity and Heavy Metal Concentrations in Agricultural Fields within the Indus Basin Region, Pakistan. Front. Microbiol. 2022, 13, 1020175. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Fan, K.; Li, G.; Gao, S.; Chang, D.; Liang, T.; Li, S.; Liang, H.; Zhang, J.; Che, Z.; et al. Synergistic Effects of Diazotrophs and Arbuscular Mycorrhizal Fungi on Soil Biological Nitrogen Fixation after Three Decades of Fertilization. iMeta 2023, 2, e81. [Google Scholar] [CrossRef]
- Su, J.-Y.; Liu, C.-H.; Tampus, K.; Lin, Y.-C.; Huang, C.-H. Organic Amendment Types Influence Soil Properties, the Soil Bacterial Microbiome, and Tomato Growth. Agronomy 2022, 12, 1236. [Google Scholar] [CrossRef]
- Novello, G.; Gamalero, E.; Bona, E.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Lingua, G.; Berta, G. The Rhizosphere Bacterial Microbiota of Vitis vinifera Cv. Pinot Noir in an Integrated Pest Management Vineyard. Front. Microbiol. 2017, 8, 1528. [Google Scholar] [CrossRef]
- Bona, E.; Massa, N.; Novello, G.; Boatti, L.; Cesaro, P.; Todeschini, V.; Magnelli, V.; Manfredi, M.; Marengo, E.; Mignone, F.; et al. Metaproteomic Characterization of Vitis vinifera Rhizosphere. FEMS Microbiol. Ecol. 2018, 95, fiy204. [Google Scholar] [CrossRef]


| Analyzed Compound | Results | Unit of Measure | Reference Ranges | Values |
|---|---|---|---|---|
| Fertility | ||||
| pH (in Water) | 8.29 | - | 6.50–7.50 | Very high |
| Electrical Conductivity | 105 | µS cm−1 | 200–400 | Low |
| Organic Matter | 1.69 | % | 1.20–2 | Normal |
| Total Nitrogen | 1.038 | mg kg−1 sms | 1.000–1.500 | Normal |
| C/N ratio | 9.44 | - | 10.0–15.0 | Low |
| Available Phosphorus | 19.3 | mg kg−1 | 20.0–40.0 | Low |
| Active lime | 1.93 | %CaCO3 | 1.50–4.00 | Normal |
| Microelements | ||||
| Boron | <0.50 | mg kg−1 | 0.60–1.00 | Low |
| Iron | 10.3 | mg kg−1 | 4.00–10.00 | High |
| Manganese | 8.02 | mg kg−1 | 1.00–5.00 | Very High |
| Copper | 6.56 | mg kg−1 | 0.40–1.00 | Very High |
| Zinc | 1.34 | mg kg−1 | 1.00–2.00 | Normal |
| Exchange complex | ||||
| Base saturation | 77.5 | % | 50.0–80.0 | Normal |
| Exchangeable Potassium | 0.26 | meq/100 g | 0.50–0.80 | Low |
| Exchangeable Calcium | 16,859 | meq/100 g | 8000–14,000 | Very High |
| Exchangeable Magnesium | 2.39 | meq/100 g | 1.50–2.50 | Normal |
| Exchangeable Sodium | <0.05 | meq/100 g | 0.25–0.50 | Very Low |
| CEC | 25.2 | meq/100 g | 10.0–20.0 | Very High |
| Texture | ||||
| Silty-Loam |
| Treatment | Marketable Fruits | Unmarketable (Green Fruits) | Total (Marketable + Green Fruits) | Unmarketable BER Fruits | |||
|---|---|---|---|---|---|---|---|
| Weight (g/plot) | Number/Plot | Weight (g/plot) | Number/Plot | Weight (g/plot) | Number/Plot | % on the Total Weight | |
| CFD | 7508.20 a | 127.87 a | 1908.00 a | 74.27 a | 9416.20 a | 202.13 a | 23.59 b |
| CRD | 3021.50 a | 56.11 b | 1770.50 a | 99.35 a | 4792.00 b | 155.46 a | 36.20 a |
| MYC | 6438.40 a | 105.85 ab | 1232.40 a | 65.54 a | 7670.80 ab | 171.39 a | 31.75 ab |
| PSBA | 5665.40 a | 104.19 ab | 1892.80 a | 86.33 a | 7558.20 ab | 190.52 a | 27.44 ab |
| PSBA + MYC | 3923.50 a | 84.57 ab | 1497.25 a | 82.80 a | 5420.75 ab | 167.36 a | 32.88 ab |
| Treatment | Firmness (cm/kg) | °Brix |
|---|---|---|
| CFD | 4.11 a | 5.37 a |
| CRD | 4.16 a | 5.43 a |
| MYC | 4.46 a | 5.29 a |
| PSBA | 3.78 a | 5.54 a |
| PSBA + MYC | 4.11 a | 5.49 a |
| Genus | p-Value | FDR | t0 | CFD_t1 | CRD_t1 | MYC_t1 | PSBA_t1 | PSBA + MYC_t1 | CFD_t2 | CRD_t2 | MYC_t2 | PSBA_t2 | PSBA + MYC_t2 | LDA Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acidibacter | 0.00014755 | 0.00072408 | 100 | 155.58 | 159.23 | 154.47 | 227.51 | 147.26 | 199.19 | 261.94 | 174.03 | 155.63 | 145.32 | 3.96 |
| Acidimicrobiia bacterium (uncultured) | 0.000018 | 0.00017647 | 100 | 53.77 | 67.65 | 52.85 | 50.3 | 56.62 | 47.63 | 41.02 | 44.1 | 55.62 | 52.27 | 4.14 |
| Acidobacteria bacterium (uncultured) | 0.025732 | 0.038263 | 100 | 97.56 | 83.72 | 79.36 | 101.81 | 68.34 | 97.13 | 107.49 | 118.89 | 111.66 | 95.36 | 4.4 |
| Acidobacteriaceae bacterium (uncultured) | 0.0044007 | 0.0094048 | 100 | 95.95 | 84.66 | 80.08 | 102.73 | 76.22 | 100.34 | 114.27 | 119.35 | 121.2 | 105.64 | 4.53 |
| Acidobacteriales bacterium (uncultured) | 0.00047137 | 0.0016655 | 100 | 70 | 64.58 | 58.75 | 74.47 | 52.8 | 69.75 | 85.74 | 94.28 | 105.67 | 83.58 | 4.39 |
| Acidovorax | 0.00000469 | 0.0000731 | 100 | 1599.78 | 1408.18 | 3162.47 | 1051.46 | 675.82 | 1094.97 | 460.25 | 494.62 | 814.36 | 664.01 | 3.38 |
| Acinetobacter | 0.0058531 | 0.011751 | 100 | 3001.09 | 4664.02 | 60.65 | 67.34 | 814.12 | 5094.17 | 3764.09 | 6443.89 | 2040.25 | 2307.61 | 4.94 |
| Actinobacterium (uncultured) | 0.0013672 | 0.0039813 | 100 | 74 | 89.46 | 70.72 | 57.44 | 59.21 | 57.29 | 58.04 | 55.38 | 60.4 | 60.55 | 4.05 |
| Actinomadura | 0.017131 | 0.028373 | 100 | 81.13 | 82.77 | 85.91 | 84.3 | 89.79 | 67.15 | 70.87 | 59.32 | 52.07 | 70.63 | 2.8 |
| Actinomycetales bacterium (uncultured) | 0.00000299 | 0.0000529 | 100 | 97.28 | 114.55 | 97.5 | 76.16 | 96.08 | 70.44 | 57.67 | 61.07 | 79.34 | 98.59 | 3.46 |
| Adhaeribacter | 0.00047029 | 0.0016655 | 100 | 127.82 | 114.9 | 177.52 | 165.5 | 208.19 | 108.57 | 113.35 | 115.39 | 92.46 | 113.79 | 3.91 |
| Aeromicrobium | 0.00028085 | 0.0011243 | 100 | 215.74 | 200.93 | 267.55 | 201.62 | 213.57 | 319.23 | 267.17 | 196.75 | 120.57 | 126.37 | 4.61 |
| Allorhizobium Neorhizobium Pararhizobium Rhizobium | 0.0021565 | 0.0052715 | 100 | 167.14 | 182.22 | 177.25 | 145.63 | 188.67 | 247.6 | 221.21 | 209.35 | 194.15 | 176.26 | 4.5 |
| Alsobacter | 0.017712 | 0.028974 | 100 | 103.68 | 107.04 | 169.83 | 155.94 | 200.54 | 116.4 | 142.08 | 132.67 | 133.07 | 135.71 | 3.11 |
| Altererythrobacter | 0.00032195 | 0.0012229 | 100 | 289.68 | 514.48 | 277.11 | 298.3 | 270.04 | 291.05 | 280.58 | 202.53 | 202.07 | 191.35 | 3.65 |
| Amaricoccus | 0.035663 | 0.050004 | 100 | 106.47 | 130.89 | 145.28 | 146.72 | 188.74 | 120.64 | 112.51 | 111.01 | 150.5 | 149.04 | 3.2 |
| Aminobacter | 0.000000015 | 0.00000199 | 100 | 197.43 | 139.68 | 1110.94 | 1061.6 | 1129.53 | 171.17 | 214.17 | 302.93 | 309.59 | 283.42 | 3.53 |
| Ammoniphilus | 0.018022 | 0.029299 | 100 | 71.76 | 99.9 | 83.32 | 86.55 | 105.78 | 80.39 | 89.8 | 60.67 | 85.71 | 56.67 | 3.02 |
| Amycolatopsis | 0.0000000459 | 0.00000304 | 100 | 370.65 | 261.97 | 264.12 | 79.03 | 161.58 | 2499.7 | 752.78 | 1067.1 | 921.05 | 1642.91 | 3.71 |
| Anaerolinea | 0.024548 | 0.036753 | 100 | 34.12 | 30.49 | 38.59 | 36.77 | 41.71 | 23.45 | 17.9 | 24.69 | 29.63 | 41.04 | 3.61 |
| Anaeromyxobacter | 0.0023108 | 0.0055669 | 100 | 56.91 | 57.54 | 47.92 | 64.27 | 64.7 | 46.92 | 44.31 | 57.26 | 82.9 | 57.24 | 3.63 |
| Arenimonas | 0.0000464 | 0.00036396 | 100 | 264.98 | 231.75 | 240.75 | 216.36 | 225.8 | 281.65 | 204.59 | 113.83 | 120.37 | 113.19 | 3.29 |
| Aridibacter | 0.0093128 | 0.017138 | 100 | 172.76 | 134.24 | 167.94 | 159.77 | 192.99 | 146.88 | 144.83 | 121.56 | 113.03 | 159.73 | 3.35 |
| Armatimonadetes bacterium (uncultured) | 0.00014735 | 0.00072408 | 100 | 81.28 | 78.4 | 88.19 | 87.42 | 98.96 | 54.57 | 64.2 | 62.56 | 64.6 | 70.69 | 3.03 |
| Arthrobacter | 0.0066158 | 0.012987 | 100 | 110.14 | 90.11 | 130.95 | 119.67 | 154.37 | 125.5 | 108.41 | 130.33 | 145.57 | 150.85 | 5.28 |
| Azohydromonas | 0.000000298 | 0.0000132 | 100 | 156.52 | 148.01 | 179.02 | 154.93 | 222.21 | 323.53 | 325.44 | 271.45 | 244.19 | 192.86 | 3.54 |
| Blastocatella | 0.0083444 | 0.015683 | 100 | 165.57 | 142.22 | 175.37 | 141.52 | 165.17 | 133.59 | 124.31 | 131 | 124.01 | 139.93 | 3.5 |
| Blastococcus | 0.0061343 | 0.012222 | 100 | 108.48 | 106.89 | 137.17 | 122.74 | 162.49 | 95.23 | 104.31 | 90.25 | 97.41 | 96.46 | 4.28 |
| Blastopirellula | 0.0017302 | 0.0047761 | 100 | 96.28 | 91.75 | 84.19 | 112.04 | 79.5 | 127.33 | 149.15 | 178.04 | 181.03 | 143.16 | 3.05 |
| Bosea | 0.0011646 | 0.0034678 | 100 | 185.33 | 196.19 | 234.64 | 209.3 | 293.26 | 229.65 | 247.79 | 216.03 | 241.81 | 154.01 | 3.7 |
| Bradyrhizobium | 0.00045919 | 0.0016655 | 100 | 129.5 | 154.15 | 118.4 | 167.45 | 123.65 | 153.75 | 162.94 | 161.36 | 140.06 | 125.84 | 3.95 |
| Burkholderia Caballeronia Paraburkholderia | 0.012425 | 0.02138 | 100 | 46.43 | 60.48 | 54.73 | 46.12 | 43.13 | 71.5 | 65.56 | 32.89 | 29.82 | 28.31 | 3.54 |
| Caenimonas | 0.0000533 | 0.00038145 | 100 | 159 | 145.17 | 204.66 | 197.78 | 244.47 | 208.96 | 198.63 | 156.15 | 123.62 | 127.8 | 4.13 |
| Candidatus Udaeobacter | 0.02707 | 0.039854 | 100 | 95.45 | 90.54 | 58.67 | 65.19 | 41.52 | 82.86 | 95.55 | 95.68 | 89.25 | 92.82 | 4.15 |
| Candidatus Xiphinematobacter | 0.0098109 | 0.017686 | 100 | 91.35 | 73.1 | 72.62 | 87.72 | 64.97 | 97.42 | 116.23 | 128.84 | 122.85 | 120.7 | 4.66 |
| Caulobacter | 0.033071 | 0.047372 | 100 | 111.4 | 301.28 | 94.01 | 83.12 | 102.03 | 95.46 | 81.41 | 70.05 | 51.29 | 41.22 | 3.31 |
| Cellulomonas | 0.0018814 | 0.0050246 | 100 | 66.57 | 88.69 | 100.53 | 102.72 | 157.41 | 65.68 | 64.37 | 68.77 | 112.36 | 64.88 | 2.98 |
| Chloroflexi bacterium (uncultured) | 0.00000138 | 0.0000304 | 100 | 86.92 | 94.22 | 99.21 | 88.22 | 96.63 | 72.5 | 64.09 | 66.27 | 72.16 | 84.66 | 4.32 |
| Chloroflexus sp (uncultured) | 0.00000908 | 0.00010461 | 100 | 101.7 | 102.45 | 82.23 | 59.29 | 62.44 | 88.91 | 73.71 | 65.16 | 53.25 | 78.24 | 3.84 |
| Chthoniobacter | 0.00082646 | 0.0027039 | 100 | 155.64 | 124.66 | 163.29 | 184.98 | 168.04 | 132.46 | 149.85 | 140.19 | 104.27 | 123.15 | 4.28 |
| Clostridium sensu stricto 1 | 0.015432 | 0.02572 | 100 | 55.73 | 63.89 | 48.1 | 56.44 | 62.09 | 44.54 | 30.51 | 60.27 | 50.98 | 48.2 | 3.25 |
| Cohnella | 0.0024156 | 0.0057671 | 100 | 64.17 | 79.14 | 73.08 | 82.54 | 97.11 | 42.83 | 52.74 | 50.9 | 82.82 | 74.89 | 3.44 |
| Comamonas | 0.00000807 | 0.00010186 | 100 | 199.52 | 199.96 | 203.55 | 221.02 | 189.06 | 422.54 | 364.9 | 324.98 | 335.35 | 229.65 | 3.18 |
| Conexibacter | 0.0039591 | 0.0088165 | 100 | 94.22 | 117.14 | 119.44 | 90.52 | 105.8 | 77.22 | 68.14 | 63.1 | 82.41 | 77.05 | 2.86 |
| Conexibacter sp (uncultured) | 0.00000683 | 0.0000905 | 100 | 68.43 | 79.04 | 74.4 | 68.73 | 76 | 47.69 | 41.99 | 46.9 | 58.86 | 63.81 | 2.98 |
| Cupriavidus | 0.00012448 | 0.0006732 | 100 | 143.59 | 151.65 | 104.49 | 128.17 | 189.52 | 199.9 | 420.69 | 237.72 | 212.86 | 201.48 | 3.53 |
| Desulfuromonadales bacterium (uncultured) | 0.033797 | 0.047965 | 100 | 65.76 | 59.63 | 61.14 | 96.03 | 76.67 | 67.29 | 65.58 | 67.82 | 92.78 | 91.59 | 3.17 |
| Devosia | 0.0000096 | 0.00010605 | 100 | 226.22 | 319.51 | 225.4 | 271.28 | 210.97 | 234.93 | 216.19 | 169.16 | 174.26 | 148.16 | 4.34 |
| Domibacillus | 0.01899 | 0.030499 | 100 | 90.7 | 78.89 | 69.59 | 81.07 | 108.49 | 160.57 | 129.7 | 152.24 | 156.85 | 235.77 | 3.37 |
| Dongia | 0.00020379 | 0.00090009 | 100 | 70.74 | 54.21 | 48.68 | 54.72 | 44.35 | 60.5 | 74.36 | 73.15 | 64.36 | 62.84 | 3.83 |
| Dyadobacter | 0.0016816 | 0.0046909 | 100 | 232.62 | 226.93 | 196.06 | 117.61 | 161.45 | 597.49 | 204.79 | 493.9 | 216.82 | 199.51 | 3.82 |
| Ensifer | 0.0000334 | 0.0002946 | 100 | 154.57 | 144.99 | 148.37 | 136.6 | 166.19 | 201.05 | 210.7 | 228.49 | 191.3 | 195.35 | 3.92 |
| Enterobacter | 0.0041214 | 0.0090263 | 100 | 508.34 | 66.44 | 68.61 | 37.4 | 107.23 | 2294.43 | 390.86 | 435.46 | 1041.92 | 391.72 | 4.28 |
| Ferruginibacter | 0.022231 | 0.033858 | 100 | 131.11 | 117.94 | 152.2 | 151.21 | 158.63 | 105.22 | 111.4 | 113.77 | 94.65 | 101.97 | 3.33 |
| Fictibacillus | 0.0000701 | 0.00048619 | 100 | 57 | 61.58 | 60.46 | 53.77 | 73.42 | 35.9 | 71.85 | 35.74 | 59.62 | 60.49 | 3.82 |
| Fimbriiglobus | 0.0021683 | 0.0052715 | 100 | 132.57 | 123.36 | 110.92 | 143.44 | 112.03 | 139.1 | 154.72 | 165.93 | 161.35 | 161.62 | 3.8 |
| Flavisolibacter | 0.0014198 | 0.0040548 | 100 | 151.91 | 131.26 | 182.15 | 163.17 | 179.1 | 132.66 | 149.47 | 119.44 | 97.85 | 111.18 | 3.84 |
| Flavitalea | 0.0000169 | 0.00017193 | 100 | 181.78 | 163.92 | 233.88 | 214.95 | 202.73 | 218.18 | 263.68 | 234.51 | 197.94 | 195.31 | 3.3 |
| Gaiella | 0.000000204 | 0.0000108 | 100 | 68.41 | 81.42 | 75.03 | 61.98 | 69.54 | 51.04 | 42.22 | 45.04 | 57.89 | 61.91 | 5.06 |
| Gaiella sp (uncultured) | 0.00000393 | 0.0000652 | 100 | 77.81 | 87.76 | 97.13 | 65.55 | 91.79 | 58.1 | 46.84 | 54.62 | 69.69 | 72.27 | 3.99 |
| Gemmata | 0.01289 | 0.022038 | 100 | 116.85 | 107.42 | 86.95 | 105.48 | 83.21 | 111.89 | 125.15 | 125.91 | 119.25 | 127.67 | 4.41 |
| Gemmatimonadalesbacterium (uncultured) | 0.0026426 | 0.0061971 | 100 | 82 | 70.69 | 51.2 | 67.43 | 41.47 | 65.43 | 75.35 | 76.76 | 83.05 | 81.29 | 3.46 |
| Gemmatimonadetesbacterium (uncultured) | 0.034256 | 0.048287 | 100 | 107.82 | 92.29 | 115.96 | 142.77 | 89.71 | 129.7 | 165.71 | 150.98 | 115.88 | 105.49 | 3.47 |
| Gemmatimonas | 0.00011975 | 0.00066221 | 100 | 167.46 | 134.25 | 176.39 | 187.47 | 155.72 | 175.34 | 190.87 | 179.69 | 140.62 | 139.7 | 3.52 |
| Haliangium | 0.019262 | 0.03075 | 100 | 142.07 | 150.98 | 178.47 | 160.7 | 100.56 | 126.82 | 138.32 | 128.47 | 70.12 | 83.26 | 3.66 |
| Herbinix | 0.010326 | 0.018366 | 100 | 59.77 | 69.48 | 54.16 | 58.89 | 75.81 | 44.86 | 44.65 | 36.5 | 68.65 | 55.66 | 2.88 |
| Herpetosiphon | 0.021416 | 0.032805 | 100 | 153 | 170.49 | 216.45 | 139.36 | 189.54 | 188.93 | 132.95 | 123.08 | 124.75 | 125.94 | 3.88 |
| Hirschia | 0.00000632 | 0.0000905 | 100 | 344.79 | 525.52 | 408.98 | 682.66 | 418.08 | 373.67 | 1232.23 | 424.42 | 210.06 | 180.65 | 3.82 |
| Hyphomicrobium | 0.01002 | 0.017942 | 100 | 73.17 | 71.68 | 66.73 | 67.52 | 58.92 | 86.64 | 78.82 | 96.11 | 108.3 | 97.81 | 3.04 |
| Iamia | 0.039147 | 0.0546 | 100 | 95.31 | 106.8 | 94.52 | 76.34 | 72.34 | 107.42 | 83.82 | 70.76 | 72.27 | 74.64 | 3.64 |
| Iamia sp (uncultured) | 0.001423 | 0.0040548 | 100 | 161.63 | 150.05 | 203.71 | 144.97 | 155.12 | 144.88 | 81.65 | 81.53 | 69.71 | 71.87 | 2.93 |
| Ideonella | 0.0040052 | 0.0088447 | 100 | 356.9 | 919.34 | 392.73 | 310.44 | 422.8 | 479.36 | 581.88 | 567.97 | 277.46 | 332.01 | 3.16 |
| Ilumatobacter | 0.047015 | 0.063566 | 100 | 78.54 | 99.56 | 70.17 | 55.72 | 60.49 | 75.65 | 50.63 | 59.65 | 68.94 | 79.38 | 3.47 |
| Knoellia | 0.0021669 | 0.0052715 | 100 | 90.33 | 87.36 | 96.59 | 90.73 | 110.48 | 81.06 | 70.72 | 69.84 | 94.59 | 80.69 | 4.4 |
| Kribbella | 0.00081255 | 0.0026916 | 100 | 71.11 | 83.71 | 70.86 | 65.83 | 71.24 | 84.09 | 70.24 | 74.46 | 79.79 | 83.96 | 3.05 |
| Lacibacter | 0.011803 | 0.020486 | 100 | 245.01 | 200.19 | 252.26 | 185.76 | 187.34 | 276.88 | 199.35 | 186.66 | 127.6 | 128.52 | 3.24 |
| Lechevalieria | 0.00000228 | 0.0000432 | 100 | 173.32 | 190.25 | 120.07 | 95.16 | 113.47 | 331.88 | 211.27 | 179.01 | 192.24 | 195.11 | 3.98 |
| Leptolyngbya EcFYyyy 00 | 0.0000139 | 0.00014736 | 100 | 2312.27 | 5119.22 | 9376.78 | 6632.3 | 7496.83 | 1967.75 | 2638.27 | 5665.63 | 1643.89 | 1494.45 | 4.59 |
| Litorilinea | 0.043669 | 0.05996 | 100 | 121.16 | 126.57 | 136.59 | 151.58 | 166.85 | 121.76 | 110.6 | 119.61 | 105.13 | 134.46 | 3.4 |
| Luedemannella | 0.0000735 | 0.00048692 | 100 | 39.99 | 59.94 | 47.15 | 51.28 | 60.53 | 39.02 | 30.94 | 39.17 | 60.62 | 45.17 | 3.45 |
| Luteitalea | 0.0052232 | 0.01073 | 100 | 118.55 | 124.73 | 140.83 | 154.58 | 167.95 | 131.97 | 144.26 | 140.13 | 193.24 | 129.56 | 3.42 |
| Luteolibacter | 0.0000482 | 0.00036526 | 100 | 190.94 | 284.65 | 204.06 | 210.41 | 190.7 | 219.9 | 179.66 | 170.08 | 127.91 | 125.81 | 3.71 |
| Lutispora sp (uncultured) | 0.0043214 | 0.0093867 | 100 | 32.62 | 31.32 | 41.91 | 40.8 | 40.63 | 23.19 | 19.5 | 29.28 | 41.92 | 37.77 | 3.64 |
| Lysobacter | 0.0045532 | 0.0095762 | 100 | 135.39 | 128.72 | 160.92 | 118.34 | 150.98 | 146.07 | 132.24 | 102.15 | 106.22 | 100.25 | 3.91 |
| Marmoricola | 0.0025191 | 0.0059603 | 100 | 80.34 | 74.33 | 82.91 | 73.03 | 85.95 | 75.16 | 82.11 | 65.41 | 68.57 | 62.57 | 4.19 |
| Massilia | 0.031063 | 0.044982 | 100 | 104.7 | 100.21 | 140.59 | 126.46 | 215.03 | 110.64 | 117.27 | 99.62 | 84.87 | 109.16 | 4.54 |
| Mesorhizobium | 0.00011549 | 0.00066221 | 100 | 184.16 | 282.23 | 208.63 | 273.02 | 212.63 | 207.48 | 276.47 | 199.8 | 196.65 | 177.18 | 4.18 |
| Methylobacillus | 0.00000104 | 0.0000251 | 100 | 1748.09 | 1936.43 | 1955.12 | 899.75 | 1931.16 | 10944.59 | 3216.56 | 5335.2 | 3268.91 | 5021.76 | 3.5 |
| Methylorosula | 0.00028426 | 0.0011243 | 100 | 134.35 | 130.99 | 182.81 | 175.05 | 196.27 | 99.66 | 113.49 | 93.63 | 86.59 | 103.86 | 2.77 |
| Methylotenera | 0.00084022 | 0.0027153 | 100 | 389.84 | 409.42 | 404.84 | 332.11 | 443.43 | 1665.5 | 553.08 | 767.07 | 496.42 | 582.81 | 3.65 |
| Microbacterium | 0.001128 | 0.0033969 | 100 | 209.17 | 180.34 | 181.68 | 122.68 | 171.04 | 426.61 | 263.56 | 226.4 | 389.73 | 240.8 | 4.02 |
| Microvirga | 0.011174 | 0.019741 | 100 | 113.12 | 98.59 | 148.99 | 132.61 | 173.22 | 129.73 | 167.03 | 130.26 | 116.6 | 130.67 | 4.68 |
| Mitsuaria | 0.000000014 | 0.00000199 | 100 | 2471.17 | 4603.04 | 1612.26 | 379.1 | 517.39 | 15332.6 | 3375.88 | 18722.49 | 6035.04 | 6936.55 | 4.18 |
| Mycobacterium | 0.0000303 | 0.00028643 | 100 | 77.44 | 87.4 | 84.17 | 77.02 | 76.92 | 76.92 | 85.46 | 66.59 | 62.03 | 72.73 | 3.82 |
| Niastella | 0.0001624 | 0.00077527 | 100 | 235.05 | 288.29 | 234.39 | 236.87 | 235.51 | 370.63 | 314.42 | 385.19 | 312.56 | 284.5 | 3.08 |
| Nitrospira | 0.0045207 | 0.0095762 | 100 | 136.34 | 128.62 | 140.07 | 170.61 | 130.4 | 121.19 | 153.19 | 159.19 | 161.09 | 158.07 | 4.33 |
| Nocardioides | 0.00032053 | 0.0012229 | 100 | 85.8 | 89.32 | 88.58 | 69.58 | 83.16 | 76.51 | 56.98 | 59.01 | 80.5 | 67.38 | 4.35 |
| Nodosilinea PCC 7104 | 0.0001395 | 0.00072068 | 100 | 2253.73 | 8243.55 | 3699.69 | 6425.11 | 12123.84 | 6449.88 | 4061.03 | 6972.14 | 3939.8 | 5529.82 | 3.94 |
| Nonomuraea | 0.025846 | 0.038263 | 100 | 60.89 | 82.72 | 78.36 | 73.94 | 91 | 57 | 47.39 | 64.38 | 77.75 | 68.59 | 3 |
| Nordella | 0.018708 | 0.030229 | 100 | 130.16 | 112.77 | 99.28 | 114.07 | 83 | 118.22 | 140.1 | 136.73 | 123.43 | 111.16 | 3.91 |
| Ohtaekwangia | 0.0010564 | 0.0032456 | 100 | 244.28 | 339.48 | 248.08 | 235.26 | 183.89 | 271.33 | 267.43 | 185.08 | 152.59 | 153.66 | 3.58 |
| Oligoflexus | 0.011473 | 0.020134 | 100 | 132.57 | 152.11 | 260.23 | 195.51 | 326.72 | 125.28 | 131.03 | 126.88 | 125.38 | 111.83 | 3.21 |
| Paenarthrobacter | 0.030718 | 0.044726 | 100 | 129.65 | 76.49 | 102.87 | 87.34 | 125.46 | 216.98 | 106.39 | 160.56 | 183.42 | 174.4 | 3.83 |
| Paenisporosarcina | 0.0077201 | 0.014613 | 100 | 84.37 | 77.24 | 75.17 | 76.1 | 102.17 | 48.46 | 73.1 | 52.54 | 79.84 | 77.1 | 3.51 |
| Parasegetibacter | 0.00018969 | 0.00085201 | 100 | 204.92 | 218.3 | 294.66 | 222.86 | 361.11 | 195.78 | 173.93 | 204.47 | 172.61 | 160.28 | 3.45 |
| Parviterribacter | 0.017589 | 0.028952 | 100 | 87.33 | 94.87 | 99.83 | 83.27 | 104.38 | 69.24 | 71.59 | 58.34 | 69.31 | 56.79 | 3.03 |
| Paucibacter | 0.014338 | 0.024202 | 100 | 284.78 | 208.85 | 265.14 | 247.19 | 257.97 | 217.79 | 224.76 | 185.15 | 171.78 | 179.84 | 2.8 |
| Pedobacter | 0.00021864 | 0.00093453 | 100 | 164.82 | 143.39 | 199.91 | 183.08 | 259.43 | 116.46 | 114.2 | 106.46 | 110.47 | 137.43 | 3.81 |
| Pedococcus Phycicoccus | 0.0087706 | 0.016368 | 100 | 85.21 | 89.04 | 86.24 | 61 | 70.52 | 82.17 | 55.39 | 54.03 | 94.25 | 77.84 | 2.75 |
| Pedomicrobium | 0.00069317 | 0.0023252 | 100 | 71.75 | 74.73 | 66.39 | 82.76 | 60.34 | 83.97 | 97.89 | 100.87 | 104.39 | 86.8 | 3.79 |
| Pelobacter sp (uncultured) | 0.011828 | 0.020486 | 100 | 122.6 | 102.05 | 122.5 | 107.26 | 59.33 | 125.71 | 192.02 | 199.04 | 110.51 | 90.47 | 2.92 |
| Phenylobacterium | 0.0045993 | 0.0095969 | 100 | 89.49 | 105.03 | 102.19 | 93.27 | 115.14 | 72.43 | 68.3 | 63.98 | 74.15 | 72.89 | 2.75 |
| Pirellula | 0.014538 | 0.024383 | 100 | 130.33 | 110.51 | 96.29 | 103.92 | 76.54 | 132.36 | 141.09 | 136.47 | 117.04 | 127.69 | 4.56 |
| Planctomicrobium | 0.0015673 | 0.0044184 | 100 | 188.68 | 156.59 | 131.19 | 142.59 | 87.73 | 264.15 | 215.38 | 197.44 | 150.77 | 200.12 | 2.97 |
| Planctomycetales bacterium (uncultured) | 0.00023737 | 0.00098821 | 100 | 192.28 | 167.13 | 173.3 | 210.72 | 122.1 | 285.77 | 298.34 | 247.74 | 193.15 | 258.19 | 2.96 |
| Planctomycete (uncultured) | 0.000033 | 0.0002946 | 100 | 199.24 | 149.86 | 189.05 | 213.02 | 166.87 | 225.66 | 248.26 | 194.23 | 134.27 | 162.06 | 4.33 |
| Polaromonas | 0.009301 | 0.017138 | 100 | 179.46 | 167.51 | 133.39 | 182.9 | 144.3 | 128.52 | 118.78 | 129.68 | 117.05 | 100.35 | 2.77 |
| Polyangium brachysporum group | 0.000000784 | 0.0000208 | 100 | 268.51 | 232.62 | 326.15 | 205.78 | 221.01 | 619.71 | 707.05 | 324.78 | 356.17 | 395.77 | 3.66 |
| Pontibacter | 0.0094018 | 0.017138 | 100 | 173.16 | 147.9 | 236.04 | 176.91 | 330.57 | 146.09 | 123.67 | 125.21 | 115.82 | 133.43 | 3.95 |
| Promicromonospora | 0.024173 | 0.036397 | 100 | 52.37 | 83.01 | 39.49 | 53.33 | 62.08 | 88.85 | 101.5 | 58.59 | 50.56 | 57.65 | 3.26 |
| Pseudarthrobacter | 0.0053672 | 0.010857 | 100 | 117.69 | 91.15 | 163.45 | 118.36 | 171.83 | 152.47 | 105.46 | 136.53 | 157.57 | 145.83 | 3.1 |
| Pseudoduganella | 0.00011 | 0.00066221 | 100 | 672.85 | 586.11 | 1138.12 | 411.03 | 310.16 | 834.46 | 670.35 | 770.86 | 412.84 | 450.85 | 3.55 |
| Pseudoflavitalea | 0.0021239 | 0.0052715 | 100 | 156.52 | 170.9 | 125.18 | 286.59 | 115.08 | 92.09 | 290.91 | 162.12 | 105.56 | 107.96 | 2.97 |
| Pseudolabrys | 0.0050874 | 0.010532 | 100 | 121.48 | 186.31 | 90.15 | 145.14 | 85.18 | 85.56 | 91.97 | 75.5 | 73.53 | 76.44 | 3.87 |
| Pseudomuriella schumacherensis | 0.00011443 | 0.00066221 | 100 | 1106.64 | 1024.92 | 1188.77 | 1364.16 | 1518.29 | 548.54 | 893.81 | 1066.76 | 659.19 | 692.24 | 3.48 |
| Pseudorhodoplanes | 0.00213 | 0.0052715 | 100 | 179.58 | 201.33 | 186.53 | 260.72 | 168.81 | 158.37 | 237.17 | 206.26 | 181.6 | 167.52 | 3.02 |
| Pseudoxanthomonas | 0.00011106 | 0.00066221 | 100 | 253.63 | 454.14 | 234.01 | 173.62 | 201.24 | 559.88 | 403.38 | 381.86 | 291.59 | 216.07 | 3.99 |
| Qipengyuania | 0.00000207 | 0.0000422 | 100 | 297.75 | 257.97 | 339.14 | 191.27 | 277.65 | 180.67 | 116.69 | 119.76 | 138.04 | 134.82 | 3.83 |
| Ramlibacter | 0.00000652 | 0.0000905 | 100 | 181.28 | 161 | 236.7 | 219.99 | 277.08 | 265.84 | 273.63 | 218.76 | 141.41 | 154.73 | 4.08 |
| Reyranella | 0.004395 | 0.0094048 | 100 | 126.84 | 137.28 | 113.72 | 150.05 | 97.42 | 137.09 | 163.7 | 166.75 | 117.23 | 122.19 | 3.35 |
| Rhizobacter | 0.0000521 | 0.00038145 | 100 | 174.57 | 197.02 | 194.21 | 191.55 | 218.94 | 223.06 | 200.44 | 179.82 | 164.11 | 150.41 | 3.96 |
| Rhodobacter | 0.0000467 | 0.00036396 | 100 | 221.61 | 342.49 | 418.73 | 396.49 | 466.8 | 204.58 | 122.81 | 136.38 | 134.34 | 109.9 | 3.52 |
| Rhodocytophaga | 0.028682 | 0.041993 | 100 | 191.82 | 167.95 | 210.05 | 187.24 | 253.2 | 196.79 | 192.47 | 238.35 | 217.3 | 225.2 | 3.21 |
| Rhodopirellula | 0.0000374 | 0.00031979 | 100 | 286.18 | 240.7 | 315.43 | 257.59 | 351.41 | 280.86 | 318.8 | 218.5 | 178.75 | 202.5 | 3.71 |
| Rhodoplanes | 0.040253 | 0.055849 | 100 | 124.38 | 115.81 | 90.29 | 108.79 | 73.07 | 125.18 | 129.23 | 146.31 | 125.73 | 116.69 | 3.69 |
| Roseimicrobium | 0.046453 | 0.063129 | 100 | 127.53 | 103.51 | 116.26 | 131.14 | 79.08 | 131.94 | 123.46 | 126.8 | 95.22 | 102.17 | 3.29 |
| Roseomonas | 0.0028822 | 0.0065843 | 100 | 152.77 | 155.58 | 216.23 | 213.38 | 317.78 | 160.55 | 177.73 | 154.02 | 144.78 | 130.02 | 3.18 |
| Rubellimicrobium | 0.00097379 | 0.0030721 | 100 | 180.34 | 153.55 | 261.22 | 232.54 | 350.73 | 187.62 | 165.95 | 162.6 | 165.33 | 146.86 | 4.16 |
| Rubrobacter | 0.0094423 | 0.017138 | 100 | 92.81 | 99.13 | 115.01 | 104.29 | 122.75 | 77.76 | 85 | 83.38 | 83.85 | 96.1 | 4.52 |
| Rubrobacterales bacterium (uncultured) | 0.0000000363 | 0.00000304 | 100 | 70.11 | 79.72 | 65.57 | 53.54 | 55.68 | 47.42 | 39.19 | 38.75 | 48.12 | 60.7 | 4.44 |
| Rubrobacteria bacterium (uncultured) | 0.000000765 | 0.0000208 | 100 | 75.13 | 92.58 | 78.42 | 61.37 | 73.87 | 54.54 | 44.77 | 48.19 | 60.06 | 68.59 | 3.93 |
| Saccharothrix | 0.031572 | 0.04547 | 100 | 176.51 | 131.74 | 138.84 | 138.58 | 199.97 | 120.79 | 80.1 | 84.76 | 73.18 | 98.96 | 3.17 |
| Shimazuella | 0.020834 | 0.032287 | 100 | 56.67 | 73.25 | 82.5 | 74.12 | 100.33 | 70.95 | 52.39 | 79.53 | 87.62 | 68.01 | 2.79 |
| Shinella | 0.00053039 | 0.0018494 | 100 | 573.73 | 766.54 | 460.95 | 311.9 | 450.67 | 722.3 | 409.51 | 561.55 | 450.4 | 332.76 | 3.6 |
| Skermanella | 0.033847 | 0.047965 | 100 | 93.88 | 103.19 | 127.89 | 151.19 | 176.97 | 98.94 | 97.63 | 100.79 | 119.95 | 111.45 | 4.41 |
| Solirubrobacter | 0.0000462 | 0.00036396 | 100 | 74.19 | 84.79 | 80.89 | 73.16 | 78.94 | 57 | 53.45 | 53.21 | 70.25 | 63.61 | 3.97 |
| Sorangium | 0.0021467 | 0.0052715 | 100 | 46.76 | 115.95 | 56.46 | 55.15 | 41.56 | 128.87 | 164.06 | 69.81 | 79.75 | 51.39 | 3.08 |
| Sphingoaurantiacus | 0.00023866 | 0.00098821 | 100 | 137.24 | 127.25 | 188.94 | 156.67 | 253.86 | 113.44 | 105.64 | 80.63 | 104.08 | 109.26 | 3.22 |
| Sphingobium | 0.00000846 | 0.00010189 | 100 | 1535.96 | 1807.61 | 2282.28 | 1206.9 | 1949.55 | 4110.56 | 2082.64 | 2135.58 | 1950.4 | 1651.48 | 4.7 |
| Sphingomonas | 0.00025341 | 0.0010331 | 100 | 113.23 | 115.85 | 128.93 | 95.94 | 124.51 | 98.88 | 96.23 | 77.57 | 84.93 | 94.5 | 4.65 |
| Sphingopyxis | 0.00011995 | 0.00066221 | 100 | 336.67 | 528.52 | 344.6 | 267.99 | 391.86 | 330.81 | 114.14 | 152.5 | 170.98 | 211.84 | 3.34 |
| Steroidobacter | 0.00032304 | 0.0012229 | 100 | 77.88 | 89.19 | 80.14 | 103.14 | 84.97 | 104.13 | 123.26 | 123.89 | 137.99 | 106.49 | 3.9 |
| Streptomyces | 0.00053812 | 0.001852 | 100 | 143.71 | 147.41 | 108.38 | 114.58 | 117.28 | 169.34 | 147.64 | 120.62 | 137.28 | 137.42 | 4.2 |
| Sumerlaea | 0.0010606 | 0.0032456 | 100 | 202.52 | 154.59 | 213.89 | 210.88 | 208.49 | 155.3 | 203.34 | 189.77 | 170.64 | 133.07 | 3.09 |
| Synechococcus IR11 | 0.0018714 | 0.0050246 | 100 | 2059.31 | 8772.95 | 7010.01 | 9186.14 | 13235.59 | 3336.4 | 9873.98 | 3522.57 | 3217.56 | 4680.08 | 3.55 |
| Tahibacter | 0.0067223 | 0.013003 | 100 | 325.35 | 250.32 | 201 | 152.21 | 171.68 | 456.93 | 513.45 | 240.03 | 191.08 | 242.91 | 3.09 |
| Tepidisphaera | 0.00308 | 0.0069668 | 100 | 192.6 | 131.89 | 222.59 | 225.5 | 186.88 | 151.91 | 164.68 | 156.13 | 83.33 | 98.18 | 3.19 |
| Terrimonas | 0.0031022 | 0.0069668 | 100 | 173.55 | 162.97 | 160.76 | 177.85 | 132.57 | 193.79 | 238.46 | 200.37 | 140.34 | 137.5 | 3.85 |
| Thermomicrobia bacterium (uncultured) | 0.041362 | 0.057088 | 100 | 92.53 | 105.02 | 86.79 | 97.95 | 114.08 | 91.75 | 71.3 | 72.09 | 74.38 | 96.51 | 2.63 |
| Truepera | 0.000000551 | 0.0000198 | 100 | 246.02 | 169.5 | 255.84 | 302.82 | 315.02 | 698.33 | 970.88 | 831.76 | 967.32 | 617.8 | 3.55 |
| Tumebacillus | 0.0018961 | 0.0050246 | 100 | 48.55 | 94.68 | 54.44 | 65.91 | 86.44 | 36.34 | 28.91 | 46.08 | 103.41 | 69.26 | 3.71 |
| Tychonema CCAP 1459 11B | 0.0066796 | 0.013003 | 100 | 369.76 | 335.37 | 240.12 | 439.63 | 398.29 | 132.46 | 88.47 | 234 | 161.77 | 141.96 | 4.09 |
| Variovorax | 0.0000987 | 0.0006228 | 100 | 164.4 | 186.5 | 126.31 | 131.17 | 138.76 | 283.49 | 193.64 | 207.47 | 174.6 | 124.95 | 3.81 |
| Yonghaparkia | 0.000000596 | 0.0000198 | 100 | 879.56 | 846.51 | 1136.14 | 574.27 | 1120.71 | 294.83 | 186.43 | 200.65 | 247.47 | 271.22 | 3.71 |
| Genus | p-Value | FDR | t0 | t1 | t2 | LDA Score |
|---|---|---|---|---|---|---|
| Acidibacter | 0.00016 | 0.00041 | 100 | 168.63 | 188.29 | 3.69 |
| Acidimicrobiia bacterium (uncultured) | 0.00000 | 0.00000 | 100 | 56.33 | 48.02 | 4.09 |
| Acidobacteria bacterium (uncultured) | 0.00959 | 0.01405 | 100 | 85.88 | 106.38 | 4.01 |
| Acidobacteriaceae bacterium (uncultured) | 0.00021 | 0.00051 | 100 | 87.83 | 112.32 | 4.27 |
| Acidobacteriales bacterium (uncultured) | 0.00002 | 0.00008 | 100 | 63.97 | 87.92 | 4.22 |
| Acidobacterium sp (uncultured) | 0.00948 | 0.01399 | 100 | 97.12 | 73.9 | 3.16 |
| Acidovorax | 0.00000 | 0.00000 | 100 | 1517.39 | 706.69 | 3.04 |
| Acinetobacter | 0.00044 | 0.00094 | 100 | 1740.27 | 3971.71 | 4.73 |
| Actinobacterium (uncultured) | 0.00011 | 0.00030 | 100 | 69.88 | 58.28 | 4.03 |
| Actinomadura | 0.00018 | 0.00043 | 100 | 84.88 | 63.84 | 2.67 |
| Actinomycetales bacterium (uncultured) | 0.00000 | 0.00002 | 100 | 96.28 | 72.78 | 3.14 |
| Adhaeribacter | 0.00009 | 0.00025 | 100 | 159.55 | 108.58 | 3.63 |
| Aeromicrobium | 0.00034 | 0.00075 | 100 | 218.53 | 208.06 | 4.34 |
| Allorhizobium Neorhizobium Pararhizobium Rhizobium | 0.00001 | 0.00005 | 100 | 172.47 | 210.57 | 4.37 |
| Altererythrobacter | 0.00000 | 0.00001 | 100 | 329.74 | 234.6 | 3.39 |
| Aminobacter | 0.00002 | 0.00008 | 100 | 728.31 | 255.57 | 3.32 |
| Ammoniphilus | 0.00586 | 0.00930 | 100 | 90.02 | 75.11 | 2.73 |
| Amycolatopsis | 0.00000 | 0.00000 | 100 | 224.91 | 1369.89 | 3.43 |
| Anaerolinea | 0.00078 | 0.00153 | 100 | 36.41 | 26.99 | 3.56 |
| Anaeromyxobacter | 0.00042 | 0.00091 | 100 | 58.69 | 57.74 | 3.51 |
| Arenimonas | 0.00001 | 0.00003 | 100 | 235.56 | 168.1 | 3.16 |
| Aridibacter | 0.00047 | 0.00099 | 100 | 166.17 | 136.63 | 3.2 |
| Armatimonadetes bacterium (uncultured) | 0.00000 | 0.00000 | 100 | 87.12 | 63.13 | 2.93 |
| Aurantisolimonas | 0.00384 | 0.00640 | 100 | 159.08 | 183.63 | 2.65 |
| Azohydromonas | 0.00000 | 0.00000 | 100 | 173.22 | 273.51 | 3.43 |
| Blastocatella | 0.00007 | 0.00020 | 100 | 157.71 | 130.33 | 3.38 |
| Blastococcus | 0.00027 | 0.00063 | 100 | 128.19 | 96.74 | 3.92 |
| Blastopirellula | 0.00001 | 0.00004 | 100 | 92.63 | 156.06 | 2.85 |
| Bosea | 0.00005 | 0.00015 | 100 | 225.21 | 219.49 | 3.52 |
| Bradyrhizobium | 0.00052 | 0.00109 | 100 | 138.76 | 149.38 | 3.81 |
| Burkholderia Caballeronia Paraburkholderia | 0.00151 | 0.00277 | 100 | 49.89 | 46.06 | 3.41 |
| Caenimonas | 0.00007 | 0.00022 | 100 | 191.21 | 163.94 | 3.93 |
| Candidatus Alysiosphaera | 0.04607 | 0.05786 | 100 | 118.65 | 99.14 | 3.23 |
| Candidatus Solibacter | 0.00510 | 0.00824 | 100 | 70.93 | 82.45 | 3.01 |
| Candidatus Udaeobacter | 0.01891 | 0.02544 | 100 | 69.85 | 91.19 | 3.86 |
| Candidatus Xiphinematobacter | 0.00013 | 0.00033 | 100 | 77.76 | 117.12 | 4.45 |
| Caulobacter | 0.00712 | 0.01103 | 100 | 138.57 | 68.57 | 2.74 |
| Cellulomonas | 0.01327 | 0.01870 | 100 | 104.61 | 75.48 | 2.47 |
| Chloroflexi bacterium (uncultured) | 0.00000 | 0.00000 | 100 | 92.98 | 71.61 | 4.22 |
| Chloroflexus sp (uncultured) | 0.00419 | 0.00695 | 100 | 81.13 | 71.69 | 3.6 |
| Chthoniobacter | 0.00013 | 0.00033 | 100 | 159.44 | 130.16 | 4.13 |
| Chthonomonas | 0.00694 | 0.01089 | 100 | 140.14 | 124.96 | 2.43 |
| Clostridium sensu stricto 1 | 0.00065 | 0.00132 | 100 | 57.6 | 46.87 | 3.13 |
| Clostridium sensu stricto 13 | 0.00240 | 0.00424 | 100 | 77.34 | 58.68 | 2.64 |
| Clostridium sensu stricto 8 | 0.00433 | 0.00712 | 100 | 66.59 | 54.98 | 2.77 |
| Cohnella | 0.00089 | 0.00168 | 100 | 79.81 | 60.48 | 3.28 |
| Comamonas | 0.00000 | 0.00000 | 100 | 202.26 | 338.2 | 3.05 |
| Conexibacter | 0.00004 | 0.00012 | 100 | 105.08 | 73.49 | 2.61 |
| Conexibacter sp (uncultured) | 0.00000 | 0.00000 | 100 | 73.36 | 51.54 | 2.9 |
| Cupriavidus | 0.00001 | 0.00003 | 100 | 145.61 | 255.89 | 3.21 |
| Cyanobacterium sp (uncultured) | 0.01007 | 0.01467 | 100 | 157.02 | 79.88 | 3.01 |
| Desmochloris halophila | 0.00000 | 0.00000 | 100 | 3208.47 | 1996.35 | 3.27 |
| Desulfuromonadales bacterium (uncultured) | 0.03356 | 0.04317 | 100 | 72.23 | 76.64 | 3.01 |
| Devosia | 0.00000 | 0.00000 | 100 | 250.31 | 189.57 | 4.18 |
| Domibacillus | 0.00023 | 0.00055 | 100 | 86.72 | 165.26 | 3.05 |
| Dongia | 0.00001 | 0.00003 | 100 | 54.43 | 67.15 | 3.74 |
| Dyadobacter | 0.00061 | 0.00124 | 100 | 186.07 | 346.17 | 3.52 |
| Ensifer | 0.00000 | 0.00000 | 100 | 150.59 | 205.63 | 3.84 |
| Enterobacter | 0.00005 | 0.00015 | 100 | 158.57 | 924.19 | 3.84 |
| Ferrimicrobium sp (uncultured) | 0.01624 | 0.02254 | 100 | 74.34 | 83.52 | 2.8 |
| Ferruginibacter | 0.00054 | 0.00112 | 100 | 142.38 | 105.49 | 3.15 |
| Fictibacillus | 0.00002 | 0.00006 | 100 | 61.57 | 52.52 | 3.69 |
| Fimbriiglobus | 0.00001 | 0.00006 | 100 | 124.49 | 156.41 | 3.73 |
| Flavisolibacter | 0.00033 | 0.00074 | 100 | 161.44 | 122.4 | 3.71 |
| Flavitalea | 0.00000 | 0.00001 | 100 | 198.67 | 222.61 | 3.17 |
| Gaiella | 0.00000 | 0.00000 | 100 | 71.14 | 51.35 | 4.99 |
| Gaiella sp (uncultured) | 0.00000 | 0.00000 | 100 | 83.87 | 60 | 3.86 |
| Galbitalea | 0.03930 | 0.04983 | 100 | 97.77 | 71.06 | 3 |
| Gemmata | 0.00113 | 0.00213 | 100 | 99.89 | 121.83 | 4.1 |
| Gemmatimonadales bacterium (uncultured) | 0.00020 | 0.00049 | 100 | 62.32 | 76.25 | 3.27 |
| Gemmatimonadetes bacterium (uncultured) | 0.02065 | 0.02763 | 100 | 109.05 | 134.27 | 3.12 |
| Gemmatimonas | 0.00003 | 0.00009 | 100 | 163.74 | 165.9 | 3.39 |
| Geodermatophilus | 0.02480 | 0.03286 | 100 | 141.78 | 120.65 | 2.4 |
| Haliangium | 0.03644 | 0.04665 | 100 | 144.6 | 110.07 | 3.28 |
| Herbinix | 0.00058 | 0.00119 | 100 | 64.16 | 49.92 | 2.78 |
| Herpetosiphon | 0.00144 | 0.00265 | 100 | 173.1 | 139.47 | 3.68 |
| Hirschia | 0.00000 | 0.00000 | 100 | 476.23 | 491.99 | 3.36 |
| Hyphomicrobium | 0.00011 | 0.00030 | 100 | 67.41 | 93.43 | 2.86 |
| Iamia sp (uncultured) | 0.00004 | 0.00013 | 100 | 161.88 | 90.39 | 2.66 |
| Ideonella | 0.00006 | 0.00017 | 100 | 481.19 | 450.69 | 2.83 |
| Ilumatobacter | 0.00477 | 0.00775 | 100 | 72.65 | 66.53 | 3.3 |
| Intrasporangium | 0.01812 | 0.02475 | 100 | 89.74 | 71.47 | 2.72 |
| Knoellia | 0.00037 | 0.00081 | 100 | 95.44 | 79.35 | 4.1 |
| Kribbella | 0.00008 | 0.00024 | 100 | 72.56 | 78.37 | 2.96 |
| Lacibacter | 0.00466 | 0.00763 | 100 | 212.49 | 185.22 | 3.05 |
| Lechevalieria | 0.00000 | 0.00000 | 100 | 138.29 | 222.59 | 3.7 |
| Leptolyngbya EcFYyyy 00 | 0.00000 | 0.00000 | 100 | 6140.49 | 2712.46 | 4.4 |
| Litorilinea | 0.00832 | 0.01254 | 100 | 141.3 | 117.9 | 3.19 |
| Luedemannella | 0.00001 | 0.00004 | 100 | 52.11 | 42.93 | 3.37 |
| Luteitalea | 0.00951 | 0.01399 | 100 | 142.01 | 148.3 | 3.14 |
| Luteolibacter | 0.00000 | 0.00001 | 100 | 215.82 | 165.67 | 3.51 |
| Lutispora sp (uncultured) | 0.00033 | 0.00074 | 100 | 37.42 | 30.14 | 3.58 |
| Lysobacter | 0.00127 | 0.00235 | 100 | 138.62 | 117.82 | 3.71 |
| Marmoricola | 0.00004 | 0.00012 | 100 | 79.39 | 70.97 | 4.08 |
| Massilia | 0.03110 | 0.04021 | 100 | 139.26 | 104.19 | 4.02 |
| Mesorhizobium | 0.00000 | 0.00001 | 100 | 232.23 | 212.4 | 4.04 |
| Methylobacillus | 0.00000 | 0.00000 | 100 | 1693.52 | 5571.14 | 3.21 |
| Methylorosula | 0.00000 | 0.00003 | 100 | 164.23 | 99.33 | 2.54 |
| Methylotenera | 0.00001 | 0.00004 | 100 | 396.89 | 818.87 | 3.31 |
| Microbacterium | 0.00001 | 0.00004 | 100 | 172.71 | 311.19 | 3.83 |
| Microvirga | 0.03040 | 0.03969 | 100 | 133.91 | 134.97 | 4.35 |
| Mitsuaria | 0.00000 | 0.00000 | 100 | 1889.22 | 10161.07 | 3.91 |
| Modestobacter | 0.01267 | 0.01804 | 100 | 131.43 | 105.42 | 2.74 |
| Mycobacterium | 0.00001 | 0.00005 | 100 | 80.41 | 72.75 | 3.67 |
| Niastella | 0.00000 | 0.00000 | 100 | 246.05 | 334.72 | 2.99 |
| Nitrosospira | 0.01857 | 0.02523 | 100 | 214 | 140.23 | 2.88 |
| Nitrospira | 0.00069 | 0.00139 | 100 | 140.96 | 150.35 | 4.18 |
| Nocardia | 0.00690 | 0.01089 | 100 | 164.31 | 195.6 | 2.73 |
| Nocardioides | 0.00001 | 0.00004 | 100 | 83.15 | 68.1 | 4.22 |
| Nodosilinea PCC 7104 | 0.00000 | 0.00001 | 100 | 6759.82 | 5387 | 3.68 |
| Nonomuraea | 0.00570 | 0.00910 | 100 | 77.7 | 62.88 | 2.84 |
| Nordella | 0.01450 | 0.02033 | 100 | 107.45 | 126.31 | 3.58 |
| Ohtaekwangia | 0.00011 | 0.00030 | 100 | 248.6 | 207.36 | 3.37 |
| Oligoflexus | 0.00231 | 0.00412 | 100 | 215.09 | 124.39 | 2.92 |
| Paenarthrobacter | 0.02146 | 0.02858 | 100 | 104.93 | 168.2 | 3.52 |
| Paenisporosarcina | 0.00367 | 0.00615 | 100 | 83.68 | 65.93 | 3.32 |
| Parasegetibacter | 0.00000 | 0.00001 | 100 | 262.03 | 181.96 | 3.24 |
| Parviterribacter | 0.00009 | 0.00025 | 100 | 94.05 | 65.27 | 2.89 |
| Paucibacter | 0.00028 | 0.00065 | 100 | 252.6 | 196.28 | 2.71 |
| Pedobacter | 0.00000 | 0.00001 | 100 | 191.62 | 116.48 | 3.57 |
| Pedococcus Phycicoccus | 0.00348 | 0.00594 | 100 | 78.01 | 72.61 | 2.53 |
| Pedomicrobium | 0.00002 | 0.00006 | 100 | 71.04 | 94.99 | 3.6 |
| Pelobacter sp (uncultured) | 0.01273 | 0.01804 | 100 | 101.17 | 144.91 | 2.43 |
| Phenylobacterium | 0.00001 | 0.00005 | 100 | 101.35 | 70.29 | 2.53 |
| Phyllobacterium | 0.00854 | 0.01279 | 100 | 142.54 | 161.39 | 3.23 |
| Pirellula | 0.00358 | 0.00609 | 100 | 103.02 | 131.02 | 4.24 |
| Planctomicrobium | 0.00024 | 0.00057 | 100 | 140.27 | 205.71 | 2.74 |
| Planctomycetales bacterium (uncultured) | 0.00000 | 0.00003 | 100 | 171.82 | 256.6 | 2.86 |
| planctomycete (uncultured) | 0.00009 | 0.00024 | 100 | 183.06 | 193.69 | 4.13 |
| Polaromonas | 0.00012 | 0.00031 | 100 | 161.78 | 119.35 | 2.64 |
| Polyangium brachysporum group | 0.00000 | 0.00000 | 100 | 248.18 | 482.87 | 3.46 |
| Polycyclovorans | 0.00710 | 0.01103 | 100 | 118.08 | 62.03 | 2.94 |
| Pontibacter | 0.00013 | 0.00034 | 100 | 215.28 | 128.73 | 3.64 |
| Promicromonospora | 0.01552 | 0.02164 | 100 | 58.62 | 71.79 | 3.09 |
| Pseudoduganella | 0.00002 | 0.00007 | 100 | 602.96 | 632.41 | 3.26 |
| Pseudolabrys | 0.00329 | 0.00570 | 100 | 125.53 | 80.71 | 3.47 |
| Pseudomuriella schumacherensis | 0.00000 | 0.00000 | 100 | 1248.8 | 774.15 | 3.38 |
| Pseudonocardia | 0.00768 | 0.01183 | 100 | 89.75 | 73.45 | 2.79 |
| Pseudorhodoplanes | 0.00008 | 0.00022 | 100 | 198.95 | 190.77 | 2.81 |
| Pseudoxanthomonas | 0.00002 | 0.00008 | 100 | 262.52 | 374.52 | 3.77 |
| Qipengyuania | 0.00000 | 0.00000 | 100 | 271.22 | 138.08 | 3.68 |
| Ramlibacter | 0.00001 | 0.00004 | 100 | 216.22 | 212.31 | 3.9 |
| Reyranella | 0.01073 | 0.01554 | 100 | 124.65 | 141.88 | 3.13 |
| Rhizobacter | 0.00000 | 0.00003 | 100 | 195.87 | 184.42 | 3.85 |
| Rhodobacter | 0.00000 | 0.00000 | 100 | 370.43 | 142.41 | 3.39 |
| Rhodocytophaga | 0.00081 | 0.00156 | 100 | 203.13 | 213.74 | 3.08 |
| Rhodopirellula | 0.00000 | 0.00001 | 100 | 291.17 | 240.84 | 3.59 |
| Rhodoplanes | 0.01165 | 0.01668 | 100 | 102.04 | 128.93 | 3.28 |
| Risungbinella | 0.04659 | 0.05824 | 100 | 87.62 | 88.28 | 2.1 |
| Roseomonas | 0.00025 | 0.00058 | 100 | 213.69 | 154.02 | 2.9 |
| Rubellimicrobium | 0.00005 | 0.00015 | 100 | 237.91 | 166.15 | 3.9 |
| Rubrobacter | 0.00160 | 0.00289 | 100 | 106.99 | 84.94 | 4.22 |
| Rubrobacterales bacterium (uncultured) | 0.00000 | 0.00000 | 100 | 64.68 | 46.48 | 4.38 |
| Rubrobacteria bacterium (uncultured) | 0.00000 | 0.00000 | 100 | 76.16 | 54.89 | 3.84 |
| Saccharothrix | 0.00049 | 0.00104 | 100 | 158.65 | 91.37 | 2.89 |
| Shinella | 0.00001 | 0.00004 | 100 | 512.5 | 499.48 | 3.39 |
| Solirubrobacter | 0.00000 | 0.00000 | 100 | 78.34 | 59.4 | 3.91 |
| Sorangium | 0.00117 | 0.00219 | 100 | 62.8 | 99.99 | 2.57 |
| Sphingoaurantiacus | 0.00000 | 0.00003 | 100 | 174.41 | 102.44 | 2.85 |
| Sphingobium | 0.00000 | 0.00001 | 100 | 1748.14 | 2404.97 | 4.46 |
| Sphingomonas | 0.00002 | 0.00008 | 100 | 115.58 | 90.32 | 4.34 |
| Sphingopyxis | 0.00002 | 0.00008 | 100 | 375.11 | 195.65 | 3.14 |
| Sporacetigenium | 0.04686 | 0.05829 | 100 | 89.43 | 76.16 | 2.54 |
| Stenotrophobacter | 0.00870 | 0.01296 | 100 | 123.55 | 97.76 | 2.6 |
| Steroidobacter | 0.00001 | 0.00005 | 100 | 87.18 | 119.48 | 3.63 |
| Streptomyces | 0.00085 | 0.00162 | 100 | 126.49 | 142.59 | 3.99 |
| Subgroup 10 | 0.00005 | 0.00015 | 100 | 184.27 | 160.54 | 3.92 |
| Sumerlaea | 0.00006 | 0.00017 | 100 | 197.94 | 171.38 | 3.02 |
| Synechococcus IR11 | 0.00003 | 0.00011 | 100 | 8208.54 | 4932.48 | 3.34 |
| Syntrophobacterales bacterium (uncultured) | 0.03018 | 0.03959 | 100 | 104.28 | 130.87 | 2.24 |
| Tahibacter | 0.00155 | 0.00282 | 100 | 219.39 | 331.07 | 2.84 |
| Tepidisphaera | 0.00362 | 0.00611 | 100 | 191 | 131.68 | 3 |
| Terrimonas | 0.00084 | 0.00162 | 100 | 160.84 | 183.24 | 3.63 |
| Thermomicrobia bacterium (uncultured) | 0.00774 | 0.01185 | 100 | 99.96 | 80.82 | 2.29 |
| Truepera | 0.00000 | 0.00000 | 100 | 259.32 | 822.33 | 3.47 |
| Tumebacillus | 0.01690 | 0.02332 | 100 | 70.8 | 56.48 | 3.48 |
| Tychonema CCAP 1459 11B | 0.00003 | 0.00011 | 100 | 360.59 | 151.98 | 3.96 |
| Variovorax | 0.00004 | 0.00012 | 100 | 149.74 | 198.67 | 3.54 |
| Yonghaparkia | 0.00000 | 0.00000 | 100 | 911.04 | 239.32 | 3.6 |
| Genus | p-Value | FDR | t0 | CFD_t1 | CRD_t1 | MYC_t1 | PSBA_t1 | PSBA + MYC_t1 | CFD_t2 | CRD_t2 | MYC_t2 | PSBA_t2 | PSBA + MYC_t2 | LDA Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acidibacter | 0.00015 | 0.00072 | 64.28 | 100 | 102.35 | 99.28 | 146.23 | 94.65 | 100 | 131.51 | 87.37 | 78.13 | 72.96 | 3.96 |
| Acidimicrobiia bacterium (uncultured) | 0.00002 | 0.00018 | 185.98 | 100 | 125.83 | 98.29 | 93.56 | 105.3 | 100 | 86.13 | 92.59 | 116.77 | 109.73 | 4.14 |
| Acidobacteria bacterium (uncultured) | 0.02573 | 0.03826 | 102.5 | 100 | 85.82 | 81.35 | 104.36 | 70.05 | 100 | 110.67 | 122.39 | 114.96 | 98.17 | 4.4 |
| Acidobacteriaceae bacterium (uncultured) | 0.00440 | 0.00940 | 104.22 | 100 | 88.24 | 83.46 | 107.07 | 79.43 | 100 | 113.88 | 118.94 | 120.78 | 105.28 | 4.53 |
| Acidobacteriales bacterium (uncultured) | 0.00047 | 0.00167 | 142.86 | 100 | 92.26 | 83.92 | 106.38 | 75.43 | 100 | 122.93 | 135.17 | 151.49 | 119.83 | 4.39 |
| Acidovorax | 0.00000 | 0.00007 | 6.25 | 100 | 88.02 | 197.68 | 65.73 | 42.24 | 100 | 42.03 | 45.17 | 74.37 | 60.64 | 3.38 |
| Acinetobacter | 0.00585 | 0.01175 | 3.33 | 100 | 155.41 | 2.02 | 2.24 | 27.13 | 100 | 73.89 | 126.5 | 40.05 | 45.3 | 4.94 |
| Actinobacterium (uncultured) | 0.00137 | 0.00398 | 135.13 | 100 | 120.88 | 95.57 | 77.62 | 80.01 | 100 | 101.31 | 96.68 | 105.43 | 105.69 | 4.05 |
| Actinomadura | 0.01713 | 0.02837 | 123.26 | 100 | 102.02 | 105.9 | 103.91 | 110.68 | 100 | 105.54 | 88.34 | 77.54 | 105.18 | 2.8 |
| Actinomycetales bacterium (uncultured) | 0.00000 | 0.00005 | 102.79 | 100 | 117.75 | 100.22 | 78.29 | 98.77 | 100 | 81.87 | 86.69 | 112.63 | 139.96 | 3.46 |
| Adhaeribacter | 0.00047 | 0.00167 | 78.24 | 100 | 89.89 | 138.89 | 129.48 | 162.88 | 100 | 104.4 | 106.29 | 85.17 | 104.81 | 3.91 |
| Aeromicrobium | 0.00028 | 0.00112 | 46.35 | 100 | 93.14 | 124.02 | 93.46 | 99 | 100 | 83.69 | 61.63 | 37.77 | 39.58 | 4.61 |
| Allorhizobium Neorhizobium Pararhizobium Rhizobium | 0.00216 | 0.00527 | 59.83 | 100 | 109.03 | 106.05 | 87.13 | 112.88 | 100 | 89.34 | 84.55 | 78.41 | 71.19 | 4.5 |
| Alsobacter | 0.01771 | 0.02897 | 96.45 | 100 | 103.24 | 163.8 | 150.41 | 193.42 | 100 | 122.06 | 113.97 | 114.31 | 116.59 | 3.11 |
| Altererythrobacter | 0.00032 | 0.00122 | 34.52 | 100 | 177.6 | 95.66 | 102.98 | 93.22 | 100 | 96.4 | 69.59 | 69.43 | 65.75 | 3.65 |
| Amaricoccus | 0.03566 | 0.05000 | 93.92 | 100 | 122.93 | 136.45 | 137.8 | 177.26 | 100 | 93.26 | 92.02 | 124.75 | 123.54 | 3.2 |
| Aminobacter | 0.00000 | 0.00000 | 50.65 | 100 | 70.75 | 562.69 | 537.69 | 572.1 | 100 | 125.13 | 176.98 | 180.87 | 165.58 | 3.53 |
| Ammoniphilus | 0.01802 | 0.02930 | 139.35 | 100 | 139.21 | 116.1 | 120.61 | 147.41 | 100 | 111.7 | 75.46 | 106.62 | 70.49 | 3.02 |
| Amycolatopsis | 0.00000 | 0.00000 | 26.98 | 100 | 70.68 | 71.26 | 21.32 | 43.59 | 100 | 30.11 | 42.69 | 36.85 | 65.72 | 3.71 |
| Anaerolinea | 0.02455 | 0.03675 | 293.06 | 100 | 89.34 | 113.08 | 107.76 | 122.25 | 100 | 76.34 | 105.3 | 126.33 | 175 | 3.61 |
| Anaeromyxobacter | 0.00231 | 0.00557 | 175.72 | 100 | 101.11 | 84.21 | 112.93 | 113.7 | 100 | 94.44 | 122.04 | 176.69 | 121.99 | 3.63 |
| Arenimonas | 0.00005 | 0.00036 | 37.74 | 100 | 87.46 | 90.86 | 81.65 | 85.21 | 100 | 72.64 | 40.42 | 42.74 | 40.19 | 3.29 |
| Aridibacter | 0.00931 | 0.01714 | 57.89 | 100 | 77.7 | 97.22 | 92.48 | 111.72 | 100 | 98.61 | 82.76 | 76.95 | 108.75 | 3.35 |
| Armatimonadetes bacterium (uncultured) | 0.00015 | 0.00072 | 123.04 | 100 | 96.46 | 108.5 | 107.56 | 121.76 | 100 | 117.64 | 114.64 | 118.38 | 129.54 | 3.03 |
| Arthrobacter | 0.00662 | 0.01299 | 90.79 | 100 | 81.82 | 118.9 | 108.65 | 140.16 | 100 | 86.38 | 103.85 | 115.99 | 120.2 | 5.28 |
| Azohydromonas | 0.00000 | 0.00001 | 63.89 | 100 | 94.56 | 114.38 | 98.98 | 141.97 | 100 | 100.59 | 83.9 | 75.48 | 59.61 | 3.54 |
| Blastocatella | 0.00834 | 0.01568 | 60.4 | 100 | 85.9 | 105.92 | 85.48 | 99.76 | 100 | 93.05 | 98.06 | 92.82 | 104.74 | 3.5 |
| Blastococcus | 0.00613 | 0.01222 | 92.19 | 100 | 98.54 | 126.45 | 113.14 | 149.79 | 100 | 109.53 | 94.77 | 102.3 | 101.3 | 4.28 |
| Blastopirellula | 0.00173 | 0.00478 | 103.86 | 100 | 95.29 | 87.44 | 116.36 | 82.57 | 100 | 117.14 | 139.83 | 142.18 | 112.43 | 3.05 |
| Bosea | 0.00116 | 0.00347 | 53.96 | 100 | 105.86 | 126.6 | 112.93 | 158.23 | 100 | 107.9 | 94.07 | 105.3 | 67.06 | 3.7 |
| Bradyrhizobium | 0.00046 | 0.00167 | 77.22 | 100 | 119.03 | 91.43 | 129.3 | 95.48 | 100 | 105.98 | 104.95 | 91.1 | 81.84 | 3.95 |
| Burkholderia Caballeronia Paraburkholderia | 0.01243 | 0.02138 | 215.37 | 100 | 130.25 | 117.86 | 99.32 | 92.88 | 100 | 91.69 | 46.01 | 41.7 | 39.6 | 3.54 |
| Caenimonas | 0.00005 | 0.00038 | 62.89 | 100 | 91.3 | 128.72 | 124.39 | 153.75 | 100 | 95.05 | 74.73 | 59.16 | 61.16 | 4.13 |
| Candidatus Udaeobacter | 0.02707 | 0.03985 | 104.76 | 100 | 94.85 | 61.47 | 68.29 | 43.49 | 100 | 115.31 | 115.47 | 107.71 | 112.02 | 4.15 |
| Candidatus Xiphinematobacter | 0.00981 | 0.01769 | 109.47 | 100 | 80.02 | 79.5 | 96.03 | 71.13 | 100 | 119.3 | 132.25 | 126.1 | 123.89 | 4.66 |
| Caulobacter | 0.03307 | 0.04737 | 89.77 | 100 | 270.46 | 84.39 | 74.62 | 91.59 | 100 | 85.27 | 73.37 | 53.73 | 43.18 | 3.31 |
| Cellulomonas | 0.00188 | 0.00502 | 150.21 | 100 | 133.23 | 151.02 | 154.3 | 236.45 | 100 | 98 | 104.7 | 171.07 | 98.77 | 2.98 |
| Chloroflexi bacterium (uncultured) | 0.00000 | 0.00003 | 115.05 | 100 | 108.39 | 114.14 | 101.5 | 111.16 | 100 | 88.41 | 91.41 | 99.54 | 116.77 | 4.32 |
| Chloroflexus sp (uncultured) | 0.00001 | 0.00010 | 98.32 | 100 | 100.73 | 80.85 | 58.3 | 61.4 | 100 | 82.9 | 73.29 | 59.88 | 87.99 | 3.84 |
| Chthoniobacter | 0.00083 | 0.00270 | 64.25 | 100 | 80.09 | 104.91 | 118.85 | 107.96 | 100 | 113.12 | 105.83 | 78.72 | 92.97 | 4.28 |
| Clostridium sensu stricto 1 | 0.01543 | 0.02572 | 179.45 | 100 | 114.66 | 86.31 | 101.28 | 111.43 | 100 | 68.49 | 135.32 | 114.45 | 108.22 | 3.25 |
| Cohnella | 0.00242 | 0.00577 | 155.83 | 100 | 123.32 | 113.88 | 128.62 | 151.32 | 100 | 123.15 | 118.85 | 193.39 | 174.87 | 3.44 |
| Comamonas | 0.00001 | 0.00010 | 50.12 | 100 | 100.22 | 102.02 | 110.78 | 94.76 | 100 | 86.36 | 76.91 | 79.37 | 54.35 | 3.18 |
| Conexibacter | 0.00396 | 0.00882 | 106.13 | 100 | 124.32 | 126.76 | 96.07 | 112.28 | 100 | 88.24 | 81.71 | 106.72 | 99.78 | 2.86 |
| Conexibacter sp (uncultured) | 0.00001 | 0.00009 | 146.13 | 100 | 115.5 | 108.72 | 100.43 | 111.05 | 100 | 88.04 | 98.33 | 123.42 | 133.81 | 2.98 |
| Cupriavidus | 0.00012 | 0.00067 | 69.64 | 100 | 105.61 | 72.77 | 89.26 | 131.98 | 100 | 210.44 | 118.92 | 106.48 | 100.79 | 3.53 |
| Desulfuromonadales bacterium (uncultured) | 0.03380 | 0.04797 | 152.06 | 100 | 90.67 | 92.97 | 146.01 | 116.58 | 100 | 97.45 | 100.78 | 137.87 | 136.11 | 3.17 |
| Devosia | 0.00001 | 0.00011 | 44.2 | 100 | 141.24 | 99.64 | 119.92 | 93.26 | 100 | 92.02 | 72 | 74.17 | 63.06 | 4.34 |
| Domibacillus | 0.01899 | 0.03050 | 110.25 | 100 | 86.97 | 76.72 | 89.38 | 119.61 | 100 | 80.78 | 94.81 | 97.69 | 146.83 | 3.37 |
| Dongia | 0.00020 | 0.00090 | 141.37 | 100 | 76.64 | 68.82 | 77.36 | 62.7 | 100 | 122.9 | 120.9 | 106.37 | 103.87 | 3.83 |
| Dyadobacter | 0.00168 | 0.00469 | 42.99 | 100 | 97.55 | 84.28 | 50.56 | 69.41 | 100 | 34.27 | 82.66 | 36.29 | 33.39 | 3.82 |
| Ensifer | 0.00003 | 0.00029 | 64.7 | 100 | 93.8 | 95.99 | 88.38 | 107.52 | 100 | 104.8 | 113.65 | 95.15 | 97.16 | 3.92 |
| Enterobacter | 0.00412 | 0.00903 | 19.67 | 100 | 13.07 | 13.5 | 7.36 | 21.09 | 100 | 17.04 | 18.98 | 45.41 | 17.07 | 4.28 |
| Ferruginibacter | 0.02223 | 0.03386 | 76.27 | 100 | 89.95 | 116.09 | 115.33 | 120.99 | 100 | 105.87 | 108.12 | 89.95 | 96.91 | 3.33 |
| Fictibacillus | 0.00007 | 0.00049 | 175.43 | 100 | 108.02 | 106.06 | 94.32 | 128.8 | 100 | 200.17 | 99.55 | 166.08 | 168.51 | 3.82 |
| Fimbriiglobus | 0.00217 | 0.00527 | 75.43 | 100 | 93.06 | 83.67 | 108.2 | 84.51 | 100 | 111.23 | 119.29 | 115.99 | 116.19 | 3.8 |
| Flavisolibacter | 0.00142 | 0.00405 | 65.83 | 100 | 86.41 | 119.91 | 107.41 | 117.9 | 100 | 112.68 | 90.04 | 73.77 | 83.81 | 3.84 |
| Flavitalea | 0.00002 | 0.00017 | 55.01 | 100 | 90.17 | 128.66 | 118.24 | 111.52 | 100 | 120.85 | 107.48 | 90.72 | 89.52 | 3.3 |
| Gaiella | 0.00000 | 0.00001 | 146.19 | 100 | 119.03 | 109.68 | 90.61 | 101.66 | 100 | 82.71 | 88.23 | 113.41 | 121.29 | 5.06 |
| Gaiella sp (uncultured) | 0.00000 | 0.00007 | 128.52 | 100 | 112.79 | 124.84 | 84.24 | 117.97 | 100 | 80.62 | 94.01 | 119.96 | 124.39 | 3.99 |
| Gemmata | 0.01289 | 0.02204 | 85.58 | 100 | 91.93 | 74.41 | 90.27 | 71.21 | 100 | 111.85 | 112.53 | 106.58 | 114.1 | 4.41 |
| Gemmatimonadalesbacterium (uncultured) | 0.00264 | 0.00620 | 121.95 | 100 | 86.21 | 62.44 | 82.23 | 50.58 | 100 | 115.15 | 117.31 | 126.92 | 124.24 | 3.46 |
| Gemmatimonadetesbacterium (uncultured) | 0.03426 | 0.04829 | 92.74 | 100 | 85.6 | 107.54 | 132.41 | 83.2 | 100 | 127.76 | 116.41 | 89.34 | 81.33 | 3.47 |
| Gemmatimonas | 0.00012 | 0.00066 | 59.72 | 100 | 80.17 | 105.33 | 111.95 | 92.99 | 100 | 108.86 | 102.49 | 80.2 | 79.67 | 3.52 |
| Haliangium | 0.01926 | 0.03075 | 70.39 | 100 | 106.27 | 125.61 | 113.11 | 70.78 | 100 | 109.07 | 101.3 | 55.29 | 65.65 | 3.66 |
| Herbinix | 0.01033 | 0.01837 | 167.31 | 100 | 116.25 | 90.61 | 98.53 | 126.84 | 100 | 99.52 | 81.36 | 153.03 | 124.08 | 2.88 |
| Herpetosiphon | 0.02142 | 0.03281 | 65.36 | 100 | 111.43 | 141.47 | 91.08 | 123.88 | 100 | 70.37 | 65.14 | 66.03 | 66.66 | 3.88 |
| Hirschia | 0.00001 | 0.00009 | 29 | 100 | 152.42 | 118.62 | 197.99 | 121.26 | 100 | 329.77 | 113.58 | 56.22 | 48.35 | 3.82 |
| Hyphomicrobium | 0.01002 | 0.01794 | 136.67 | 100 | 97.96 | 91.2 | 92.28 | 80.52 | 100 | 90.97 | 110.93 | 125 | 112.89 | 3.04 |
| Iamia | 0.03915 | 0.05460 | 104.92 | 100 | 112.06 | 99.18 | 80.1 | 75.9 | 100 | 78.03 | 65.87 | 67.28 | 69.49 | 3.64 |
| Iamia sp (uncultured) | 0.00142 | 0.00405 | 61.87 | 100 | 92.84 | 126.04 | 89.69 | 95.97 | 100 | 56.36 | 56.27 | 48.11 | 49.61 | 2.93 |
| Ideonella | 0.00401 | 0.00884 | 28.02 | 100 | 257.59 | 110.04 | 86.98 | 118.47 | 100 | 121.39 | 118.48 | 57.88 | 69.26 | 3.16 |
| Ilumatobacter | 0.04702 | 0.06357 | 127.33 | 100 | 126.77 | 89.35 | 70.95 | 77.02 | 100 | 66.93 | 78.85 | 91.13 | 104.93 | 3.47 |
| Knoellia | 0.00217 | 0.00527 | 110.71 | 100 | 96.72 | 106.93 | 100.44 | 122.31 | 100 | 87.25 | 86.16 | 116.69 | 99.55 | 4.4 |
| Kribbella | 0.00081 | 0.00269 | 140.63 | 100 | 117.72 | 99.66 | 92.58 | 100.19 | 100 | 83.53 | 88.55 | 94.88 | 99.85 | 3.05 |
| Lacibacter | 0.01180 | 0.02049 | 40.81 | 100 | 81.71 | 102.96 | 75.82 | 76.46 | 100 | 72 | 67.42 | 46.08 | 46.42 | 3.24 |
| Lechevalieria | 0.00000 | 0.00004 | 57.7 | 100 | 109.76 | 69.28 | 54.9 | 65.47 | 100 | 63.66 | 53.94 | 57.93 | 58.79 | 3.98 |
| Leptolyngbya EcFYyyy 00 | 0.00001 | 0.00015 | 4.32 | 100 | 221.39 | 405.52 | 286.83 | 324.22 | 100 | 134.08 | 287.92 | 83.54 | 75.95 | 4.59 |
| Litorilinea | 0.04367 | 0.05996 | 82.54 | 100 | 104.47 | 112.74 | 125.11 | 137.71 | 100 | 90.84 | 98.23 | 86.34 | 110.44 | 3.4 |
| Luedemannella | 0.00007 | 0.00049 | 250.08 | 100 | 149.9 | 117.91 | 128.24 | 151.38 | 100 | 79.31 | 100.38 | 155.36 | 115.77 | 3.45 |
| Luteitalea | 0.00522 | 0.01073 | 84.35 | 100 | 105.21 | 118.79 | 130.39 | 141.67 | 100 | 109.31 | 106.19 | 146.43 | 98.18 | 3.42 |
| Luteolibacter | 0.00005 | 0.00037 | 52.37 | 100 | 149.08 | 106.88 | 110.2 | 99.88 | 100 | 81.7 | 77.34 | 58.17 | 57.21 | 3.71 |
| Lutispora sp (uncultured) | 0.00432 | 0.00939 | 306.56 | 100 | 96.01 | 128.46 | 125.08 | 124.57 | 100 | 84.12 | 126.3 | 180.79 | 162.91 | 3.64 |
| Lysobacter | 0.00455 | 0.00958 | 73.86 | 100 | 95.08 | 118.86 | 87.41 | 111.52 | 100 | 90.53 | 69.93 | 72.72 | 68.63 | 3.91 |
| Marmoricola | 0.00252 | 0.00596 | 124.47 | 100 | 92.53 | 103.21 | 90.9 | 106.99 | 100 | 109.25 | 87.03 | 91.23 | 83.24 | 4.19 |
| Massilia | 0.03106 | 0.04498 | 95.51 | 100 | 95.71 | 134.28 | 120.78 | 205.38 | 100 | 105.99 | 90.04 | 76.71 | 98.66 | 4.54 |
| Mesorhizobium | 0.00012 | 0.00066 | 54.3 | 100 | 153.25 | 113.28 | 148.25 | 115.46 | 100 | 133.25 | 96.3 | 94.78 | 85.39 | 4.18 |
| Methylobacillus | 0.00000 | 0.00003 | 5.72 | 100 | 110.77 | 111.84 | 51.47 | 110.47 | 100 | 29.39 | 48.75 | 29.87 | 45.88 | 3.5 |
| Methylorosula | 0.00028 | 0.00112 | 74.43 | 100 | 97.5 | 136.08 | 130.3 | 146.09 | 100 | 113.88 | 93.94 | 86.89 | 104.21 | 2.77 |
| Methylotenera | 0.00084 | 0.00272 | 25.65 | 100 | 105.02 | 103.85 | 85.19 | 113.75 | 100 | 33.21 | 46.06 | 29.81 | 34.99 | 3.65 |
| Microbacterium | 0.00113 | 0.00340 | 47.81 | 100 | 86.21 | 86.86 | 58.65 | 81.77 | 100 | 61.78 | 53.07 | 91.35 | 56.44 | 4.02 |
| Microvirga | 0.01117 | 0.01974 | 88.4 | 100 | 87.16 | 131.72 | 117.23 | 153.13 | 100 | 128.75 | 100.41 | 89.88 | 100.73 | 4.68 |
| Mitsuaria | 0.00000 | 0.00000 | 4.05 | 100 | 186.27 | 65.24 | 15.34 | 20.94 | 100 | 22.02 | 122.11 | 39.36 | 45.24 | 4.18 |
| Mycobacterium | 0.00003 | 0.00029 | 129.13 | 100 | 112.86 | 108.69 | 99.46 | 99.32 | 100 | 111.11 | 86.57 | 80.64 | 94.55 | 3.82 |
| Niastella | 0.00016 | 0.00078 | 42.54 | 100 | 122.65 | 99.72 | 100.77 | 100.19 | 100 | 84.83 | 103.93 | 84.33 | 76.76 | 3.08 |
| Nitrospira | 0.00452 | 0.00958 | 73.35 | 100 | 94.34 | 102.74 | 125.14 | 95.65 | 100 | 126.41 | 131.36 | 132.93 | 130.44 | 4.33 |
| Nocardioides | 0.00032 | 0.00122 | 116.55 | 100 | 104.1 | 103.25 | 81.1 | 96.92 | 100 | 74.47 | 77.12 | 105.21 | 88.06 | 4.35 |
| Nodosilinea PCC 7104 | 0.00014 | 0.00072 | 4.44 | 100 | 365.77 | 164.16 | 285.09 | 537.94 | 100 | 62.96 | 108.1 | 61.08 | 85.74 | 3.94 |
| Nonomuraea | 0.02585 | 0.03826 | 164.23 | 100 | 135.86 | 128.69 | 121.44 | 149.45 | 100 | 83.14 | 112.95 | 136.41 | 120.34 | 3 |
| Nordella | 0.01871 | 0.03023 | 76.83 | 100 | 86.64 | 76.27 | 87.63 | 63.77 | 100 | 118.51 | 115.66 | 104.41 | 94.03 | 3.91 |
| Ohtaekwangia | 0.00106 | 0.00325 | 40.94 | 100 | 138.97 | 101.56 | 96.31 | 75.28 | 100 | 98.56 | 68.21 | 56.24 | 56.63 | 3.58 |
| Oligoflexus | 0.01147 | 0.02013 | 75.43 | 100 | 114.74 | 196.29 | 147.47 | 246.44 | 100 | 104.58 | 101.28 | 100.08 | 89.26 | 3.21 |
| Paenarthrobacter | 0.03072 | 0.04473 | 77.13 | 100 | 59 | 79.34 | 67.36 | 96.77 | 100 | 49.03 | 74 | 84.53 | 80.38 | 3.83 |
| Paenisporosarcina | 0.00772 | 0.01461 | 118.53 | 100 | 91.56 | 89.1 | 90.2 | 121.1 | 100 | 150.84 | 108.43 | 164.75 | 159.11 | 3.51 |
| Parasegetibacter | 0.00019 | 0.00085 | 48.8 | 100 | 106.53 | 143.79 | 108.75 | 176.22 | 100 | 88.84 | 104.44 | 88.17 | 81.87 | 3.45 |
| Parviterribacter | 0.01759 | 0.02895 | 114.51 | 100 | 108.64 | 114.32 | 95.35 | 119.53 | 100 | 103.39 | 84.25 | 100.1 | 82.02 | 3.03 |
| Paucibacter | 0.01434 | 0.02420 | 35.11 | 100 | 73.34 | 93.1 | 86.8 | 90.59 | 100 | 103.2 | 85.01 | 78.87 | 82.57 | 2.8 |
| Pedobacter | 0.00022 | 0.00093 | 60.67 | 100 | 87 | 121.29 | 111.08 | 157.41 | 100 | 98.06 | 91.41 | 94.85 | 118 | 3.81 |
| Pedococcus Phycicoccus | 0.00877 | 0.01637 | 117.36 | 100 | 104.5 | 101.22 | 71.6 | 82.76 | 100 | 67.41 | 65.76 | 114.7 | 94.73 | 2.75 |
| Pedomicrobium | 0.00069 | 0.00233 | 139.37 | 100 | 104.16 | 92.52 | 115.34 | 84.09 | 100 | 116.58 | 120.13 | 124.33 | 103.38 | 3.79 |
| Pelobacter sp (uncultured) | 0.01183 | 0.02049 | 81.57 | 100 | 83.24 | 99.92 | 87.49 | 48.4 | 100 | 152.75 | 158.34 | 87.91 | 71.97 | 2.92 |
| Phenylobacterium | 0.00460 | 0.00960 | 111.74 | 100 | 117.36 | 114.19 | 104.22 | 128.67 | 100 | 94.3 | 88.33 | 102.38 | 100.64 | 2.75 |
| Pirellula | 0.01454 | 0.02438 | 76.73 | 100 | 84.79 | 73.88 | 79.74 | 58.73 | 100 | 106.6 | 103.1 | 88.43 | 96.47 | 4.56 |
| Planctomicrobium | 0.00157 | 0.00442 | 53 | 100 | 82.99 | 69.53 | 75.57 | 46.5 | 100 | 81.54 | 74.74 | 57.08 | 75.76 | 2.97 |
| Planctomycetales bacterium (uncultured) | 0.00024 | 0.00099 | 52.01 | 100 | 86.92 | 90.13 | 109.59 | 63.5 | 100 | 104.4 | 86.69 | 67.59 | 90.35 | 2.96 |
| Planctomycete (uncultured) | 0.00003 | 0.00029 | 50.19 | 100 | 75.22 | 94.88 | 106.91 | 83.75 | 100 | 110.02 | 86.07 | 59.5 | 71.82 | 4.33 |
| Polaromonas | 0.00930 | 0.01714 | 55.72 | 100 | 93.34 | 74.33 | 101.92 | 80.41 | 100 | 92.42 | 100.91 | 91.08 | 78.08 | 2.77 |
| Polyangium brachysporum group | 0.00000 | 0.00002 | 37.24 | 100 | 86.63 | 121.47 | 76.64 | 82.31 | 100 | 114.09 | 52.41 | 57.47 | 63.86 | 3.66 |
| Pontibacter | 0.00940 | 0.01714 | 57.75 | 100 | 85.41 | 136.31 | 102.16 | 190.91 | 100 | 84.65 | 85.7 | 79.28 | 91.33 | 3.95 |
| Promicromonospora | 0.02417 | 0.03640 | 190.96 | 100 | 158.51 | 75.41 | 101.83 | 118.54 | 100 | 114.24 | 65.95 | 56.91 | 64.89 | 3.26 |
| Pseudarthrobacter | 0.00537 | 0.01086 | 84.97 | 100 | 77.45 | 138.88 | 100.57 | 146 | 100 | 69.17 | 89.54 | 103.35 | 95.64 | 3.1 |
| Pseudoduganella | 0.00011 | 0.00066 | 14.86 | 100 | 87.11 | 169.15 | 61.09 | 46.1 | 100 | 80.33 | 92.38 | 49.47 | 54.03 | 3.55 |
| Pseudoflavitalea | 0.00212 | 0.00527 | 63.89 | 100 | 109.19 | 79.98 | 183.11 | 73.52 | 100 | 315.91 | 176.06 | 114.63 | 117.23 | 2.97 |
| Pseudolabrys | 0.00509 | 0.01053 | 82.32 | 100 | 153.37 | 74.21 | 119.47 | 70.11 | 100 | 107.49 | 88.24 | 85.94 | 89.34 | 3.87 |
| Pseudomuriella schumacherensis | 0.00011 | 0.00066 | 9.04 | 100 | 92.62 | 107.42 | 123.27 | 137.2 | 100 | 162.94 | 194.47 | 120.17 | 126.2 | 3.48 |
| Pseudorhodoplanes | 0.00213 | 0.00527 | 55.69 | 100 | 112.11 | 103.87 | 145.19 | 94.01 | 100 | 149.76 | 130.24 | 114.67 | 105.78 | 3.02 |
| Pseudoxanthomonas | 0.00011 | 0.00066 | 39.43 | 100 | 179.05 | 92.26 | 68.45 | 79.34 | 100 | 72.05 | 68.2 | 52.08 | 38.59 | 3.99 |
| Qipengyuania | 0.00000 | 0.00004 | 33.59 | 100 | 86.64 | 113.9 | 64.24 | 93.25 | 100 | 64.59 | 66.28 | 76.4 | 74.62 | 3.83 |
| Ramlibacter | 0.00001 | 0.00009 | 55.16 | 100 | 88.81 | 130.57 | 121.36 | 152.85 | 100 | 102.93 | 82.29 | 53.2 | 58.2 | 4.08 |
| Reyranella | 0.00440 | 0.00940 | 78.84 | 100 | 108.23 | 89.66 | 118.3 | 76.81 | 100 | 119.41 | 121.64 | 85.51 | 89.13 | 3.35 |
| Rhizobacter | 0.00005 | 0.00038 | 57.28 | 100 | 112.86 | 111.25 | 109.72 | 125.42 | 100 | 89.86 | 80.62 | 73.57 | 67.43 | 3.96 |
| Rhodobacter | 0.00005 | 0.00036 | 45.12 | 100 | 154.54 | 188.95 | 178.91 | 210.64 | 100 | 60.03 | 66.67 | 65.67 | 53.72 | 3.52 |
| Rhodocytophaga | 0.02868 | 0.04199 | 52.13 | 100 | 87.56 | 109.5 | 97.61 | 132 | 100 | 97.81 | 121.12 | 110.42 | 114.44 | 3.21 |
| Rhodopirellula | 0.00004 | 0.00032 | 34.94 | 100 | 84.11 | 110.22 | 90.01 | 122.8 | 100 | 113.5 | 77.79 | 63.64 | 72.1 | 3.71 |
| Rhodoplanes | 0.04025 | 0.05585 | 80.4 | 100 | 93.11 | 72.59 | 87.46 | 58.75 | 100 | 103.24 | 116.88 | 100.44 | 93.22 | 3.69 |
| Roseimicrobium | 0.04645 | 0.06313 | 78.41 | 100 | 81.16 | 91.17 | 102.83 | 62.01 | 100 | 93.57 | 96.1 | 72.17 | 77.43 | 3.29 |
| Roseomonas | 0.00288 | 0.00658 | 65.46 | 100 | 101.84 | 141.54 | 139.67 | 208.01 | 100 | 110.7 | 95.93 | 90.18 | 80.99 | 3.18 |
| Rubellimicrobium | 0.00097 | 0.00307 | 55.45 | 100 | 85.14 | 144.84 | 128.94 | 194.48 | 100 | 88.45 | 86.67 | 88.12 | 78.28 | 4.16 |
| Rubrobacter | 0.00944 | 0.01714 | 107.74 | 100 | 106.81 | 123.91 | 112.37 | 132.26 | 100 | 109.31 | 107.23 | 107.84 | 123.59 | 4.52 |
| Rubrobacterales bacterium (uncultured) | 0.00000 | 0.00000 | 142.64 | 100 | 113.72 | 93.52 | 76.37 | 79.43 | 100 | 82.64 | 81.7 | 101.47 | 127.99 | 4.44 |
| Rubrobacteria bacterium (uncultured) | 0.00000 | 0.00002 | 133.1 | 100 | 123.22 | 104.38 | 81.68 | 98.33 | 100 | 82.09 | 88.35 | 110.12 | 125.76 | 3.93 |
| Saccharothrix | 0.03157 | 0.04547 | 56.66 | 100 | 74.64 | 78.66 | 78.51 | 113.29 | 100 | 66.31 | 70.17 | 60.58 | 81.93 | 3.17 |
| Shimazuella | 0.02083 | 0.03229 | 176.45 | 100 | 129.24 | 145.57 | 130.79 | 177.02 | 100 | 73.85 | 112.1 | 123.5 | 95.85 | 2.79 |
| Shinella | 0.00053 | 0.00185 | 17.43 | 100 | 133.61 | 80.34 | 54.36 | 78.55 | 100 | 56.69 | 77.75 | 62.36 | 46.07 | 3.6 |
| Skermanella | 0.03385 | 0.04797 | 106.51 | 100 | 109.91 | 136.22 | 161.04 | 188.5 | 100 | 98.68 | 101.87 | 121.24 | 112.65 | 4.41 |
| Solirubrobacter | 0.00005 | 0.00036 | 134.79 | 100 | 114.29 | 109.03 | 98.61 | 106.4 | 100 | 93.78 | 93.35 | 123.24 | 111.6 | 3.97 |
| Sorangium | 0.00215 | 0.00527 | 213.85 | 100 | 247.96 | 120.74 | 117.95 | 88.89 | 100 | 127.31 | 54.17 | 61.89 | 39.88 | 3.08 |
| Sphingoaurantiacus | 0.00024 | 0.00099 | 72.87 | 100 | 92.72 | 137.67 | 114.16 | 184.98 | 100 | 93.12 | 71.07 | 91.75 | 96.31 | 3.22 |
| Sphingobium | 0.00001 | 0.00010 | 6.51 | 100 | 117.69 | 148.59 | 78.58 | 126.93 | 100 | 50.67 | 51.95 | 47.45 | 40.18 | 4.7 |
| Sphingomonas | 0.00025 | 0.00103 | 88.32 | 100 | 102.32 | 113.86 | 84.73 | 109.96 | 100 | 97.32 | 78.45 | 85.9 | 95.57 | 4.65 |
| Sphingopyxis | 0.00012 | 0.00066 | 29.7 | 100 | 156.98 | 102.35 | 79.6 | 116.39 | 100 | 34.5 | 46.1 | 51.69 | 64.04 | 3.34 |
| Steroidobacter | 0.00032 | 0.00122 | 128.4 | 100 | 114.51 | 102.9 | 132.42 | 109.09 | 100 | 118.38 | 118.98 | 132.53 | 102.27 | 3.9 |
| Streptomyces | 0.00054 | 0.00185 | 69.59 | 100 | 102.58 | 75.42 | 79.73 | 81.61 | 100 | 87.19 | 71.23 | 81.07 | 81.15 | 4.2 |
| Sumerlaea | 0.00106 | 0.00325 | 49.38 | 100 | 76.33 | 105.62 | 104.13 | 102.95 | 100 | 130.93 | 122.19 | 109.87 | 85.69 | 3.09 |
| Synechococcus IR11 | 0.00187 | 0.00502 | 4.86 | 100 | 426.01 | 340.41 | 446.08 | 642.72 | 100 | 295.95 | 105.58 | 96.44 | 140.27 | 3.55 |
| Tahibacter | 0.00672 | 0.01300 | 30.74 | 100 | 76.94 | 61.78 | 46.78 | 52.77 | 100 | 112.37 | 52.53 | 41.82 | 53.16 | 3.09 |
| Tepidisphaera | 0.00308 | 0.00697 | 51.92 | 100 | 68.48 | 115.57 | 117.08 | 97.03 | 100 | 108.41 | 102.77 | 54.86 | 64.63 | 3.19 |
| Terrimonas | 0.00310 | 0.00697 | 57.62 | 100 | 93.9 | 92.63 | 102.48 | 76.39 | 100 | 123.05 | 103.4 | 72.42 | 70.95 | 3.85 |
| Thermomicrobia bacterium (uncultured) | 0.04136 | 0.05709 | 108.08 | 100 | 113.5 | 93.8 | 105.86 | 123.29 | 100 | 77.72 | 78.57 | 81.08 | 105.2 | 2.63 |
| Truepera | 0.00000 | 0.00002 | 40.65 | 100 | 68.9 | 103.99 | 123.09 | 128.05 | 100 | 139.03 | 119.11 | 138.52 | 88.47 | 3.55 |
| Tumebacillus | 0.00190 | 0.00502 | 205.98 | 100 | 195.02 | 112.13 | 135.76 | 178.05 | 100 | 79.54 | 126.8 | 284.58 | 190.6 | 3.71 |
| Tychonema CCAP 1459 11B | 0.00668 | 0.01300 | 27.04 | 100 | 90.7 | 64.94 | 118.9 | 107.72 | 100 | 66.79 | 176.66 | 122.13 | 107.17 | 4.09 |
| Variovorax | 0.00010 | 0.00062 | 60.83 | 100 | 113.44 | 76.83 | 79.79 | 84.4 | 100 | 68.31 | 73.19 | 61.59 | 44.08 | 3.81 |
| Yonghaparkia | 0.00000 | 0.00002 | 11.37 | 100 | 96.24 | 129.17 | 65.29 | 127.42 | 100 | 63.24 | 68.06 | 83.94 | 91.99 | 3.71 |
| Genus | p-Value | FDR | t0 | CFD_t1 | CRD_t1 | MYC_t1 | PSBA_t1 | PSBA + MYC_t1 | CFD_t2 | CRD_t2 | MYC_t2 | PSBA_t2 | PSBA + MYC_t2 | LDA Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acidibacter | 0.00015 | 0.00072 | 62.8 | 97.71 | 100 | 97.01 | 142.88 | 92.48 | 76.04 | 100 | 66.44 | 59.42 | 55.48 | 3.96 |
| Acidimicrobiia bacterium (uncultured) | 0.00002 | 0.00018 | 147.81 | 79.47 | 100 | 78.12 | 74.35 | 83.68 | 116.1 | 100 | 107.5 | 135.58 | 127.4 | 4.14 |
| Acidobacteria bacterium (uncultured) | 0.02573 | 0.03826 | 119.44 | 116.53 | 100 | 94.79 | 121.61 | 81.63 | 90.36 | 100 | 110.6 | 103.88 | 88.71 | 4.4 |
| Acidobacteriaceae bacterium (uncultured) | 0.00440 | 0.00940 | 118.11 | 113.33 | 100 | 94.58 | 121.34 | 90.02 | 87.81 | 100 | 104.44 | 106.06 | 92.45 | 4.53 |
| Acidobacteriales bacterium (uncultured) | 0.00047 | 0.00167 | 154.84 | 108.39 | 100 | 90.96 | 115.3 | 81.76 | 81.35 | 100 | 109.96 | 123.24 | 97.48 | 4.39 |
| Acidovorax | 0.00000 | 0.00007 | 7.1 | 113.61 | 100 | 224.58 | 74.67 | 47.99 | 237.91 | 100 | 107.47 | 176.94 | 144.27 | 3.38 |
| Acinetobacter | 0.00585 | 0.01175 | 2.14 | 64.35 | 100 | 1.3 | 1.44 | 17.46 | 135.34 | 100 | 171.19 | 54.2 | 61.31 | 4.94 |
| Actinobacterium (uncultured) | 0.00137 | 0.00398 | 111.79 | 82.73 | 100 | 79.06 | 64.21 | 66.19 | 98.71 | 100 | 95.42 | 104.07 | 104.32 | 4.05 |
| Actinomadura | 0.01713 | 0.02837 | 120.82 | 98.02 | 100 | 103.8 | 101.85 | 108.49 | 94.75 | 100 | 83.71 | 73.47 | 99.66 | 2.8 |
| Actinomycetales bacterium (uncultured) | 0.00000 | 0.00005 | 87.3 | 84.93 | 100 | 85.12 | 66.49 | 83.88 | 122.14 | 100 | 105.88 | 137.57 | 170.94 | 3.46 |
| Adhaeribacter | 0.00047 | 0.00167 | 87.04 | 111.25 | 100 | 154.51 | 144.04 | 181.2 | 95.78 | 100 | 101.81 | 81.57 | 100.39 | 3.91 |
| Aeromicrobium | 0.00028 | 0.00112 | 49.77 | 107.37 | 100 | 133.16 | 100.34 | 106.29 | 119.49 | 100 | 73.64 | 45.13 | 47.3 | 4.61 |
| Allorhizobium Neorhizobium Pararhizobium Rhizobium | 0.00216 | 0.00527 | 54.88 | 91.72 | 100 | 97.27 | 79.92 | 103.54 | 111.93 | 100 | 94.64 | 87.77 | 79.68 | 4.5 |
| Alsobacter | 0.01771 | 0.02897 | 93.43 | 96.86 | 100 | 158.66 | 145.69 | 187.36 | 81.93 | 100 | 93.37 | 93.65 | 95.52 | 3.11 |
| Altererythrobacter | 0.00032 | 0.00122 | 19.44 | 56.31 | 100 | 53.86 | 57.98 | 52.49 | 103.73 | 100 | 72.18 | 72.02 | 68.2 | 3.65 |
| Amaricoccus | 0.03566 | 0.05000 | 76.4 | 81.34 | 100 | 110.99 | 112.09 | 144.19 | 107.23 | 100 | 98.67 | 133.77 | 132.47 | 3.2 |
| Aminobacter | 0.00000 | 0.00000 | 71.59 | 141.35 | 100 | 795.33 | 760.01 | 808.64 | 79.92 | 100 | 141.44 | 144.55 | 132.33 | 3.53 |
| Ammoniphilus | 0.01802 | 0.02930 | 100.1 | 71.83 | 100 | 83.4 | 86.64 | 105.89 | 89.53 | 100 | 67.56 | 95.45 | 63.11 | 3.02 |
| Amycolatopsis | 0.00000 | 0.00000 | 38.17 | 141.49 | 100 | 100.82 | 30.17 | 61.68 | 332.06 | 100 | 141.75 | 122.35 | 218.25 | 3.71 |
| Anaerolinea | 0.02455 | 0.03675 | 328.02 | 111.93 | 100 | 126.57 | 120.62 | 136.83 | 130.99 | 100 | 137.94 | 165.48 | 229.24 | 3.61 |
| Anaeromyxobacter | 0.00231 | 0.00557 | 173.8 | 98.91 | 100 | 83.29 | 111.7 | 112.45 | 105.88 | 100 | 129.22 | 187.09 | 129.17 | 3.63 |
| Arenimonas | 0.00005 | 0.00036 | 43.15 | 114.34 | 100 | 103.88 | 93.36 | 97.43 | 137.67 | 100 | 55.64 | 58.83 | 55.32 | 3.29 |
| Aridibacter | 0.00931 | 0.01714 | 74.49 | 128.69 | 100 | 125.11 | 119.01 | 143.77 | 101.41 | 100 | 83.93 | 78.04 | 110.28 | 3.35 |
| Armatimonadetes bacterium (uncultured) | 0.00015 | 0.00072 | 127.55 | 103.67 | 100 | 112.49 | 111.51 | 126.23 | 85.01 | 100 | 97.45 | 100.63 | 110.12 | 3.03 |
| Arthrobacter | 0.00662 | 0.01299 | 110.97 | 122.23 | 100 | 145.32 | 132.79 | 171.31 | 115.77 | 100 | 120.23 | 134.28 | 139.16 | 5.28 |
| Azohydromonas | 0.00000 | 0.00001 | 67.56 | 105.75 | 100 | 120.95 | 104.68 | 150.13 | 99.41 | 100 | 83.41 | 75.04 | 59.26 | 3.54 |
| Blastocatella | 0.00834 | 0.01568 | 70.31 | 116.42 | 100 | 123.31 | 99.51 | 116.14 | 107.47 | 100 | 105.39 | 99.76 | 112.57 | 3.5 |
| Blastococcus | 0.00613 | 0.01222 | 93.55 | 101.48 | 100 | 128.33 | 114.82 | 152.01 | 91.3 | 100 | 86.52 | 93.39 | 92.48 | 4.28 |
| Blastopirellula | 0.00173 | 0.00478 | 108.99 | 104.94 | 100 | 91.76 | 122.11 | 86.64 | 85.37 | 100 | 119.37 | 121.37 | 95.98 | 3.05 |
| Bosea | 0.00116 | 0.00347 | 50.97 | 94.47 | 100 | 119.6 | 106.69 | 149.48 | 92.68 | 100 | 87.18 | 97.58 | 62.15 | 3.7 |
| Bradyrhizobium | 0.00046 | 0.00167 | 64.87 | 84.01 | 100 | 76.81 | 108.63 | 80.21 | 94.36 | 100 | 99.03 | 85.96 | 77.23 | 3.95 |
| Burkholderia Caballeronia Paraburkholderia | 0.01243 | 0.02138 | 165.35 | 76.78 | 100 | 90.49 | 76.26 | 71.31 | 109.07 | 100 | 50.18 | 45.48 | 43.19 | 3.54 |
| Caenimonas | 0.00005 | 0.00038 | 68.88 | 109.53 | 100 | 140.98 | 136.24 | 168.41 | 105.2 | 100 | 78.61 | 62.23 | 64.34 | 4.13 |
| Candidatus Udaeobacter | 0.02707 | 0.03985 | 110.45 | 105.43 | 100 | 64.8 | 72 | 45.85 | 86.72 | 100 | 100.13 | 93.4 | 97.14 | 4.15 |
| Candidatus Xiphinematobacter | 0.00981 | 0.01769 | 136.81 | 124.97 | 100 | 99.35 | 120 | 88.89 | 83.82 | 100 | 110.85 | 105.7 | 103.84 | 4.66 |
| Caulobacter | 0.03307 | 0.04737 | 33.19 | 36.97 | 100 | 31.2 | 27.59 | 33.86 | 117.27 | 100 | 86.05 | 63.01 | 50.64 | 3.31 |
| Cellulomonas | 0.00188 | 0.00502 | 112.75 | 75.06 | 100 | 113.35 | 115.82 | 177.48 | 102.04 | 100 | 106.84 | 174.56 | 100.79 | 2.98 |
| Chloroflexi bacterium (uncultured) | 0.00000 | 0.00003 | 106.14 | 92.26 | 100 | 105.3 | 93.64 | 102.55 | 113.11 | 100 | 103.39 | 112.59 | 132.08 | 4.32 |
| Chloroflexus sp (uncultured) | 0.00001 | 0.00010 | 97.61 | 99.27 | 100 | 80.27 | 57.87 | 60.95 | 120.63 | 100 | 88.41 | 72.24 | 106.15 | 3.84 |
| Chthoniobacter | 0.00083 | 0.00270 | 80.22 | 124.86 | 100 | 130.99 | 148.39 | 134.8 | 88.4 | 100 | 93.55 | 69.58 | 82.19 | 4.28 |
| Clostridium sensu stricto 1 | 0.01543 | 0.02572 | 156.51 | 87.22 | 100 | 75.28 | 88.33 | 97.18 | 146 | 100 | 197.57 | 167.09 | 157.99 | 3.25 |
| Cohnella | 0.00242 | 0.00577 | 126.36 | 81.09 | 100 | 92.34 | 104.3 | 122.71 | 81.2 | 100 | 96.51 | 157.04 | 141.99 | 3.44 |
| Comamonas | 0.00001 | 0.00010 | 50.01 | 99.78 | 100 | 101.8 | 110.54 | 94.55 | 115.8 | 100 | 89.06 | 91.9 | 62.94 | 3.18 |
| Conexibacter | 0.00396 | 0.00882 | 85.37 | 80.44 | 100 | 101.96 | 77.27 | 90.32 | 113.33 | 100 | 92.6 | 120.94 | 113.08 | 2.86 |
| Conexibacter sp (uncultured) | 0.00001 | 0.00009 | 126.52 | 86.58 | 100 | 94.13 | 86.95 | 96.15 | 113.58 | 100 | 111.69 | 140.18 | 151.98 | 2.98 |
| Cupriavidus | 0.00012 | 0.00067 | 65.94 | 94.69 | 100 | 68.9 | 84.52 | 124.97 | 47.52 | 100 | 56.51 | 50.6 | 47.89 | 3.53 |
| Desulfuromonadales bacterium (uncultured) | 0.03380 | 0.04797 | 167.71 | 110.29 | 100 | 102.54 | 161.05 | 128.58 | 102.61 | 100 | 103.42 | 141.48 | 139.67 | 3.17 |
| Devosia | 0.00001 | 0.00011 | 31.3 | 70.8 | 100 | 70.55 | 84.91 | 66.03 | 108.67 | 100 | 78.25 | 80.6 | 68.53 | 4.34 |
| Domibacillus | 0.01899 | 0.03050 | 126.77 | 114.98 | 100 | 88.22 | 102.77 | 137.53 | 123.8 | 100 | 117.38 | 120.94 | 181.78 | 3.37 |
| Dongia | 0.00020 | 0.00090 | 184.46 | 130.49 | 100 | 89.8 | 100.94 | 81.81 | 81.36 | 100 | 98.37 | 86.55 | 84.51 | 3.83 |
| Dyadobacter | 0.00168 | 0.00469 | 44.07 | 102.51 | 100 | 86.4 | 51.83 | 71.15 | 291.76 | 100 | 241.18 | 105.88 | 97.42 | 3.82 |
| Ensifer | 0.00003 | 0.00029 | 68.97 | 106.61 | 100 | 102.34 | 94.22 | 114.62 | 95.42 | 100 | 108.45 | 90.79 | 92.72 | 3.92 |
| Enterobacter | 0.00412 | 0.00903 | 150.52 | 765.15 | 100 | 103.27 | 56.3 | 161.41 | 587.02 | 100 | 111.41 | 266.57 | 100.22 | 4.28 |
| Ferruginibacter | 0.02223 | 0.03386 | 84.79 | 111.17 | 100 | 129.05 | 128.21 | 134.5 | 94.45 | 100 | 102.12 | 84.96 | 91.53 | 3.33 |
| Fictibacillus | 0.00007 | 0.00049 | 162.4 | 92.57 | 100 | 98.18 | 87.32 | 119.23 | 49.96 | 100 | 49.73 | 82.97 | 84.19 | 3.82 |
| Fimbriiglobus | 0.00217 | 0.00527 | 81.06 | 107.46 | 100 | 89.92 | 116.28 | 90.81 | 89.9 | 100 | 107.24 | 104.28 | 104.46 | 3.8 |
| Flavisolibacter | 0.00142 | 0.00405 | 76.18 | 115.73 | 100 | 138.77 | 124.31 | 136.44 | 88.75 | 100 | 79.91 | 65.47 | 74.38 | 3.84 |
| Flavitalea | 0.00002 | 0.00017 | 61.01 | 110.9 | 100 | 142.68 | 131.13 | 123.68 | 82.75 | 100 | 88.94 | 75.07 | 74.07 | 3.3 |
| Gaiella | 0.00000 | 0.00001 | 122.81 | 84.01 | 100 | 92.14 | 76.12 | 85.4 | 120.9 | 100 | 106.67 | 137.11 | 146.64 | 5.06 |
| Gaiella sp (uncultured) | 0.00000 | 0.00007 | 113.95 | 88.66 | 100 | 110.68 | 74.69 | 104.59 | 124.04 | 100 | 116.6 | 148.8 | 154.29 | 3.99 |
| Gemmata | 0.01289 | 0.02204 | 93.09 | 108.77 | 100 | 80.94 | 98.19 | 77.46 | 89.41 | 100 | 100.61 | 95.29 | 102.02 | 4.41 |
| Gemmatimonadalesbacterium (uncultured) | 0.00264 | 0.00620 | 141.46 | 115.99 | 100 | 72.43 | 95.38 | 58.67 | 86.84 | 100 | 101.88 | 110.22 | 107.89 | 3.46 |
| Gemmatimonadetesbacterium (uncultured) | 0.03426 | 0.04829 | 108.35 | 116.83 | 100 | 125.64 | 154.69 | 97.2 | 78.27 | 100 | 91.11 | 69.93 | 63.66 | 3.47 |
| Gemmatimonas | 0.00012 | 0.00066 | 74.49 | 124.74 | 100 | 131.39 | 139.65 | 115.99 | 91.86 | 100 | 94.15 | 73.67 | 73.19 | 3.52 |
| Haliangium | 0.01926 | 0.03075 | 66.24 | 94.1 | 100 | 118.21 | 106.44 | 66.61 | 91.69 | 100 | 92.88 | 50.7 | 60.19 | 3.66 |
| Herbinix | 0.01033 | 0.01837 | 143.92 | 86.02 | 100 | 77.94 | 84.76 | 109.11 | 100.48 | 100 | 81.75 | 153.76 | 124.68 | 2.88 |
| Herpetosiphon | 0.02142 | 0.03281 | 58.65 | 89.74 | 100 | 126.96 | 81.74 | 111.17 | 142.11 | 100 | 92.58 | 93.83 | 94.73 | 3.88 |
| Hirschia | 0.00001 | 0.00009 | 19.03 | 65.61 | 100 | 77.82 | 129.9 | 79.55 | 30.32 | 100 | 34.44 | 17.05 | 14.66 | 3.82 |
| Hyphomicrobium | 0.01002 | 0.01794 | 139.51 | 102.08 | 100 | 93.1 | 94.2 | 82.2 | 109.93 | 100 | 121.94 | 137.41 | 124.1 | 3.04 |
| Iamia | 0.03915 | 0.05460 | 93.63 | 89.24 | 100 | 88.5 | 71.48 | 67.73 | 128.16 | 100 | 84.42 | 86.22 | 89.05 | 3.64 |
| Iamia sp (uncultured) | 0.00142 | 0.00405 | 66.64 | 107.71 | 100 | 135.76 | 96.61 | 103.38 | 177.43 | 100 | 99.85 | 85.37 | 88.02 | 2.93 |
| Ideonella | 0.00401 | 0.00884 | 10.88 | 38.82 | 100 | 42.72 | 33.77 | 45.99 | 82.38 | 100 | 97.61 | 47.68 | 57.06 | 3.16 |
| Ilumatobacter | 0.04702 | 0.06357 | 100.44 | 78.89 | 100 | 70.48 | 55.97 | 60.76 | 149.42 | 100 | 117.81 | 136.16 | 156.78 | 3.47 |
| Knoellia | 0.00217 | 0.00527 | 114.47 | 103.4 | 100 | 110.56 | 103.85 | 126.46 | 114.61 | 100 | 98.75 | 133.74 | 114.09 | 4.4 |
| Kribbella | 0.00081 | 0.00269 | 119.46 | 84.95 | 100 | 84.66 | 78.65 | 85.11 | 119.71 | 100 | 106.01 | 113.59 | 119.53 | 3.05 |
| Lacibacter | 0.01180 | 0.02049 | 49.95 | 122.39 | 100 | 126.01 | 92.79 | 93.58 | 138.89 | 100 | 93.63 | 64.01 | 64.47 | 3.24 |
| Lechevalieria | 0.00000 | 0.00004 | 52.56 | 91.1 | 100 | 63.11 | 50.02 | 59.65 | 157.09 | 100 | 84.73 | 90.99 | 92.35 | 3.98 |
| Leptolyngbya EcFYyyy 00 | 0.00001 | 0.00015 | 1.95 | 45.17 | 100 | 183.17 | 129.56 | 146.44 | 74.58 | 100 | 214.75 | 62.31 | 56.64 | 4.59 |
| Litorilinea | 0.04367 | 0.05996 | 79.01 | 95.72 | 100 | 107.92 | 119.76 | 131.82 | 110.08 | 100 | 108.14 | 95.05 | 121.57 | 3.4 |
| Luedemannella | 0.00007 | 0.00049 | 166.82 | 66.71 | 100 | 78.66 | 85.55 | 100.98 | 126.09 | 100 | 126.57 | 195.9 | 145.98 | 3.45 |
| Luteitalea | 0.00522 | 0.01073 | 80.17 | 95.05 | 100 | 112.9 | 123.93 | 134.65 | 91.48 | 100 | 97.14 | 133.96 | 89.81 | 3.42 |
| Luteolibacter | 0.00005 | 0.00037 | 35.13 | 67.08 | 100 | 71.69 | 73.92 | 66.99 | 122.4 | 100 | 94.67 | 71.2 | 70.03 | 3.71 |
| Lutispora sp (uncultured) | 0.00432 | 0.00939 | 319.31 | 104.16 | 100 | 133.81 | 130.28 | 129.75 | 118.88 | 100 | 150.14 | 214.92 | 193.66 | 3.64 |
| Lysobacter | 0.00455 | 0.00958 | 77.69 | 105.18 | 100 | 125.01 | 91.94 | 117.29 | 110.46 | 100 | 77.25 | 80.32 | 75.81 | 3.91 |
| Marmoricola | 0.00252 | 0.00596 | 134.53 | 108.08 | 100 | 111.54 | 98.25 | 115.63 | 91.53 | 100 | 79.66 | 83.51 | 76.19 | 4.19 |
| Massilia | 0.03106 | 0.04498 | 99.79 | 104.48 | 100 | 140.29 | 126.19 | 214.58 | 94.35 | 100 | 84.95 | 72.37 | 93.09 | 4.54 |
| Mesorhizobium | 0.00012 | 0.00066 | 35.43 | 65.25 | 100 | 73.92 | 96.74 | 75.34 | 75.05 | 100 | 72.27 | 71.13 | 64.08 | 4.18 |
| Methylobacillus | 0.00000 | 0.00003 | 5.16 | 90.27 | 100 | 100.97 | 46.46 | 99.73 | 340.26 | 100 | 165.87 | 101.63 | 156.12 | 3.5 |
| Methylorosula | 0.00028 | 0.00112 | 76.34 | 102.56 | 100 | 139.56 | 133.64 | 149.84 | 87.81 | 100 | 82.5 | 76.3 | 91.52 | 2.77 |
| Methylotenera | 0.00084 | 0.00272 | 24.43 | 95.22 | 100 | 98.88 | 81.12 | 108.31 | 301.13 | 100 | 138.69 | 89.76 | 105.38 | 3.65 |
| Microbacterium | 0.00113 | 0.00340 | 55.45 | 115.99 | 100 | 100.75 | 68.03 | 94.84 | 161.87 | 100 | 85.9 | 147.87 | 91.36 | 4.02 |
| Microvirga | 0.01117 | 0.01974 | 101.43 | 114.73 | 100 | 151.12 | 134.5 | 175.69 | 77.67 | 100 | 77.99 | 69.81 | 78.23 | 4.68 |
| Mitsuaria | 0.00000 | 0.00000 | 2.17 | 53.69 | 100 | 35.03 | 8.24 | 11.24 | 454.18 | 100 | 554.6 | 178.77 | 205.47 | 4.18 |
| Mycobacterium | 0.00003 | 0.00029 | 114.42 | 88.61 | 100 | 96.31 | 88.13 | 88 | 90 | 100 | 77.91 | 72.58 | 85.1 | 3.82 |
| Niastella | 0.00016 | 0.00078 | 34.69 | 81.53 | 100 | 81.3 | 82.17 | 81.69 | 117.88 | 100 | 122.51 | 99.41 | 90.49 | 3.08 |
| Nitrospira | 0.00452 | 0.00958 | 77.75 | 106 | 100 | 108.9 | 132.65 | 101.38 | 79.11 | 100 | 103.91 | 105.16 | 103.19 | 4.33 |
| Nocardioides | 0.00032 | 0.00122 | 111.96 | 96.06 | 100 | 99.18 | 77.9 | 93.11 | 134.28 | 100 | 103.56 | 141.28 | 118.24 | 4.35 |
| Nodosilinea PCC 7104 | 0.00014 | 0.00072 | 1.21 | 27.34 | 100 | 44.88 | 77.94 | 147.07 | 158.82 | 100 | 171.68 | 97.01 | 136.17 | 3.94 |
| Nonomuraea | 0.02585 | 0.03826 | 120.89 | 73.61 | 100 | 94.72 | 89.38 | 110.01 | 120.28 | 100 | 135.85 | 164.07 | 144.75 | 3 |
| Nordella | 0.01871 | 0.03023 | 88.67 | 115.42 | 100 | 88.03 | 101.14 | 73.6 | 84.38 | 100 | 97.59 | 88.1 | 79.34 | 3.91 |
| Ohtaekwangia | 0.00106 | 0.00325 | 29.46 | 71.96 | 100 | 73.08 | 69.3 | 54.17 | 101.46 | 100 | 69.21 | 57.06 | 57.46 | 3.58 |
| Oligoflexus | 0.01147 | 0.02013 | 65.74 | 87.15 | 100 | 171.08 | 128.53 | 214.79 | 95.62 | 100 | 96.84 | 95.69 | 85.35 | 3.21 |
| Paenarthrobacter | 0.03072 | 0.04473 | 130.73 | 169.49 | 100 | 134.48 | 114.18 | 164.02 | 203.95 | 100 | 150.92 | 172.4 | 163.93 | 3.83 |
| Paenisporosarcina | 0.00772 | 0.01461 | 129.46 | 109.22 | 100 | 97.31 | 98.52 | 132.27 | 66.29 | 100 | 71.88 | 109.22 | 105.48 | 3.51 |
| Parasegetibacter | 0.00019 | 0.00085 | 45.81 | 93.87 | 100 | 134.98 | 102.09 | 165.42 | 112.56 | 100 | 117.55 | 99.24 | 92.15 | 3.45 |
| Parviterribacter | 0.01759 | 0.02895 | 105.41 | 92.05 | 100 | 105.23 | 87.77 | 110.02 | 96.72 | 100 | 81.49 | 96.82 | 79.33 | 3.03 |
| Paucibacter | 0.01434 | 0.02420 | 47.88 | 136.36 | 100 | 126.95 | 118.36 | 123.52 | 96.9 | 100 | 82.38 | 76.43 | 80.01 | 2.8 |
| Pedobacter | 0.00022 | 0.00093 | 69.74 | 114.94 | 100 | 139.41 | 127.68 | 180.92 | 101.98 | 100 | 93.22 | 96.73 | 120.34 | 3.81 |
| Pedococcus Phycicoccus | 0.00877 | 0.01637 | 112.31 | 95.7 | 100 | 96.86 | 68.52 | 79.2 | 148.35 | 100 | 97.55 | 170.15 | 140.53 | 2.75 |
| Pedomicrobium | 0.00069 | 0.00233 | 133.81 | 96.01 | 100 | 88.83 | 110.74 | 80.74 | 85.78 | 100 | 103.05 | 106.64 | 88.67 | 3.79 |
| Pelobacter sp (uncultured) | 0.01183 | 0.02049 | 97.99 | 120.13 | 100 | 120.03 | 105.1 | 58.14 | 65.47 | 100 | 103.66 | 57.55 | 47.12 | 2.92 |
| Phenylobacterium | 0.00460 | 0.00960 | 95.21 | 85.21 | 100 | 97.3 | 88.8 | 109.63 | 106.04 | 100 | 93.67 | 108.57 | 106.72 | 2.75 |
| Pirellula | 0.01454 | 0.02438 | 90.49 | 117.94 | 100 | 87.13 | 94.04 | 69.26 | 93.81 | 100 | 96.72 | 82.95 | 90.5 | 4.56 |
| Planctomicrobium | 0.00157 | 0.00442 | 63.86 | 120.49 | 100 | 83.78 | 91.06 | 56.02 | 122.64 | 100 | 91.67 | 70 | 92.91 | 2.97 |
| Planctomycetales bacterium (uncultured) | 0.00024 | 0.00099 | 59.83 | 115.05 | 100 | 103.7 | 126.08 | 73.06 | 95.79 | 100 | 83.04 | 64.74 | 86.54 | 2.96 |
| Planctomycete (uncultured) | 0.00003 | 0.00029 | 66.73 | 132.95 | 100 | 126.15 | 142.15 | 111.35 | 90.89 | 100 | 78.24 | 54.09 | 65.28 | 4.33 |
| Polaromonas | 0.00930 | 0.01714 | 59.7 | 107.13 | 100 | 79.63 | 109.19 | 86.14 | 108.2 | 100 | 109.18 | 98.55 | 84.48 | 2.77 |
| Polyangium brachysporum group | 0.00000 | 0.00002 | 42.99 | 115.43 | 100 | 140.21 | 88.46 | 95.01 | 87.65 | 100 | 45.94 | 50.37 | 55.97 | 3.66 |
| Pontibacter | 0.00940 | 0.01714 | 67.62 | 117.08 | 100 | 159.6 | 119.62 | 223.52 | 118.13 | 100 | 101.24 | 93.65 | 107.89 | 3.95 |
| Promicromonospora | 0.02417 | 0.03640 | 120.47 | 63.09 | 100 | 47.58 | 64.24 | 74.79 | 87.54 | 100 | 57.73 | 49.82 | 56.8 | 3.26 |
| Pseudarthrobacter | 0.00537 | 0.01086 | 109.71 | 129.12 | 100 | 179.32 | 129.85 | 188.51 | 144.57 | 100 | 129.45 | 149.41 | 138.27 | 3.1 |
| Pseudoduganella | 0.00011 | 0.00066 | 17.06 | 114.8 | 100 | 194.18 | 70.13 | 52.92 | 124.48 | 100 | 114.99 | 61.59 | 67.26 | 3.55 |
| Pseudoflavitalea | 0.00212 | 0.00527 | 58.51 | 91.58 | 100 | 73.25 | 167.7 | 67.34 | 31.65 | 100 | 55.73 | 36.29 | 37.11 | 2.97 |
| Pseudolabrys | 0.00509 | 0.01053 | 53.67 | 65.2 | 100 | 48.39 | 77.9 | 45.72 | 93.04 | 100 | 82.1 | 79.96 | 83.12 | 3.87 |
| Pseudomuriella schumacherensis | 0.00011 | 0.00066 | 9.76 | 107.97 | 100 | 115.99 | 133.1 | 148.14 | 61.37 | 100 | 119.35 | 73.75 | 77.45 | 3.48 |
| Pseudorhodoplanes | 0.00213 | 0.00527 | 49.67 | 89.2 | 100 | 92.65 | 129.5 | 83.85 | 66.77 | 100 | 86.97 | 76.57 | 70.63 | 3.02 |
| Pseudoxanthomonas | 0.00011 | 0.00066 | 22.02 | 55.85 | 100 | 51.53 | 38.23 | 44.31 | 138.8 | 100 | 94.66 | 72.29 | 53.56 | 3.99 |
| Qipengyuania | 0.00000 | 0.00004 | 38.76 | 115.42 | 100 | 131.47 | 74.14 | 107.63 | 154.83 | 100 | 102.63 | 118.29 | 115.54 | 3.83 |
| Ramlibacter | 0.00001 | 0.00009 | 62.11 | 112.6 | 100 | 147.02 | 136.65 | 172.1 | 97.16 | 100 | 79.95 | 51.68 | 56.55 | 4.08 |
| Reyranella | 0.00440 | 0.00940 | 72.85 | 92.39 | 100 | 82.84 | 109.3 | 70.97 | 83.74 | 100 | 101.86 | 71.61 | 74.64 | 3.35 |
| Rhizobacter | 0.00005 | 0.00038 | 50.76 | 88.61 | 100 | 98.57 | 97.22 | 111.13 | 111.29 | 100 | 89.72 | 81.87 | 75.04 | 3.96 |
| Rhodobacter | 0.00005 | 0.00036 | 29.2 | 64.71 | 100 | 122.26 | 115.77 | 136.3 | 166.58 | 100 | 111.05 | 109.39 | 89.49 | 3.52 |
| Rhodocytophaga | 0.02868 | 0.04199 | 59.54 | 114.21 | 100 | 125.07 | 111.49 | 150.76 | 102.24 | 100 | 123.84 | 112.9 | 117.01 | 3.21 |
| Rhodopirellula | 0.00004 | 0.00032 | 41.55 | 118.9 | 100 | 131.05 | 107.02 | 146 | 88.1 | 100 | 68.54 | 56.07 | 63.52 | 3.71 |
| Rhodoplanes | 0.04025 | 0.05585 | 86.35 | 107.4 | 100 | 77.97 | 93.94 | 63.1 | 96.86 | 100 | 113.22 | 97.29 | 90.3 | 3.69 |
| Roseimicrobium | 0.04645 | 0.06313 | 96.61 | 123.21 | 100 | 112.32 | 126.69 | 76.4 | 106.87 | 100 | 102.7 | 77.13 | 82.75 | 3.29 |
| Roseomonas | 0.00288 | 0.00658 | 64.27 | 98.19 | 100 | 138.98 | 137.15 | 204.25 | 90.33 | 100 | 86.66 | 81.46 | 73.16 | 3.18 |
| Rubellimicrobium | 0.00097 | 0.00307 | 65.13 | 117.45 | 100 | 170.12 | 151.44 | 228.41 | 113.06 | 100 | 97.98 | 99.63 | 88.5 | 4.16 |
| Rubrobacter | 0.00944 | 0.01714 | 100.87 | 93.62 | 100 | 116.01 | 105.2 | 123.83 | 91.48 | 100 | 98.1 | 98.65 | 113.06 | 4.52 |
| Rubrobacterales bacterium (uncultured) | 0.00000 | 0.00000 | 125.43 | 87.94 | 100 | 82.24 | 67.16 | 69.84 | 121.01 | 100 | 98.86 | 122.78 | 154.88 | 4.44 |
| Rubrobacteria bacterium (uncultured) | 0.00000 | 0.00002 | 108.02 | 81.15 | 100 | 84.71 | 66.29 | 79.79 | 121.82 | 100 | 107.63 | 134.16 | 153.21 | 3.93 |
| Saccharothrix | 0.03157 | 0.04547 | 75.91 | 133.98 | 100 | 105.39 | 105.19 | 151.79 | 150.8 | 100 | 105.82 | 91.36 | 123.54 | 3.17 |
| Shimazuella | 0.02083 | 0.03229 | 136.53 | 77.37 | 100 | 112.63 | 101.2 | 136.97 | 135.41 | 100 | 151.8 | 167.24 | 129.8 | 2.79 |
| Shinella | 0.00053 | 0.00185 | 13.05 | 74.85 | 100 | 60.13 | 40.69 | 58.79 | 176.38 | 100 | 137.13 | 109.99 | 81.26 | 3.6 |
| Skermanella | 0.03385 | 0.04797 | 96.91 | 90.98 | 100 | 123.94 | 146.52 | 171.5 | 101.33 | 100 | 103.23 | 122.86 | 114.15 | 4.41 |
| Solirubrobacter | 0.00005 | 0.00036 | 117.94 | 87.5 | 100 | 95.4 | 86.28 | 93.1 | 106.64 | 100 | 99.55 | 131.42 | 119.01 | 3.97 |
| Sorangium | 0.00215 | 0.00527 | 86.25 | 40.33 | 100 | 48.69 | 47.57 | 35.85 | 78.55 | 100 | 42.55 | 48.61 | 31.32 | 3.08 |
| Sphingoaurantiacus | 0.00024 | 0.00099 | 78.59 | 107.85 | 100 | 148.48 | 123.13 | 199.51 | 107.38 | 100 | 76.32 | 98.52 | 103.43 | 3.22 |
| Sphingobium | 0.00001 | 0.00010 | 5.53 | 84.97 | 100 | 126.26 | 66.77 | 107.85 | 197.37 | 100 | 102.54 | 93.65 | 79.3 | 4.7 |
| Sphingomonas | 0.00025 | 0.00103 | 86.32 | 97.74 | 100 | 111.29 | 82.81 | 107.47 | 102.75 | 100 | 80.61 | 88.26 | 98.2 | 4.65 |
| Sphingopyxis | 0.00012 | 0.00066 | 18.92 | 63.7 | 100 | 65.2 | 50.71 | 74.14 | 289.82 | 100 | 133.61 | 149.8 | 185.6 | 3.34 |
| Steroidobacter | 0.00032 | 0.00122 | 112.12 | 87.33 | 100 | 89.86 | 115.64 | 95.27 | 84.48 | 100 | 100.51 | 111.95 | 86.39 | 3.9 |
| Streptomyces | 0.00054 | 0.00185 | 67.84 | 97.49 | 100 | 73.52 | 77.73 | 79.56 | 114.7 | 100 | 81.7 | 92.98 | 93.07 | 4.2 |
| Sumerlaea | 0.00106 | 0.00325 | 64.69 | 131.01 | 100 | 138.36 | 136.41 | 134.87 | 76.38 | 100 | 93.33 | 83.92 | 65.45 | 3.09 |
| Synechococcus IR11 | 0.00187 | 0.00502 | 1.14 | 23.47 | 100 | 79.9 | 104.71 | 150.87 | 33.79 | 100 | 35.68 | 32.59 | 47.4 | 3.55 |
| Tahibacter | 0.00672 | 0.01300 | 39.95 | 129.97 | 100 | 80.3 | 60.81 | 68.59 | 88.99 | 100 | 46.75 | 37.21 | 47.31 | 3.09 |
| Tepidisphaera | 0.00308 | 0.00697 | 75.82 | 146.03 | 100 | 168.77 | 170.98 | 141.7 | 92.25 | 100 | 94.8 | 50.6 | 59.62 | 3.19 |
| Terrimonas | 0.00310 | 0.00697 | 61.36 | 106.49 | 100 | 98.64 | 109.13 | 81.35 | 81.27 | 100 | 84.03 | 58.85 | 57.66 | 3.85 |
| Thermomicrobia bacterium (uncultured) | 0.04136 | 0.05709 | 95.22 | 88.1 | 100 | 82.64 | 93.26 | 108.62 | 128.68 | 100 | 101.1 | 104.33 | 135.36 | 2.63 |
| Truepera | 0.00000 | 0.00002 | 59 | 145.14 | 100 | 150.94 | 178.65 | 185.85 | 71.93 | 100 | 85.67 | 99.63 | 63.63 | 3.55 |
| Tumebacillus | 0.00190 | 0.00502 | 105.62 | 51.28 | 100 | 57.5 | 69.61 | 91.3 | 125.72 | 100 | 159.41 | 357.77 | 239.62 | 3.71 |
| Tychonema CCAP 1459 11B | 0.00668 | 0.01300 | 29.82 | 110.25 | 100 | 71.6 | 131.09 | 118.76 | 149.73 | 100 | 264.51 | 182.86 | 160.47 | 4.09 |
| Variovorax | 0.00010 | 0.00062 | 53.62 | 88.15 | 100 | 67.73 | 70.33 | 74.4 | 146.4 | 100 | 107.14 | 90.17 | 64.53 | 3.81 |
| Yonghaparkia | 0.00000 | 0.00002 | 11.81 | 103.9 | 100 | 134.21 | 67.84 | 132.39 | 158.14 | 100 | 107.62 | 132.74 | 145.48 | 3.71 |
| Genus | p-Value | FDR | t0 | CFD | CRD | MYC | PSBA | PSBA + MYC | LDA Score |
|---|---|---|---|---|---|---|---|---|---|
| Acidibacter | 0.00026 | 0.00196 | 100 | 177.39 | 210.58 | 164.9 | 191.57 | 146.41 | 3.79 |
| Acidimicrobiia bacterium (uncultured) | 0.00034 | 0.00241 | 100 | 50.7 | 54.34 | 48.18 | 52.96 | 54.71 | 4.09 |
| Acidobacteriales bacterium (uncultured) | 0.01264 | 0.02937 | 100 | 69.88 | 75.16 | 77.7 | 90.07 | 66.27 | 4.19 |
| Acidovorax | 0.00006 | 0.00104 | 100 | 1347.41 | 934.23 | 1739.6 | 932.94 | 670.62 | 3.11 |
| Actinobacterium (uncultured) | 0.00567 | 0.01549 | 100 | 65.65 | 73.75 | 62.54 | 58.92 | 59.79 | 4.02 |
| Aeromicrobium | 0.00018 | 0.00155 | 100 | 267.48 | 234.05 | 229.79 | 161.1 | 175.42 | 4.49 |
| Allorhizobium Neorhizobium Pararhizobium Rhizobium | 0.00113 | 0.00474 | 100 | 207.37 | 201.71 | 194.37 | 169.89 | 183.24 | 4.36 |
| Alsobacter | 0.01434 | 0.03257 | 100 | 110.04 | 124.56 | 150.01 | 144.5 | 172.18 | 2.97 |
| Altererythrobacter | 0.00014 | 0.00146 | 100 | 290.37 | 397.53 | 237.33 | 250.19 | 235.61 | 3.51 |
| Amaricoccus | 0.00851 | 0.02128 | 100 | 113.56 | 121.7 | 127 | 148.61 | 171.37 | 3.1 |
| Aminobacter | 0.00000 | 0.00000 | 100 | 184.31 | 176.93 | 680 | 685.6 | 759.35 | 3.34 |
| Amycolatopsis | 0.00103 | 0.00456 | 100 | 1435.17 | 507.39 | 692.37 | 500.04 | 809.66 | 3.45 |
| Anaerolinea | 0.01738 | 0.03871 | 100 | 28.79 | 24.19 | 31.18 | 33.2 | 41.42 | 3.58 |
| Anaeromyxobacter | 0.00021 | 0.00165 | 100 | 51.91 | 50.93 | 52.91 | 73.59 | 61.44 | 3.58 |
| Arenimonas | 0.00057 | 0.00307 | 100 | 273.32 | 218.17 | 173.06 | 168.36 | 176.53 | 3.27 |
| Aridibacter | 0.01313 | 0.03025 | 100 | 159.82 | 139.54 | 143.2 | 136.39 | 178.44 | 3.27 |
| Armatimonadetes bacterium (uncultured) | 0.01982 | 0.04340 | 100 | 67.92 | 71.3 | 74.52 | 76.01 | 86.59 | 2.87 |
| Arthrobacter | 0.00067 | 0.00325 | 100 | 117.82 | 99.26 | 130.62 | 132.62 | 152.83 | 5.2 |
| Azohydromonas | 0.00193 | 0.00683 | 100 | 240.03 | 236.72 | 228.32 | 199.56 | 209.37 | 3.34 |
| Blastocatella | 0.00530 | 0.01498 | 100 | 149.57 | 133.26 | 151.71 | 132.76 | 154.13 | 3.35 |
| Bosea | 0.00107 | 0.00466 | 100 | 207.49 | 222 | 224.71 | 225.56 | 232.34 | 3.54 |
| Bradyrhizobium | 0.00047 | 0.00273 | 100 | 141.63 | 158.55 | 141.31 | 153.76 | 124.6 | 3.89 |
| Burkholderia Caballeronia Paraburkholderia | 0.00876 | 0.02171 | 100 | 58.97 | 63.02 | 43.08 | 37.97 | 36.65 | 3.48 |
| Caenimonas | 0.00377 | 0.01188 | 100 | 183.98 | 171.9 | 178.79 | 160.7 | 193.43 | 3.94 |
| Cellulomonas | 0.04245 | 0.08045 | 100 | 66.13 | 76.53 | 83.6 | 107.54 | 116.93 | 2.71 |
| Chloroflexi bacterium (uncultured) | 0.00771 | 0.01984 | 100 | 79.71 | 79.16 | 81.64 | 80.19 | 91.38 | 4.09 |
| Chloroflexus sp (uncultured) | 0.00001 | 0.00045 | 100 | 95.31 | 88.08 | 73.13 | 56.27 | 69.35 | 3.79 |
| Chthonomonas | 0.02807 | 0.05767 | 100 | 130.07 | 130.32 | 120.33 | 131.04 | 150.69 | 2.53 |
| Clostridium sensu stricto 1 | 0.01438 | 0.03257 | 100 | 50.13 | 47.2 | 54.59 | 53.71 | 56.02 | 3.13 |
| Clostridium sensu stricto 8 | 0.02672 | 0.05533 | 100 | 53.39 | 58.16 | 56.83 | 67.68 | 67.99 | 2.78 |
| Cohnella | 0.00127 | 0.00509 | 100 | 53.5 | 65.94 | 61.25 | 82.68 | 87.39 | 3.35 |
| Comamonas | 0.00042 | 0.00272 | 100 | 311.03 | 282.43 | 268.31 | 278.19 | 206.81 | 3 |
| Conexibacter sp (uncultured) | 0.00047 | 0.00273 | 100 | 58.06 | 60.52 | 59.73 | 63.8 | 70.67 | 2.84 |
| Cupriavidus | 0.01615 | 0.03628 | 100 | 171.75 | 286.17 | 175.54 | 170.52 | 194.75 | 3.29 |
| Defluviicoccus | 0.01052 | 0.02557 | 100 | 73.67 | 70.43 | 75.95 | 98.44 | 107.26 | 2.72 |
| Desmochloris halophila | 0.00010 | 0.00136 | 100 | 3389.26 | 2325.21 | 2344.77 | 3150.88 | 1823.76 | 3.3 |
| Desulfuromonadales bacterium (uncultured) | 0.00284 | 0.00970 | 100 | 66.53 | 62.6 | 64.7 | 94.4 | 83.2 | 3.14 |
| Devosia | 0.00002 | 0.00064 | 100 | 230.58 | 267.85 | 195.41 | 222.77 | 183.48 | 4.23 |
| Dongia | 0.00144 | 0.00529 | 100 | 65.62 | 64.28 | 61.73 | 59.54 | 52.44 | 3.76 |
| Dyadobacter | 0.00067 | 0.00325 | 100 | 415.05 | 215.86 | 354.92 | 167.22 | 178.1 | 3.62 |
| Ensifer | 0.00133 | 0.00518 | 100 | 177.81 | 177.84 | 191.1 | 163.95 | 178.95 | 3.77 |
| Ferrimicrobium sp (uncultured) | 0.03729 | 0.07311 | 100 | 79.64 | 79.49 | 81.54 | 86.81 | 67.03 | 2.91 |
| Fictibacillus | 0.00011 | 0.00136 | 100 | 46.45 | 66.72 | 47.27 | 56.69 | 67.76 | 3.74 |
| Flavitalea | 0.00003 | 0.00067 | 100 | 199.98 | 213.8 | 234.21 | 206.44 | 199.48 | 3.21 |
| Gaiella | 0.00065 | 0.00325 | 100 | 59.73 | 61.82 | 59.03 | 59.93 | 66.2 | 4.91 |
| Gaiella sp (uncultured) | 0.00782 | 0.01994 | 100 | 67.95 | 67.3 | 74.46 | 67.62 | 83.25 | 3.78 |
| Gemmatimonadales bacterium (uncultured) | 0.04590 | 0.08447 | 100 | 73.72 | 73.02 | 64.83 | 75.24 | 58.89 | 3.31 |
| Gemmatimonas | 0.00017 | 0.00147 | 100 | 171.4 | 162.56 | 178.15 | 164.04 | 148.71 | 3.46 |
| Herbidospora | 0.04913 | 0.08761 | 100 | 84.66 | 143.88 | 89.56 | 151.54 | 132.11 | 2.75 |
| Herbinix | 0.00344 | 0.01114 | 100 | 52.32 | 57.07 | 44.74 | 63.77 | 67 | 2.82 |
| Hirschia | 0.00028 | 0.00207 | 100 | 359.23 | 878.92 | 417.22 | 446.36 | 314.2 | 3.65 |
| Iamia | 0.01109 | 0.02647 | 100 | 101.36 | 95.31 | 81.85 | 74.31 | 73.34 | 3.52 |
| Ideonella | 0.00021 | 0.00165 | 100 | 418.12 | 750.61 | 486.18 | 293.94 | 383.1 | 3.06 |
| Knoellia | 0.03739 | 0.07311 | 100 | 85.69 | 79.04 | 82.32 | 92.66 | 97.45 | 4.11 |
| Kribbella | 0.00435 | 0.01326 | 100 | 77.6 | 76.97 | 72.78 | 72.81 | 76.81 | 2.95 |
| Lacibacter | 0.00211 | 0.00736 | 100 | 260.94 | 199.77 | 217.27 | 156.68 | 161.61 | 3.2 |
| Lechevalieria | 0.00046 | 0.00273 | 100 | 252.6 | 200.75 | 151.51 | 143.7 | 149.2 | 3.79 |
| Leptolyngbya EcFYyyy 00 | 0.00002 | 0.00064 | 100 | 2140.01 | 3878.75 | 7397.56 | 4138.1 | 4870.86 | 4.48 |
| Litorilinea | 0.03752 | 0.07311 | 100 | 121.46 | 118.59 | 127.53 | 128.35 | 152.68 | 3.29 |
| Luedemannella | 0.00007 | 0.00108 | 100 | 39.5 | 45.44 | 42.89 | 55.95 | 53.81 | 3.39 |
| Luteitalea | 0.00135 | 0.00518 | 100 | 125.26 | 134.49 | 140.46 | 173.91 | 151.16 | 3.32 |
| Luteolibacter | 0.00039 | 0.00258 | 100 | 205.42 | 232.16 | 185.95 | 169.16 | 162.31 | 3.56 |
| Lutispora sp (uncultured) | 0.00067 | 0.00325 | 100 | 27.9 | 25.41 | 35.17 | 41.36 | 39.38 | 3.61 |
| Marmoricola | 0.00515 | 0.01487 | 100 | 77.75 | 78.22 | 73.58 | 70.8 | 75.72 | 4.08 |
| Mesorhizobium | 0.00002 | 0.00067 | 100 | 195.82 | 279.35 | 203.92 | 234.84 | 197.12 | 4.17 |
| metagenome | 0.00406 | 0.01251 | 100 | 78.96 | 83.84 | 81.02 | 82.88 | 79.27 | 4.28 |
| Methylobacillus | 0.00011 | 0.00136 | 100 | 6346.25 | 2576.58 | 3757.78 | 2084.31 | 3283.36 | 3.26 |
| Methylotenera | 0.00009 | 0.00136 | 100 | 1027.66 | 481.25 | 598.02 | 414.26 | 504.4 | 3.42 |
| Microbacterium | 0.00372 | 0.01186 | 100 | 317.9 | 221.95 | 205.53 | 256.2 | 201.55 | 3.84 |
| Microlunatus | 0.04250 | 0.08045 | 100 | 88.37 | 91.93 | 97.19 | 106.18 | 117.57 | 3.74 |
| Microvirga | 0.04487 | 0.08314 | 100 | 121.42 | 132.82 | 139 | 124.61 | 154.6 | 4.55 |
| Mitsuaria | 0.00001 | 0.00045 | 100 | 8902 | 3989.43 | 10737.74 | 3207.06 | 3325.77 | 3.94 |
| Mycobacterium | 0.00012 | 0.00136 | 100 | 77.18 | 86.43 | 74.79 | 69.53 | 75.09 | 3.72 |
| Nakamurella | 0.04455 | 0.08314 | 100 | 106.58 | 111.13 | 124.89 | 130.79 | 164.98 | 2.53 |
| Niastella | 0.00023 | 0.00182 | 100 | 302.84 | 301.36 | 314.82 | 274.72 | 256.95 | 2.96 |
| Nitrosospira | 0.04718 | 0.08622 | 100 | 128.25 | 153.5 | 165.62 | 188.07 | 251.72 | 3.01 |
| Nitrospira | 0.00112 | 0.00474 | 100 | 128.76 | 140.9 | 150.26 | 165.85 | 142.5 | 4.3 |
| Nocardia | 0.02207 | 0.04605 | 100 | 173.32 | 232.51 | 171.13 | 141.82 | 179.46 | 2.87 |
| Nocardioides | 0.04227 | 0.08045 | 100 | 81.16 | 73.15 | 72.81 | 75.04 | 76.25 | 4.15 |
| Nodosilinea PCC 7104 | 0.00001 | 0.00046 | 100 | 4351.84 | 6152.39 | 5445.13 | 5182.46 | 9239.08 | 3.82 |
| Nonomuraea | 0.02141 | 0.04575 | 100 | 58.94 | 65.05 | 70.9 | 75.85 | 81.19 | 2.89 |
| Nordella | 0.02831 | 0.05771 | 100 | 124.19 | 126.44 | 119.25 | 118.75 | 95.32 | 3.65 |
| Ohtaekwangia | 0.00037 | 0.00258 | 100 | 257.81 | 303.46 | 214.48 | 193.93 | 170.66 | 3.51 |
| Paenisporosarcina | 0.00535 | 0.01498 | 100 | 66.41 | 75.17 | 63.1 | 77.97 | 91.2 | 3.35 |
| Parasegetibacter | 0.00067 | 0.00325 | 100 | 200.35 | 196.12 | 246.56 | 197.73 | 273.25 | 3.27 |
| Paucibacter | 0.02168 | 0.04597 | 100 | 251.29 | 216.81 | 222.48 | 209.49 | 223.8 | 2.71 |
| Pelobacter sp (uncultured) | 0.03482 | 0.06990 | 100 | 124.15 | 147.03 | 163.32 | 108.89 | 72.96 | 2.73 |
| Planctomicrobium | 0.02204 | 0.04605 | 100 | 226.42 | 185.98 | 166.52 | 146.69 | 136.9 | 2.82 |
| Planctomycetales bacterium (uncultured) | 0.00765 | 0.01984 | 100 | 239.03 | 232.73 | 213.01 | 201.93 | 181.64 | 2.81 |
| planctomycete (uncultured) | 0.00016 | 0.00147 | 100 | 212.45 | 199.06 | 191.81 | 173.65 | 164.77 | 4.21 |
| Polyangium brachysporum group | 0.00006 | 0.00104 | 100 | 444.11 | 469.82 | 325.42 | 280.97 | 297.46 | 3.44 |
| Promicromonospora | 0.00656 | 0.01757 | 100 | 70.61 | 92.25 | 49.68 | 51.94 | 60.14 | 3.17 |
| Pseudarthrobacter | 0.00094 | 0.00421 | 100 | 135.08 | 98.31 | 149.09 | 137.97 | 160.45 | 2.99 |
| Pseudoduganella | 0.00000 | 0.00045 | 100 | 753.66 | 628.22 | 942.24 | 411.93 | 371.72 | 3.46 |
| Pseudoflavitalea | 0.01219 | 0.02860 | 100 | 124.3 | 230.9 | 144.89 | 196.08 | 111.96 | 2.79 |
| Pseudomuriella schumacherensis | 0.00013 | 0.00138 | 100 | 827.59 | 959.36 | 1123.69 | 1011.69 | 1156.88 | 3.35 |
| Pseudorhodoplanes | 0.00011 | 0.00136 | 100 | 168.97 | 219.25 | 197.05 | 221.16 | 168.25 | 2.89 |
| Pseudoxanthomonas | 0.00003 | 0.00067 | 100 | 406.76 | 428.75 | 312.86 | 232.6 | 207.73 | 3.85 |
| Qipengyuania | 0.01122 | 0.02654 | 100 | 239.22 | 187.33 | 222.14 | 164.66 | 215.17 | 3.59 |
| Ramlibacter | 0.00010 | 0.00136 | 100 | 223.56 | 217.31 | 227.14 | 180.7 | 223.55 | 3.94 |
| Reyranella | 0.01046 | 0.02557 | 100 | 131.96 | 150.49 | 142 | 133.64 | 108.26 | 3.21 |
| Rhizobacter | 0.00015 | 0.00146 | 100 | 198.81 | 198.73 | 186.53 | 177.83 | 188.96 | 3.87 |
| Rhodocytophaga | 0.00182 | 0.00653 | 100 | 194.31 | 180.21 | 225.14 | 202.27 | 240.95 | 3.18 |
| Rhodopirellula | 0.00006 | 0.00104 | 100 | 283.52 | 279.76 | 263.72 | 218.17 | 286.28 | 3.58 |
| Roseimicrobium | 0.03548 | 0.07069 | 100 | 129.74 | 113.48 | 121.88 | 113.18 | 89.18 | 3.18 |
| Roseomonas | 0.02030 | 0.04374 | 100 | 156.66 | 166.66 | 183.05 | 179.08 | 235.63 | 2.97 |
| Rubellimicrobium | 0.00140 | 0.00529 | 100 | 183.98 | 159.75 | 208.63 | 198.94 | 261.53 | 3.97 |
| Rubrobacterales bacterium (uncultured) | 0.00053 | 0.00299 | 100 | 58.77 | 59.46 | 51.26 | 50.83 | 57.88 | 4.34 |
| Rubrobacteria bacterium (uncultured) | 0.00345 | 0.01114 | 100 | 64.84 | 68.67 | 62.29 | 60.71 | 71.56 | 3.78 |
| Shimazuella | 0.02890 | 0.05845 | 100 | 63.81 | 62.82 | 80.92 | 80.87 | 86.18 | 2.68 |
| Shinella | 0.00003 | 0.00067 | 100 | 648.01 | 588.03 | 514.6 | 381.15 | 399.09 | 3.52 |
| Skermanella | 0.04926 | 0.08761 | 100 | 96.41 | 100.41 | 113.44 | 135.57 | 148.3 | 4.21 |
| Solirubrobacter | 0.02018 | 0.04374 | 100 | 65.59 | 69.12 | 66.12 | 71.7 | 72.23 | 3.84 |
| Sorangium | 0.00618 | 0.01672 | 100 | 87.82 | 140.01 | 63.58 | 67.45 | 45.86 | 2.97 |
| Sphingobium | 0.00001 | 0.00045 | 100 | 2823.28 | 1945.12 | 2204.04 | 1578.67 | 1819.13 | 4.53 |
| Sphingopyxis | 0.00457 | 0.01377 | 100 | 333.74 | 321.33 | 242.15 | 219.49 | 313.11 | 3.07 |
| Streptomyces | 0.00012 | 0.00136 | 100 | 156.52 | 147.53 | 114.91 | 125.93 | 126.09 | 4.11 |
| Sumerlaea | 0.00135 | 0.00518 | 100 | 178.91 | 178.96 | 201.03 | 190.76 | 175.5 | 3.03 |
| Synechococcus IR11 | 0.00015 | 0.00146 | 100 | 2697.85 | 9323.56 | 5150.12 | 6201.85 | 9492.62 | 3.41 |
| Tahibacter | 0.00313 | 0.01051 | 100 | 391.14 | 381.88 | 221.81 | 171.65 | 202.85 | 2.94 |
| Terrimonas | 0.00067 | 0.00325 | 100 | 183.68 | 200.71 | 181.88 | 159.1 | 134.73 | 3.71 |
| Truepera | 0.00123 | 0.00503 | 100 | 472.17 | 570.19 | 563.01 | 635.07 | 447.48 | 3.34 |
| Tumebacillus | 0.00814 | 0.02054 | 100 | 42.44 | 61.79 | 49.98 | 84.66 | 78.93 | 3.6 |
| Variovorax | 0.00038 | 0.00258 | 100 | 223.94 | 190.06 | 169.6 | 152.89 | 132.72 | 3.64 |
| Yonghaparkia | 0.00682 | 0.01806 | 100 | 587.2 | 516.48 | 637.19 | 410.86 | 749.05 | 3.5 |
| Genus | p-Value | FDR | t0 | CFD | CRD | MYC | PSBA | PSBA + MYC | LDA Score |
|---|---|---|---|---|---|---|---|---|---|
| Acidibacter | 0.00026 | 0.00196 | 56.37 | 100 | 118.71 | 92.96 | 107.99 | 82.54 | 3.79 |
| Acidimicrobiia bacterium (uncultured) | 0.00034 | 0.00241 | 197.24 | 100 | 107.18 | 95.04 | 104.46 | 107.92 | 4.09 |
| Acidobacteriales bacterium (uncultured) | 0.01264 | 0.02937 | 143.11 | 100 | 107.57 | 111.19 | 128.9 | 94.84 | 4.19 |
| Acidovorax | 0.00006 | 0.00104 | 7.42 | 100 | 69.34 | 129.11 | 69.24 | 49.77 | 3.11 |
| Actinobacterium (uncultured) | 0.00567 | 0.01549 | 152.33 | 100 | 112.34 | 95.27 | 89.76 | 91.09 | 4.02 |
| Aeromicrobium | 0.00018 | 0.00155 | 37.39 | 100 | 87.5 | 85.91 | 60.23 | 65.58 | 4.49 |
| Allorhizobium Neorhizobium Pararhizobium Rhizobium | 0.00113 | 0.00474 | 48.22 | 100 | 97.27 | 93.73 | 81.93 | 88.36 | 4.36 |
| Alsobacter | 0.01434 | 0.03257 | 90.88 | 100 | 113.2 | 136.32 | 131.32 | 156.47 | 2.97 |
| Altererythrobacter | 0.00014 | 0.00146 | 34.44 | 100 | 136.91 | 81.74 | 86.16 | 81.14 | 3.51 |
| Amaricoccus | 0.00851 | 0.02128 | 88.06 | 100 | 107.17 | 111.84 | 130.87 | 150.91 | 3.1 |
| Aminobacter | 0.00000 | 0.00000 | 54.26 | 100 | 96 | 368.95 | 371.99 | 412 | 3.34 |
| Amycolatopsis | 0.00103 | 0.00456 | 6.97 | 100 | 35.35 | 48.24 | 34.84 | 56.42 | 3.45 |
| Anaerolinea | 0.01738 | 0.03871 | 347.38 | 100 | 84.05 | 108.3 | 115.32 | 143.88 | 3.58 |
| Anaeromyxobacter | 0.00021 | 0.00165 | 192.62 | 100 | 98.1 | 101.91 | 141.74 | 118.35 | 3.58 |
| Arenimonas | 0.00057 | 0.00307 | 36.59 | 100 | 79.82 | 63.32 | 61.6 | 64.59 | 3.27 |
| Aridibacter | 0.01313 | 0.03025 | 62.57 | 100 | 87.31 | 89.6 | 85.34 | 111.65 | 3.27 |
| Armatimonadetes bacterium (uncultured) | 0.01982 | 0.04340 | 147.22 | 100 | 104.97 | 109.71 | 111.91 | 127.49 | 2.87 |
| Arthrobacter | 0.00067 | 0.00325 | 84.87 | 100 | 84.25 | 110.86 | 112.56 | 129.71 | 5.2 |
| Azohydromonas | 0.00193 | 0.00683 | 41.66 | 100 | 98.62 | 95.12 | 83.14 | 87.23 | 3.34 |
| Blastocatella | 0.00530 | 0.01498 | 66.86 | 100 | 89.09 | 101.43 | 88.76 | 103.05 | 3.35 |
| Bosea | 0.00107 | 0.00466 | 48.2 | 100 | 106.99 | 108.3 | 108.71 | 111.98 | 3.54 |
| Bradyrhizobium | 0.00047 | 0.00273 | 70.61 | 100 | 111.95 | 99.78 | 108.56 | 87.98 | 3.89 |
| Burkholderia Caballeronia Paraburkholderia | 0.00876 | 0.02171 | 169.59 | 100 | 106.87 | 73.07 | 64.39 | 62.15 | 3.48 |
| Caenimonas | 0.00377 | 0.01188 | 54.35 | 100 | 93.43 | 97.18 | 87.34 | 105.13 | 3.94 |
| Cellulomonas | 0.04245 | 0.08045 | 151.22 | 100 | 115.73 | 126.41 | 162.62 | 176.81 | 2.71 |
| Chloroflexi bacterium (uncultured) | 0.00771 | 0.01984 | 125.46 | 100 | 99.31 | 102.43 | 100.61 | 114.65 | 4.09 |
| Chloroflexus sp (uncultured) | 0.00001 | 0.00045 | 104.92 | 100 | 92.41 | 76.73 | 59.04 | 72.77 | 3.79 |
| Chthonomonas | 0.02807 | 0.05767 | 76.88 | 100 | 100.19 | 92.51 | 100.74 | 115.85 | 2.53 |
| Clostridium sensu stricto 1 | 0.01438 | 0.03257 | 199.47 | 100 | 94.15 | 108.89 | 107.13 | 111.74 | 3.13 |
| Clostridium sensu stricto 8 | 0.02672 | 0.05533 | 187.32 | 100 | 108.95 | 106.46 | 126.78 | 127.36 | 2.78 |
| Cohnella | 0.00127 | 0.00509 | 186.91 | 100 | 123.25 | 114.49 | 154.54 | 163.34 | 3.35 |
| Comamonas | 0.00042 | 0.00272 | 32.15 | 100 | 90.8 | 86.27 | 89.44 | 66.49 | 3 |
| Conexibacter sp (uncultured) | 0.00047 | 0.00273 | 172.23 | 100 | 104.23 | 102.87 | 109.87 | 121.71 | 2.84 |
| Cupriavidus | 0.01615 | 0.03628 | 58.23 | 100 | 166.62 | 102.21 | 99.28 | 113.39 | 3.29 |
| Defluviicoccus | 0.01052 | 0.02557 | 135.73 | 100 | 95.59 | 103.09 | 133.61 | 145.59 | 2.72 |
| Desmochloris halophila | 0.00010 | 0.00136 | 2.95 | 100 | 68.61 | 69.18 | 92.97 | 53.81 | 3.3 |
| Desulfuromonadales bacterium (uncultured) | 0.00284 | 0.00970 | 150.31 | 100 | 94.1 | 97.25 | 141.9 | 125.06 | 3.14 |
| Devosia | 0.00002 | 0.00064 | 43.37 | 100 | 116.16 | 84.75 | 96.61 | 79.58 | 4.23 |
| Dongia | 0.00144 | 0.00529 | 152.4 | 100 | 97.97 | 94.07 | 90.74 | 79.92 | 3.76 |
| Dyadobacter | 0.00067 | 0.00325 | 24.09 | 100 | 52.01 | 85.51 | 40.29 | 42.91 | 3.62 |
| Ensifer | 0.00133 | 0.00518 | 56.24 | 100 | 100.02 | 107.48 | 92.2 | 100.64 | 3.77 |
| Ferrimicrobium sp (uncultured) | 0.03729 | 0.07311 | 125.56 | 100 | 99.81 | 102.37 | 109 | 84.16 | 2.91 |
| Fictibacillus | 0.00011 | 0.00136 | 215.28 | 100 | 143.63 | 101.77 | 122.05 | 145.88 | 3.74 |
| Flavitalea | 0.00003 | 0.00067 | 50 | 100 | 106.91 | 117.12 | 103.23 | 99.75 | 3.21 |
| Gaiella | 0.00065 | 0.00325 | 167.43 | 100 | 103.51 | 98.84 | 100.35 | 110.84 | 4.91 |
| Gaiella sp (uncultured) | 0.00782 | 0.01994 | 147.16 | 100 | 99.04 | 109.57 | 99.51 | 122.51 | 3.78 |
| Gemmatimonadales bacterium (uncultured) | 0.04590 | 0.08447 | 135.66 | 100 | 99.06 | 87.95 | 102.07 | 79.89 | 3.31 |
| Gemmatimonas | 0.00017 | 0.00147 | 58.34 | 100 | 94.84 | 103.94 | 95.71 | 86.76 | 3.46 |
| Herbidospora | 0.04913 | 0.08761 | 118.11 | 100 | 169.95 | 105.78 | 178.99 | 156.05 | 2.75 |
| Herbinix | 0.00344 | 0.01114 | 191.15 | 100 | 109.08 | 85.52 | 121.9 | 128.06 | 2.82 |
| Hirschia | 0.00028 | 0.00207 | 27.84 | 100 | 244.67 | 116.14 | 124.26 | 87.46 | 3.65 |
| Iamia | 0.01109 | 0.02647 | 98.66 | 100 | 94.03 | 80.75 | 73.31 | 72.36 | 3.52 |
| Ideonella | 0.00021 | 0.00165 | 23.92 | 100 | 179.52 | 116.28 | 70.3 | 91.63 | 3.06 |
| Knoellia | 0.03739 | 0.07311 | 116.7 | 100 | 92.24 | 96.06 | 108.13 | 113.72 | 4.11 |
| Kribbella | 0.00435 | 0.01326 | 128.87 | 100 | 99.2 | 93.79 | 93.83 | 98.98 | 2.95 |
| Lacibacter | 0.00211 | 0.00736 | 38.32 | 100 | 76.56 | 83.26 | 60.05 | 61.93 | 3.2 |
| Lechevalieria | 0.00046 | 0.00273 | 39.59 | 100 | 79.48 | 59.98 | 56.89 | 59.06 | 3.79 |
| Leptolyngbya EcFYyyy 00 | 0.00002 | 0.00064 | 4.67 | 100 | 181.25 | 345.68 | 193.37 | 227.61 | 4.48 |
| Litorilinea | 0.03752 | 0.07311 | 82.33 | 100 | 97.64 | 105 | 105.67 | 125.7 | 3.29 |
| Luedemannella | 0.00007 | 0.00108 | 253.14 | 100 | 115.04 | 108.58 | 141.63 | 136.22 | 3.39 |
| Luteitalea | 0.00135 | 0.00518 | 79.83 | 100 | 107.37 | 112.13 | 138.84 | 120.68 | 3.32 |
| Luteolibacter | 0.00039 | 0.00258 | 48.68 | 100 | 113.02 | 90.52 | 82.35 | 79.01 | 3.56 |
| Lutispora sp (uncultured) | 0.00067 | 0.00325 | 358.38 | 100 | 91.07 | 126.06 | 148.23 | 141.14 | 3.61 |
| Marmoricola | 0.00515 | 0.01487 | 128.62 | 100 | 100.61 | 94.63 | 91.06 | 97.39 | 4.08 |
| Mesorhizobium | 0.00002 | 0.00067 | 51.07 | 100 | 142.66 | 104.14 | 119.92 | 100.66 | 4.17 |
| metagenome | 0.00406 | 0.01251 | 126.64 | 100 | 106.18 | 102.6 | 104.97 | 100.4 | 4.28 |
| Methylobacillus | 0.00011 | 0.00136 | 1.58 | 100 | 40.6 | 59.21 | 32.84 | 51.74 | 3.26 |
| Methylotenera | 0.00009 | 0.00136 | 9.73 | 100 | 46.83 | 58.19 | 40.31 | 49.08 | 3.42 |
| Microbacterium | 0.00372 | 0.01186 | 31.46 | 100 | 69.82 | 64.65 | 80.59 | 63.4 | 3.84 |
| Microlunatus | 0.04250 | 0.08045 | 113.16 | 100 | 104.03 | 109.98 | 120.16 | 133.05 | 3.74 |
| Microvirga | 0.04487 | 0.08314 | 82.36 | 100 | 109.38 | 114.48 | 102.62 | 127.32 | 4.55 |
| Mitsuaria | 0.00001 | 0.00045 | 1.12 | 100 | 44.81 | 120.62 | 36.03 | 37.36 | 3.94 |
| Mycobacterium | 0.00012 | 0.00136 | 129.56 | 100 | 111.99 | 96.9 | 90.08 | 97.28 | 3.72 |
| Nakamurella | 0.04455 | 0.08314 | 93.82 | 100 | 104.26 | 117.18 | 122.71 | 154.79 | 2.53 |
| Niastella | 0.00023 | 0.00182 | 33.02 | 100 | 99.51 | 103.95 | 90.71 | 84.85 | 2.96 |
| Nitrosospira | 0.04718 | 0.08622 | 77.97 | 100 | 119.68 | 129.14 | 146.64 | 196.27 | 3.01 |
| Nitrospira | 0.00112 | 0.00474 | 77.66 | 100 | 109.43 | 116.7 | 128.8 | 110.67 | 4.3 |
| Nocardia | 0.02207 | 0.04605 | 57.7 | 100 | 134.15 | 98.74 | 81.83 | 103.54 | 2.87 |
| Nocardioides | 0.04227 | 0.08045 | 123.22 | 100 | 90.13 | 89.72 | 92.46 | 93.96 | 4.15 |
| Nodosilinea PCC 7104 | 0.00001 | 0.00046 | 2.3 | 100 | 141.37 | 125.12 | 119.09 | 212.3 | 3.82 |
| Nonomuraea | 0.02141 | 0.04575 | 169.66 | 100 | 110.37 | 120.29 | 128.68 | 137.75 | 2.89 |
| Nordella | 0.02831 | 0.05771 | 80.52 | 100 | 101.81 | 96.02 | 95.62 | 76.75 | 3.65 |
| Ohtaekwangia | 0.00037 | 0.00258 | 38.79 | 100 | 117.71 | 83.19 | 75.22 | 66.2 | 3.51 |
| Paenisporosarcina | 0.00535 | 0.01498 | 150.58 | 100 | 113.19 | 95.02 | 117.4 | 137.32 | 3.35 |
| Parasegetibacter | 0.00067 | 0.00325 | 49.91 | 100 | 97.89 | 123.06 | 98.69 | 136.38 | 3.27 |
| Paucibacter | 0.02168 | 0.04597 | 39.8 | 100 | 86.28 | 88.54 | 83.37 | 89.06 | 2.71 |
| Pelobacter sp (uncultured) | 0.03482 | 0.06990 | 80.54 | 100 | 118.43 | 131.55 | 87.71 | 58.76 | 2.73 |
| Planctomicrobium | 0.02204 | 0.04605 | 44.17 | 100 | 82.14 | 73.55 | 64.79 | 60.46 | 2.82 |
| Planctomycetales bacterium (uncultured) | 0.00765 | 0.01984 | 41.84 | 100 | 97.37 | 89.11 | 84.48 | 75.99 | 2.81 |
| planctomycete (uncultured) | 0.00016 | 0.00147 | 47.07 | 100 | 93.7 | 90.29 | 81.74 | 77.56 | 4.21 |
| Polyangium brachysporum group | 0.00006 | 0.00104 | 22.52 | 100 | 105.79 | 73.28 | 63.27 | 66.98 | 3.44 |
| Promicromonospora | 0.00656 | 0.01757 | 141.62 | 100 | 130.66 | 70.36 | 73.57 | 85.18 | 3.17 |
| Pseudarthrobacter | 0.00094 | 0.00421 | 74.03 | 100 | 72.78 | 110.37 | 102.13 | 118.78 | 2.99 |
| Pseudoduganella | 0.00000 | 0.00045 | 13.27 | 100 | 83.36 | 125.02 | 54.66 | 49.32 | 3.46 |
| Pseudoflavitalea | 0.01219 | 0.02860 | 80.45 | 100 | 185.76 | 116.56 | 157.75 | 90.08 | 2.79 |
| Pseudomuriella schumacherensis | 0.00013 | 0.00138 | 12.08 | 100 | 115.92 | 135.78 | 122.24 | 139.79 | 3.35 |
| Pseudorhodoplanes | 0.00011 | 0.00136 | 59.18 | 100 | 129.76 | 116.62 | 130.89 | 99.57 | 2.89 |
| Pseudoxanthomonas | 0.00003 | 0.00067 | 24.58 | 100 | 105.41 | 76.92 | 57.18 | 51.07 | 3.85 |
| Qipengyuania | 0.01122 | 0.02654 | 41.8 | 100 | 78.31 | 92.86 | 68.83 | 89.95 | 3.59 |
| Ramlibacter | 0.00010 | 0.00136 | 44.73 | 100 | 97.21 | 101.6 | 80.83 | 100 | 3.94 |
| Reyranella | 0.01046 | 0.02557 | 75.78 | 100 | 114.04 | 107.61 | 101.27 | 82.04 | 3.21 |
| Rhizobacter | 0.00015 | 0.00146 | 50.3 | 100 | 99.96 | 93.82 | 89.44 | 95.04 | 3.87 |
| Rhodocytophaga | 0.00182 | 0.00653 | 51.47 | 100 | 92.75 | 115.87 | 104.1 | 124.01 | 3.18 |
| Rhodopirellula | 0.00006 | 0.00104 | 35.27 | 100 | 98.67 | 93.02 | 76.95 | 100.97 | 3.58 |
| Roseimicrobium | 0.03548 | 0.07069 | 77.08 | 100 | 87.47 | 93.95 | 87.24 | 68.74 | 3.18 |
| Roseomonas | 0.02030 | 0.04374 | 63.83 | 100 | 106.38 | 116.85 | 114.31 | 150.41 | 2.97 |
| Rubellimicrobium | 0.00140 | 0.00529 | 54.35 | 100 | 86.83 | 113.4 | 108.13 | 142.15 | 3.97 |
| Rubrobacterales bacterium (uncultured) | 0.00053 | 0.00299 | 170.17 | 100 | 101.18 | 87.23 | 86.5 | 98.49 | 4.34 |
| Rubrobacteria bacterium (uncultured) | 0.00345 | 0.01114 | 154.24 | 100 | 105.92 | 96.08 | 93.64 | 110.38 | 3.78 |
| Shimazuella | 0.02890 | 0.05845 | 156.72 | 100 | 98.45 | 126.81 | 126.74 | 135.07 | 2.68 |
| Shinella | 0.00003 | 0.00067 | 15.43 | 100 | 90.74 | 79.41 | 58.82 | 61.59 | 3.52 |
| Skermanella | 0.04926 | 0.08761 | 103.72 | 100 | 104.15 | 117.66 | 140.62 | 153.82 | 4.21 |
| Solirubrobacter | 0.02018 | 0.04374 | 152.45 | 100 | 105.37 | 100.81 | 109.31 | 110.12 | 3.84 |
| Sorangium | 0.00618 | 0.01672 | 113.87 | 100 | 159.43 | 72.4 | 76.81 | 52.23 | 2.97 |
| Sphingobium | 0.00001 | 0.00045 | 3.54 | 100 | 68.9 | 78.07 | 55.92 | 64.43 | 4.53 |
| Sphingopyxis | 0.00457 | 0.01377 | 29.96 | 100 | 96.28 | 72.55 | 65.76 | 93.82 | 3.07 |
| Streptomyces | 0.00012 | 0.00136 | 63.89 | 100 | 94.25 | 73.41 | 80.46 | 80.56 | 4.11 |
| Sumerlaea | 0.00135 | 0.00518 | 55.89 | 100 | 100.03 | 112.36 | 106.62 | 98.09 | 3.03 |
| Synechococcus IR11 | 0.00015 | 0.00146 | 3.71 | 100 | 345.59 | 190.9 | 229.88 | 351.86 | 3.41 |
| Tahibacter | 0.00313 | 0.01051 | 25.57 | 100 | 97.63 | 56.71 | 43.88 | 51.86 | 2.94 |
| Terrimonas | 0.00067 | 0.00325 | 54.44 | 100 | 109.27 | 99.02 | 86.62 | 73.35 | 3.71 |
| Truepera | 0.00123 | 0.00503 | 21.18 | 100 | 120.76 | 119.24 | 134.5 | 94.77 | 3.34 |
| Tumebacillus | 0.00814 | 0.02054 | 235.61 | 100 | 145.58 | 117.75 | 199.46 | 185.96 | 3.6 |
| Variovorax | 0.00038 | 0.00258 | 44.65 | 100 | 84.87 | 75.73 | 68.27 | 59.26 | 3.64 |
| Yonghaparkia | 0.00682 | 0.01806 | 17.03 | 100 | 87.96 | 108.51 | 69.97 | 127.56 | 3.5 |
| Genus | p-Value | FDR | t0 | CFD | CRD | MYC | PSBA | PSBA + MYC | LDA Score |
|---|---|---|---|---|---|---|---|---|---|
| Acidibacter | 0.00026 | 0.00196 | 47.49 | 84.24 | 100 | 78.31 | 90.97 | 69.53 | 3.79 |
| Acidimicrobiia bacterium (uncultured) | 0.00034 | 0.00241 | 184.03 | 93.3 | 100 | 88.67 | 97.47 | 100.69 | 4.09 |
| Acidobacteriales bacterium (uncultured) | 0.01264 | 0.02937 | 133.04 | 92.97 | 100 | 103.37 | 119.83 | 88.16 | 4.19 |
| Acidovorax | 0.00006 | 0.00104 | 10.7 | 144.23 | 100 | 186.21 | 99.86 | 71.78 | 3.11 |
| Actinobacterium (uncultured) | 0.00567 | 0.01549 | 135.6 | 89.02 | 100 | 84.81 | 79.9 | 81.08 | 4.02 |
| Aeromicrobium | 0.00018 | 0.00155 | 42.73 | 114.29 | 100 | 98.18 | 68.83 | 74.95 | 4.49 |
| Allorhizobium Neorhizobium Pararhizobium Rhizobium | 0.00113 | 0.00474 | 49.58 | 102.8 | 100 | 96.36 | 84.22 | 90.84 | 4.36 |
| Alsobacter | 0.01434 | 0.03257 | 80.28 | 88.34 | 100 | 120.43 | 116.01 | 138.23 | 2.97 |
| Altererythrobacter | 0.00014 | 0.00146 | 25.16 | 73.04 | 100 | 59.7 | 62.94 | 59.27 | 3.51 |
| Amaricoccus | 0.00851 | 0.02128 | 82.17 | 93.31 | 100 | 104.36 | 122.11 | 140.81 | 3.1 |
| Aminobacter | 0.00000 | 0.00000 | 56.52 | 104.17 | 100 | 384.33 | 387.49 | 429.17 | 3.34 |
| Amycolatopsis | 0.00103 | 0.00456 | 19.71 | 282.86 | 100 | 136.46 | 98.55 | 159.57 | 3.45 |
| Anaerolinea | 0.01738 | 0.03871 | 413.32 | 118.98 | 100 | 128.86 | 137.22 | 171.19 | 3.58 |
| Anaeromyxobacter | 0.00021 | 0.00165 | 196.36 | 101.94 | 100 | 103.89 | 144.5 | 120.64 | 3.58 |
| Arenimonas | 0.00057 | 0.00307 | 45.84 | 125.28 | 100 | 79.32 | 77.17 | 80.92 | 3.27 |
| Aridibacter | 0.01313 | 0.03025 | 71.67 | 114.53 | 100 | 102.63 | 97.75 | 127.88 | 3.27 |
| Armatimonadetes bacterium (uncultured) | 0.01982 | 0.04340 | 140.26 | 95.27 | 100 | 104.52 | 106.61 | 121.45 | 2.87 |
| Arthrobacter | 0.00067 | 0.00325 | 100.74 | 118.7 | 100 | 131.59 | 133.61 | 153.97 | 5.2 |
| Azohydromonas | 0.00193 | 0.00683 | 42.24 | 101.4 | 100 | 96.45 | 84.3 | 88.45 | 3.34 |
| Blastocatella | 0.00530 | 0.01498 | 75.04 | 112.25 | 100 | 113.85 | 99.63 | 115.67 | 3.35 |
| Bosea | 0.00107 | 0.00466 | 45.05 | 93.46 | 100 | 101.22 | 101.6 | 104.66 | 3.54 |
| Bradyrhizobium | 0.00047 | 0.00273 | 63.07 | 89.33 | 100 | 89.13 | 96.98 | 78.59 | 3.89 |
| Burkholderia Caballeronia Paraburkholderia | 0.00876 | 0.02171 | 158.69 | 93.57 | 100 | 68.37 | 60.25 | 58.15 | 3.48 |
| Caenimonas | 0.00377 | 0.01188 | 58.17 | 107.03 | 100 | 104.01 | 93.48 | 112.53 | 3.94 |
| Cellulomonas | 0.04245 | 0.08045 | 130.66 | 86.41 | 100 | 109.23 | 140.52 | 152.78 | 2.71 |
| Chloroflexi bacterium (uncultured) | 0.00771 | 0.01984 | 126.33 | 100.7 | 100 | 103.14 | 101.31 | 115.45 | 4.09 |
| Chloroflexus sp (uncultured) | 0.00001 | 0.00045 | 113.54 | 108.21 | 100 | 83.03 | 63.88 | 78.74 | 3.79 |
| Chthonomonas | 0.02807 | 0.05767 | 76.73 | 99.81 | 100 | 92.33 | 100.55 | 115.63 | 2.53 |
| Clostridium sensu stricto 1 | 0.01438 | 0.03257 | 211.86 | 106.21 | 100 | 115.65 | 113.78 | 118.68 | 3.13 |
| Clostridium sensu stricto 8 | 0.02672 | 0.05533 | 171.93 | 91.79 | 100 | 97.72 | 116.37 | 116.9 | 2.78 |
| Cohnella | 0.00127 | 0.00509 | 151.65 | 81.14 | 100 | 92.89 | 125.39 | 132.53 | 3.35 |
| Comamonas | 0.00042 | 0.00272 | 35.41 | 110.13 | 100 | 95 | 98.5 | 73.23 | 3 |
| Conexibacter sp (uncultured) | 0.00047 | 0.00273 | 165.24 | 95.95 | 100 | 98.7 | 105.42 | 116.78 | 2.84 |
| Cupriavidus | 0.01615 | 0.03628 | 34.94 | 60.02 | 100 | 61.34 | 59.59 | 68.05 | 3.29 |
| Defluviicoccus | 0.01052 | 0.02557 | 141.99 | 104.61 | 100 | 107.84 | 139.77 | 152.29 | 2.72 |
| Desmochloris halophila | 0.00010 | 0.00136 | 4.3 | 145.76 | 100 | 100.84 | 135.51 | 78.43 | 3.3 |
| Desulfuromonadales bacterium (uncultured) | 0.00284 | 0.00970 | 159.74 | 106.27 | 100 | 103.35 | 150.8 | 132.9 | 3.14 |
| Devosia | 0.00002 | 0.00064 | 37.33 | 86.08 | 100 | 72.95 | 83.17 | 68.5 | 4.23 |
| Dongia | 0.00144 | 0.00529 | 155.56 | 102.08 | 100 | 96.03 | 92.62 | 81.58 | 3.76 |
| Dyadobacter | 0.00067 | 0.00325 | 46.33 | 192.28 | 100 | 164.42 | 77.47 | 82.51 | 3.62 |
| Ensifer | 0.00133 | 0.00518 | 56.23 | 99.98 | 100 | 107.46 | 92.19 | 100.62 | 3.77 |
| Ferrimicrobium sp (uncultured) | 0.03729 | 0.07311 | 125.8 | 100.19 | 100 | 102.57 | 109.2 | 84.32 | 2.91 |
| Fictibacillus | 0.00011 | 0.00136 | 149.89 | 69.62 | 100 | 70.86 | 84.97 | 101.57 | 3.74 |
| Flavitalea | 0.00003 | 0.00067 | 46.77 | 93.54 | 100 | 109.55 | 96.56 | 93.3 | 3.21 |
| Gaiella | 0.00065 | 0.00325 | 161.75 | 96.61 | 100 | 95.49 | 96.94 | 107.08 | 4.91 |
| Gaiella sp (uncultured) | 0.00782 | 0.01994 | 148.59 | 100.97 | 100 | 110.63 | 100.48 | 123.7 | 3.78 |
| Gemmatimonadales bacterium (uncultured) | 0.04590 | 0.08447 | 136.95 | 100.95 | 100 | 88.79 | 103.04 | 80.65 | 3.31 |
| Gemmatimonas | 0.00017 | 0.00147 | 61.52 | 105.44 | 100 | 109.59 | 100.91 | 91.48 | 3.46 |
| Herbidospora | 0.04913 | 0.08761 | 69.5 | 58.84 | 100 | 62.24 | 105.32 | 91.82 | 2.75 |
| Herbinix | 0.00344 | 0.01114 | 175.23 | 91.67 | 100 | 78.4 | 111.75 | 117.4 | 2.82 |
| Hirschia | 0.00028 | 0.00207 | 11.38 | 40.87 | 100 | 47.47 | 50.79 | 35.75 | 3.65 |
| Iamia | 0.01109 | 0.02647 | 104.92 | 106.35 | 100 | 85.88 | 77.96 | 76.95 | 3.52 |
| Ideonella | 0.00021 | 0.00165 | 13.32 | 55.7 | 100 | 64.77 | 39.16 | 51.04 | 3.06 |
| Knoellia | 0.03739 | 0.07311 | 126.51 | 108.41 | 100 | 104.15 | 117.23 | 123.29 | 4.11 |
| Kribbella | 0.00435 | 0.01326 | 129.91 | 100.81 | 100 | 94.55 | 94.59 | 99.78 | 2.95 |
| Lacibacter | 0.00211 | 0.00736 | 50.06 | 130.62 | 100 | 108.76 | 78.43 | 80.89 | 3.2 |
| Lechevalieria | 0.00046 | 0.00273 | 49.81 | 125.82 | 100 | 75.47 | 71.58 | 74.32 | 3.79 |
| Leptolyngbya EcFYyyy 00 | 0.00002 | 0.00064 | 2.58 | 55.17 | 100 | 190.72 | 106.69 | 125.58 | 4.48 |
| Litorilinea | 0.03752 | 0.07311 | 84.33 | 102.42 | 100 | 107.54 | 108.23 | 128.75 | 3.29 |
| Luedemannella | 0.00007 | 0.00108 | 220.05 | 86.93 | 100 | 94.38 | 123.12 | 118.41 | 3.39 |
| Luteitalea | 0.00135 | 0.00518 | 74.35 | 93.13 | 100 | 104.43 | 129.31 | 112.39 | 3.32 |
| Luteolibacter | 0.00039 | 0.00258 | 43.07 | 88.48 | 100 | 80.09 | 72.86 | 69.91 | 3.56 |
| Lutispora sp (uncultured) | 0.00067 | 0.00325 | 393.53 | 109.81 | 100 | 138.42 | 162.76 | 154.98 | 3.61 |
| Marmoricola | 0.00515 | 0.01487 | 127.84 | 99.39 | 100 | 94.06 | 90.51 | 96.8 | 4.08 |
| Mesorhizobium | 0.00002 | 0.00067 | 35.8 | 70.1 | 100 | 73 | 84.06 | 70.56 | 4.17 |
| metagenome | 0.00406 | 0.01251 | 119.27 | 94.18 | 100 | 96.63 | 98.86 | 94.55 | 4.28 |
| Methylobacillus | 0.00011 | 0.00136 | 3.88 | 246.31 | 100 | 145.84 | 80.89 | 127.43 | 3.26 |
| Methylotenera | 0.00009 | 0.00136 | 20.78 | 213.54 | 100 | 124.26 | 86.08 | 104.81 | 3.42 |
| Microbacterium | 0.00372 | 0.01186 | 45.06 | 143.23 | 100 | 92.6 | 115.43 | 90.81 | 3.84 |
| Microlunatus | 0.04250 | 0.08045 | 108.78 | 96.12 | 100 | 105.72 | 115.5 | 127.89 | 3.74 |
| Microvirga | 0.04487 | 0.08314 | 75.29 | 91.42 | 100 | 104.66 | 93.82 | 116.4 | 4.55 |
| Mitsuaria | 0.00001 | 0.00045 | 2.51 | 223.14 | 100 | 269.15 | 80.39 | 83.36 | 3.94 |
| Mycobacterium | 0.00012 | 0.00136 | 115.7 | 89.3 | 100 | 86.53 | 80.44 | 86.87 | 3.72 |
| Nakamurella | 0.04455 | 0.08314 | 89.99 | 95.91 | 100 | 112.39 | 117.69 | 148.46 | 2.53 |
| Niastella | 0.00023 | 0.00182 | 33.18 | 100.49 | 100 | 104.47 | 91.16 | 85.26 | 2.96 |
| Nitrosospira | 0.04718 | 0.08622 | 65.15 | 83.55 | 100 | 107.9 | 122.53 | 163.99 | 3.01 |
| Nitrospira | 0.00112 | 0.00474 | 70.97 | 91.38 | 100 | 106.64 | 117.7 | 101.14 | 4.3 |
| Nocardia | 0.02207 | 0.04605 | 43.01 | 74.54 | 100 | 73.6 | 61 | 77.19 | 2.87 |
| Nocardioides | 0.04227 | 0.08045 | 136.71 | 110.95 | 100 | 99.54 | 102.59 | 104.24 | 4.15 |
| Nodosilinea PCC 7104 | 0.00001 | 0.00046 | 1.63 | 70.73 | 100 | 88.5 | 84.23 | 150.17 | 3.82 |
| Nonomuraea | 0.02141 | 0.04575 | 153.72 | 90.61 | 100 | 108.99 | 116.59 | 124.81 | 2.89 |
| Nordella | 0.02831 | 0.05771 | 79.09 | 98.22 | 100 | 94.32 | 93.92 | 75.39 | 3.65 |
| Ohtaekwangia | 0.00037 | 0.00258 | 32.95 | 84.95 | 100 | 70.68 | 63.9 | 56.24 | 3.51 |
| Paenisporosarcina | 0.00535 | 0.01498 | 133.03 | 88.35 | 100 | 83.95 | 103.72 | 121.32 | 3.35 |
| Parasegetibacter | 0.00067 | 0.00325 | 50.99 | 102.16 | 100 | 125.72 | 100.82 | 139.33 | 3.27 |
| Paucibacter | 0.02168 | 0.04597 | 46.12 | 115.9 | 100 | 102.61 | 96.62 | 103.22 | 2.71 |
| Pelobacter sp (uncultured) | 0.03482 | 0.06990 | 68.01 | 84.44 | 100 | 111.08 | 74.06 | 49.62 | 2.73 |
| Planctomicrobium | 0.02204 | 0.04605 | 53.77 | 121.74 | 100 | 89.54 | 78.87 | 73.61 | 2.82 |
| Planctomycetales bacterium (uncultured) | 0.00765 | 0.01984 | 42.97 | 102.71 | 100 | 91.52 | 86.76 | 78.05 | 2.81 |
| planctomycete (uncultured) | 0.00016 | 0.00147 | 50.24 | 106.73 | 100 | 96.36 | 87.23 | 82.77 | 4.21 |
| Polyangium brachysporum group | 0.00006 | 0.00104 | 21.28 | 94.53 | 100 | 69.26 | 59.8 | 63.31 | 3.44 |
| Promicromonospora | 0.00656 | 0.01757 | 108.4 | 76.54 | 100 | 53.85 | 56.31 | 65.19 | 3.17 |
| Pseudarthrobacter | 0.00094 | 0.00421 | 101.72 | 137.41 | 100 | 151.66 | 140.34 | 163.22 | 2.99 |
| Pseudoduganella | 0.00000 | 0.00045 | 15.92 | 119.97 | 100 | 149.98 | 65.57 | 59.17 | 3.46 |
| Pseudoflavitalea | 0.01219 | 0.02860 | 43.31 | 53.83 | 100 | 62.75 | 84.92 | 48.49 | 2.79 |
| Pseudomuriella schumacherensis | 0.00013 | 0.00138 | 10.42 | 86.26 | 100 | 117.13 | 105.45 | 120.59 | 3.35 |
| Pseudorhodoplanes | 0.00011 | 0.00136 | 45.61 | 77.07 | 100 | 89.87 | 100.87 | 76.74 | 2.89 |
| Pseudoxanthomonas | 0.00003 | 0.00067 | 23.32 | 94.87 | 100 | 72.97 | 54.25 | 48.45 | 3.85 |
| Qipengyuania | 0.01122 | 0.02654 | 53.38 | 127.7 | 100 | 118.58 | 87.89 | 114.86 | 3.59 |
| Ramlibacter | 0.00010 | 0.00136 | 46.02 | 102.88 | 100 | 104.52 | 83.15 | 102.87 | 3.94 |
| Reyranella | 0.01046 | 0.02557 | 66.45 | 87.69 | 100 | 94.36 | 88.8 | 71.94 | 3.21 |
| Rhizobacter | 0.00015 | 0.00146 | 50.32 | 100.04 | 100 | 93.86 | 89.48 | 95.08 | 3.87 |
| Rhodocytophaga | 0.00182 | 0.00653 | 55.49 | 107.82 | 100 | 124.93 | 112.24 | 133.7 | 3.18 |
| Rhodopirellula | 0.00006 | 0.00104 | 35.75 | 101.35 | 100 | 94.27 | 77.99 | 102.33 | 3.58 |
| Roseimicrobium | 0.03548 | 0.07069 | 88.12 | 114.32 | 100 | 107.4 | 99.73 | 78.58 | 3.18 |
| Roseomonas | 0.02030 | 0.04374 | 60 | 94 | 100 | 109.84 | 107.45 | 141.39 | 2.97 |
| Rubellimicrobium | 0.00140 | 0.00529 | 62.6 | 115.16 | 100 | 130.59 | 124.53 | 163.71 | 3.97 |
| Rubrobacterales bacterium (uncultured) | 0.00053 | 0.00299 | 168.19 | 98.83 | 100 | 86.21 | 85.49 | 97.34 | 4.34 |
| Rubrobacteria bacterium (uncultured) | 0.00345 | 0.01114 | 145.62 | 94.41 | 100 | 90.71 | 88.41 | 104.21 | 3.78 |
| Shimazuella | 0.02890 | 0.05845 | 159.19 | 101.58 | 100 | 128.81 | 128.74 | 137.2 | 2.68 |
| Shinella | 0.00003 | 0.00067 | 17.01 | 110.2 | 100 | 87.51 | 64.82 | 67.87 | 3.52 |
| Skermanella | 0.04926 | 0.08761 | 99.59 | 96.01 | 100 | 112.97 | 135.01 | 147.69 | 4.21 |
| Solirubrobacter | 0.02018 | 0.04374 | 144.68 | 94.9 | 100 | 95.67 | 103.74 | 104.5 | 3.84 |
| Sorangium | 0.00618 | 0.01672 | 71.42 | 62.72 | 100 | 45.41 | 48.18 | 32.76 | 2.97 |
| Sphingobium | 0.00001 | 0.00045 | 5.14 | 145.15 | 100 | 113.31 | 81.16 | 93.52 | 4.53 |
| Sphingopyxis | 0.00457 | 0.01377 | 31.12 | 103.86 | 100 | 75.36 | 68.31 | 97.44 | 3.07 |
| Streptomyces | 0.00012 | 0.00136 | 67.78 | 106.1 | 100 | 77.89 | 85.36 | 85.47 | 4.11 |
| Sumerlaea | 0.00135 | 0.00518 | 55.88 | 99.97 | 100 | 112.33 | 106.59 | 98.06 | 3.03 |
| Synechococcus IR11 | 0.00015 | 0.00146 | 1.07 | 28.94 | 100 | 55.24 | 66.52 | 101.81 | 3.41 |
| Tahibacter | 0.00313 | 0.01051 | 26.19 | 102.42 | 100 | 58.08 | 44.95 | 53.12 | 2.94 |
| Terrimonas | 0.00067 | 0.00325 | 49.82 | 91.51 | 100 | 90.62 | 79.27 | 67.13 | 3.71 |
| Truepera | 0.00123 | 0.00503 | 17.54 | 82.81 | 100 | 98.74 | 111.38 | 78.48 | 3.34 |
| Tumebacillus | 0.00814 | 0.02054 | 161.84 | 68.69 | 100 | 80.88 | 137.01 | 127.73 | 3.6 |
| Variovorax | 0.00038 | 0.00258 | 52.61 | 117.83 | 100 | 89.24 | 80.44 | 69.83 | 3.64 |
| Yonghaparkia | 0.00682 | 0.01806 | 19.36 | 113.69 | 100 | 123.37 | 79.55 | 145.03 | 3.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasuelli, M.; Novello, G.; Gamalero, E.; Massa, N.; Gorrasi, S.; Sudiro, C.; Hochart, M.; Altissimo, A.; Vuolo, F.; Bona, E. PGPB and/or AM Fungi Consortia Affect Tomato Native Rhizosphere Microbiota. Microorganisms 2023, 11, 1891. https://doi.org/10.3390/microorganisms11081891
Nasuelli M, Novello G, Gamalero E, Massa N, Gorrasi S, Sudiro C, Hochart M, Altissimo A, Vuolo F, Bona E. PGPB and/or AM Fungi Consortia Affect Tomato Native Rhizosphere Microbiota. Microorganisms. 2023; 11(8):1891. https://doi.org/10.3390/microorganisms11081891
Chicago/Turabian StyleNasuelli, Martina, Giorgia Novello, Elisa Gamalero, Nadia Massa, Susanna Gorrasi, Cristina Sudiro, Marie Hochart, Adriano Altissimo, Francesco Vuolo, and Elisa Bona. 2023. "PGPB and/or AM Fungi Consortia Affect Tomato Native Rhizosphere Microbiota" Microorganisms 11, no. 8: 1891. https://doi.org/10.3390/microorganisms11081891
APA StyleNasuelli, M., Novello, G., Gamalero, E., Massa, N., Gorrasi, S., Sudiro, C., Hochart, M., Altissimo, A., Vuolo, F., & Bona, E. (2023). PGPB and/or AM Fungi Consortia Affect Tomato Native Rhizosphere Microbiota. Microorganisms, 11(8), 1891. https://doi.org/10.3390/microorganisms11081891










