Characterization of Host-Associated Microbiota and Isolation of Antagonistic Bacteria from Greater Amberjack (Seriola dumerili, Risso, 1810) Larvae
Abstract
1. Introduction
2. Materials and Methods
2.1. Rearing of Greater Amberjack Larvae
2.2. Amberjack Larvae Microbiota Characterization
2.3. Bacteria Isolation from Greater Amberjack Larvae and Live Feed
2.4. Bacterial DNA Extraction and PCR Amplification
2.5. Inhibition Tests
3. Results
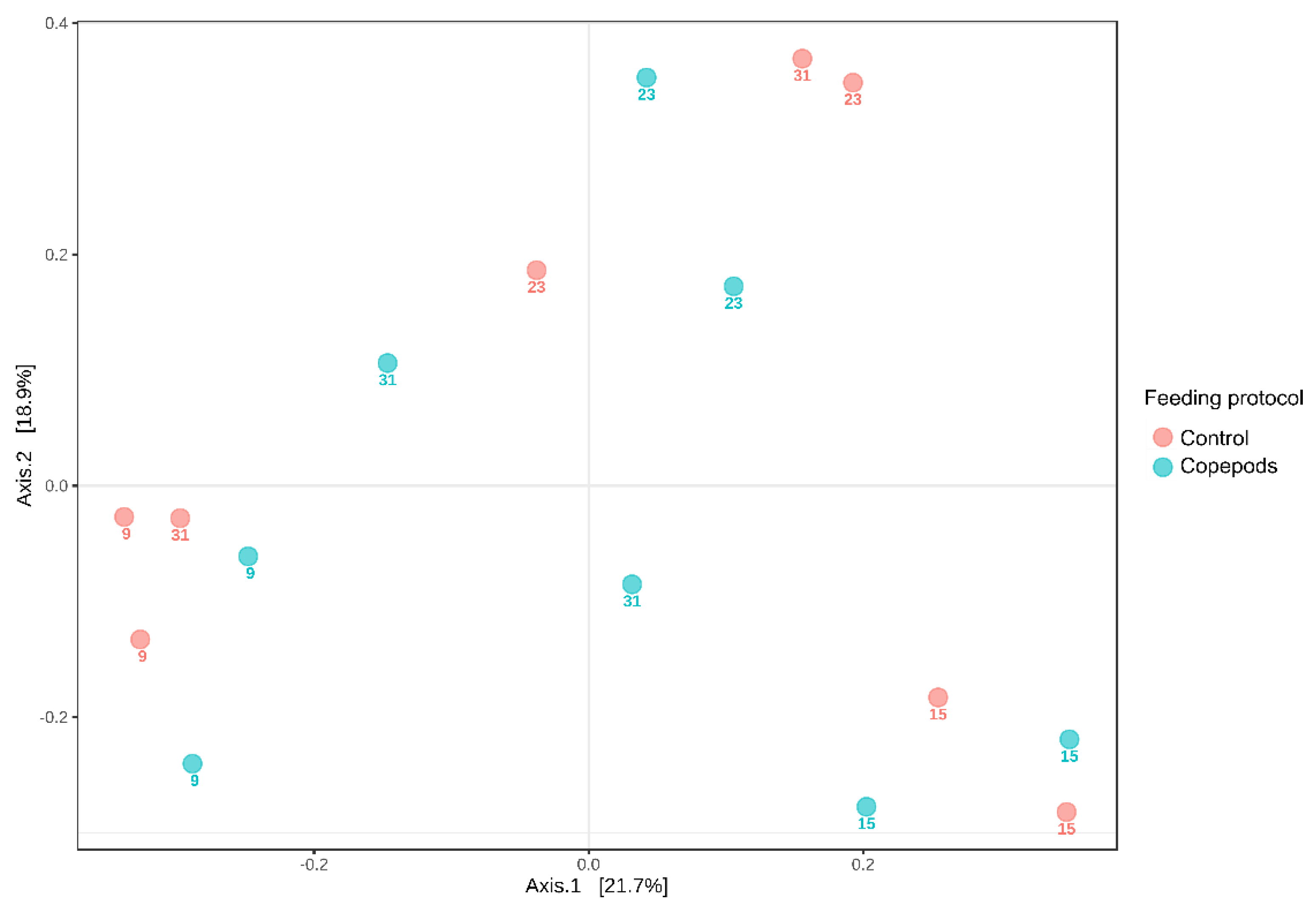
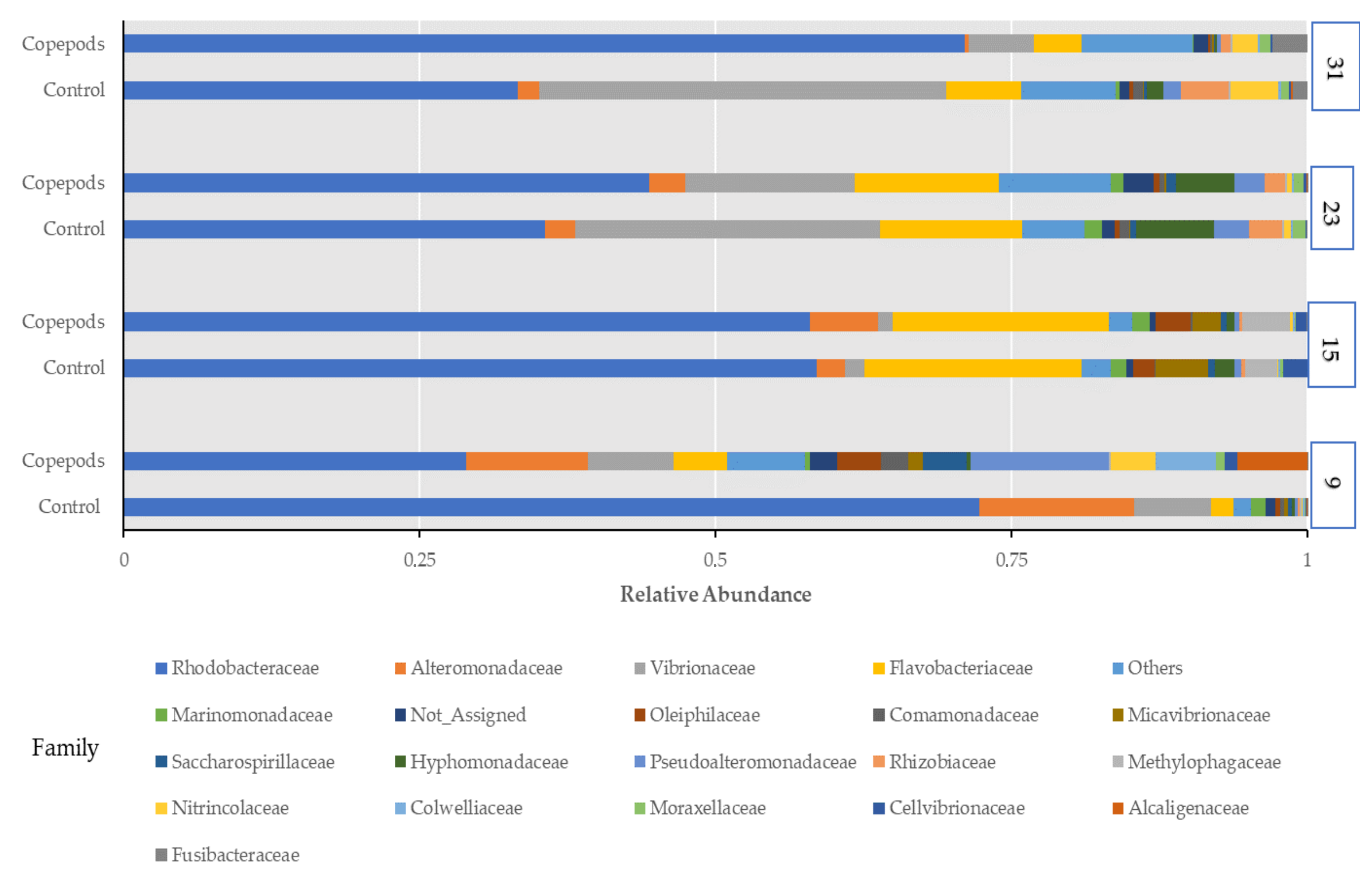
3.1. Microbiota Characterization of the Greater Amberjack Larvae
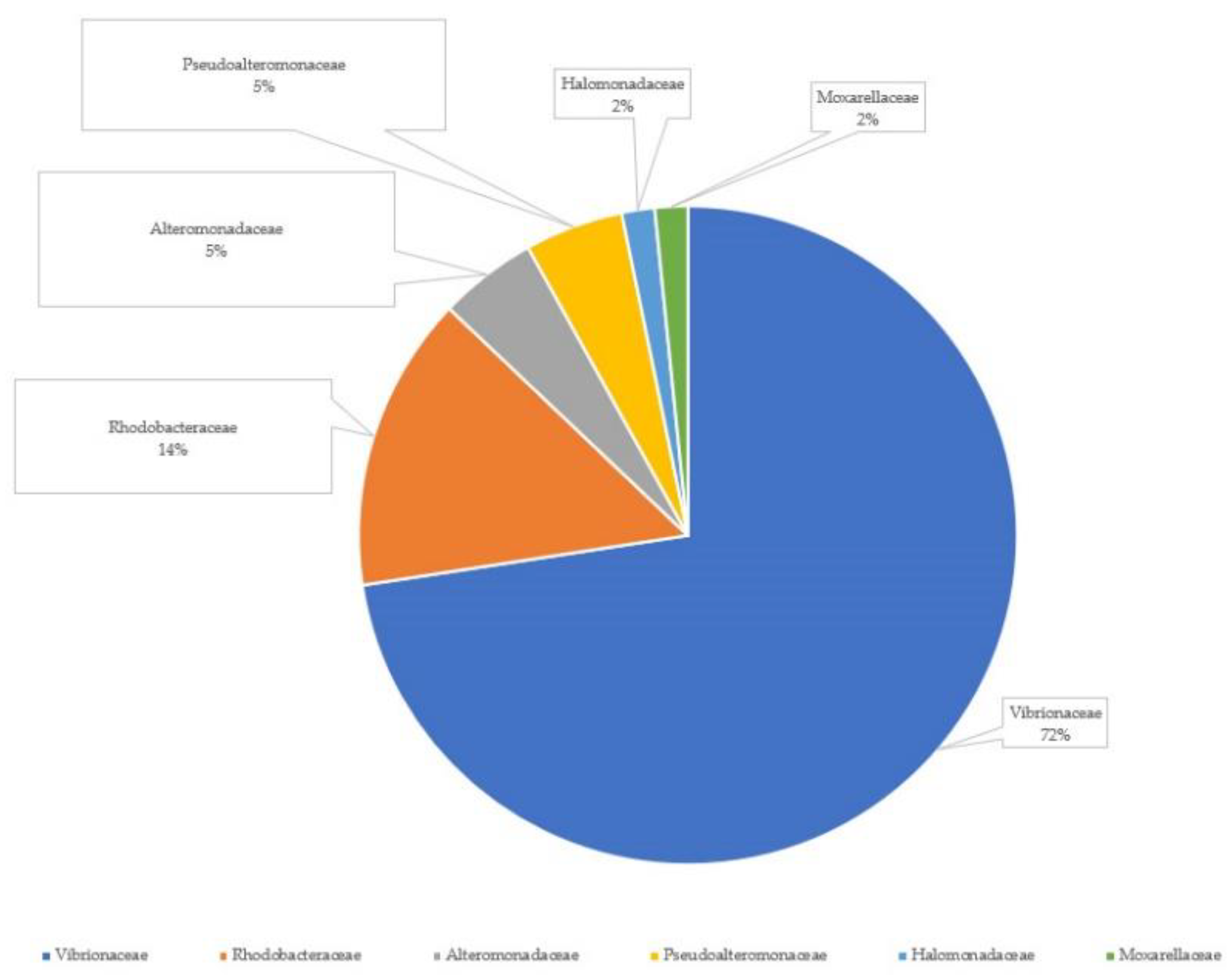
3.2. Bacterial Strains Isolated from Greater Amberjack Larvae and Live Feed
3.3. In Vitro Inhibition Tests against Pathogens
4. Discussion
4.1. Bacterial Dynamics Associated with the Early Developmental Stages of Greater Amberjack Larvae
4.2. Bacterial Composition and Functionality Associated with Different Feeding Protocols in Greater Amberjack Larvae
4.3. Isolation of Cultivable Antagonistic Bacteria with Probiotic Effect
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vadstein, O.; Øie, G.; Olsen, Y.; Salvesen, I.; Skermo, J. A Strategy to Obtain Microbial Control during Larval Development of Marine Fish; Fish Farming Technology, Reinertsen, H., Dahle, L.A., Jorgensen, L., Eds.; Balkema: Rotterdam, The Netherlands, 1993; ISBN 90 5410 3264. [Google Scholar]
- Borges, N.; Keller-Costa, T.; Sanches-Fernandes, G.M.M.; Louvado, A.; Gomes, N.C.M.; Costa, R. Bacteriome structure, function, and probiotics in fish larviculture: The good, the bad, and the gaps. Annu. Rev. Anim. Biosci. 2021, 9, 423–452. [Google Scholar] [CrossRef]
- Tarnecki, A.M.; Burgos, F.A.; Ray, C.L.; Arias, C.R. Fish intestinal microbiome: Diversity and symbiosis unravelled by metagenomics. J. Appl. Microbiol. 2017, 123, 2–17. [Google Scholar] [CrossRef]
- Vadstein, O.; Bergh, Ø.; Gatesoupe, F.-J.; Galindo-Villegas, J.; Mulero, V.; Picchietti, S.; Scapigliati, G.; Makridis, P.; Olsen, Y.; Dierckens, K.; et al. Microbiology and immunology of fish larvae. Rev. Aquacult. 2013, 5, S1–S25. [Google Scholar] [CrossRef]
- Nikouli, E.; Meziti, A.; Antonopoulou, E.; Mente, E.; Kormas, K.A. Host-associated bacterial succession during the early embryonic stages and first feeding in farmed gilthead sea bream (Sparus aurata). Genes 2019, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Califano, G.; Castanho, S.; Soares, F.; Ribeiro, L.; Cox, C.J.; Mata, L.; Costa, R. Molecular taxonomic profiling of bacterial communities in a gilthead seabream (Sparus aurata) hatchery. Front. Microbiol. 2017, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Le, D.; Nguyen, P.; Nguyen, D.; Dierckens, K.; Boon, N.; Lacoere, T.; Kerckhof, F.M.; De Vrieze, J.; Vadstein, O.; Bossier, P. Gut microbiota of migrating wild rabbit fish (Siganus guttatus) larvae have low spatial and temporal variability. Microb. Ecol. 2020, 79, 539–551. [Google Scholar] [CrossRef]
- Wilkes Walburn, J.; Wemheuer, B.; Thomas, T.; Copeland, E.; O’Connor, W.; Booth, M.; Fielder, S.; Egan, S. Diet and diet associated bacteria shape early microbiome development in yellowtail kingfish (Seriola lalandi). Microb. Biotechnol. 2019, 12, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Ingerslev, H.C.; von Gersdorff, J.; Lenz Strube, M.; Larsen, N.; Dalsgaard, I.; Boye, M.; Madsen, L. The development of the gut microbiota in rainbow trout (Oncorhynchus mykiss) is affected by first feeding and diet type. Aquaculture 2014, 424–425, 24–34. [Google Scholar] [CrossRef]
- Giatsis, C.; Sipkema, D.; Smidt, H.; Heilig, H.; Benvenuti, G.; Verreth, J.; Verdegem, M. The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci. Rep. 2015, 5, 18206. [Google Scholar] [CrossRef]
- Bakke, I.; Coward, E.; Andersen, T.; Vadstein, O. Selection in the host structures the microbiota associated with developing cod larvae (Gadus morhua). Environ. Microbiol. 2015, 17, 3914–3924. [Google Scholar] [CrossRef]
- Bledsoe, J.W.; Peterson, B.C.; Swanson, K.S.; Small, B.C. Ontogenetic characterization of the intestinal microbiota of channel catfish through 16S rRNA gene sequencing reveals insights on temporal shifts and the influence of environmental microbes. PLoS ONE 2016, 11, e0166379. [Google Scholar] [CrossRef] [PubMed]
- Lokesh, J.; Kiron, V.; Sipkema, D.; Fernandes, J.M.O.; Moum, T. Succession of embryonic and the intestinal bacterial communities of Atlantic salmon (Salmo salar) reveals stage-specific microbial signatures. Microbiol. Open 2019, 8, e00672. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.G.; Tian, J.Y.; Gatesoupe, F.J.; Li, W.X.; Zou, H.; Yang, B.J.; Wang, G.T. Intestinal microbiota of gibel carp (Carassius auratus gibelio) and its origin as revealed by 454 pyrosequencing. World J. Microbiol. Biotechnol. 2013, 29, 1585–1595. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, Y.; Feng, W.; Yan, Q.; and Gong, Y. Host species as a strong determinant of the intestinal microbiota of fish larvae. J. Microbiol. 2012, 50, 29–37. [Google Scholar] [CrossRef]
- Hansen, G.H.; Olafsen, J.A. Bacterial interactions in early life stages of marine cold water fish. Microb. Ecol. 1999, 38, 1–26. [Google Scholar] [CrossRef]
- Larsen, A.M.; Mohammed, H.H.; Arias, C.R. Characterization of the gut microbiota of three commercially valuable warmwater fish species. J. Appl. Microbiol. 2014, 116, 1396–1404. [Google Scholar] [CrossRef]
- Givens, C.E.; Ransom, B.; Bano, N.; Hollibaugh, J.T. A fish tale: Comparison of the gut microbiomes of 12 finfish and 3 shark species. Mar. Ecol. Prog. Ser. 2014, 518, 209–223. [Google Scholar] [CrossRef]
- Roeselers, G.; Mittge, E.K.; Stephens, W.Z.; Parichy, D.M.; Cavanaugh, C.M.; Guillemin, K.; Rawls, J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011, 5, 1595–1608. [Google Scholar] [CrossRef]
- Blanch, A.R.; Alsina, M.; Simon, M.; Jofre, J. Determination of bacteria associated with reared turbot (Scophthalmus maximus) larvae. J. Appl. Microbiol. 1997, 82, 729–734. [Google Scholar] [CrossRef]
- Lauzon, H.L.; Gudmundsdottir, S.; Petursdottir, S.K.; Reynisson, E.; Steinarsson, A.; Oddgeirsson, M.; Bjornsdottir, R.; Gudmundsdottir, B.K. Microbiota of Atlantic cod (Gadus morhua L.) rearing systems at pre- and posthatch stages and the effect of different treatments. J. Appl. Microbiol. 2010, 109, 1775–1789. [Google Scholar] [CrossRef]
- McIntosh, D.; Ji, B.; Forward, B.S.; Puvanendran, V.; Boyce, D.; Ritchie, R. Culture-independent characterization of the bacterial populations associated with cod (Gadus morhua L.) and live feed at an experimental hatchery facility using denaturing gradient gel electrophoresis. Aquaculture 2008, 275, 42–50. [Google Scholar] [CrossRef]
- Zapata, A.; Diez, B.; Cejalvo, T.; Gutiérrez-de Frías, C.; Cortés, A. Ontogeny of the immune system of fish. Fish Shellfish Immun. 2006, 20, 126–136. [Google Scholar] [CrossRef]
- Makridis, P.; Kokou, F.; Bournakas, C.; Papandroulakis, N.; Sarropoulou, E. Isolation of Phaeobacter sp. from larvae of Atlantic bonito (Sarda sarda) in a mesocosmos unit, and its use for the rearing of European seabass larvae (Dicentrarchus labrax L.). Microorganisms 2021, 9, 128. [Google Scholar] [CrossRef]
- Verschuere, L.; Rombaut, G.; Sorgeloos, P.; Verstraete, W. Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. R. 2000, 64, 655–671. [Google Scholar] [CrossRef]
- Mohapatra, S.; Chakraborty, T.; Kumar, V.; DeBoeck, G.; Mohanta, K.N. Aquaculture and stress management: A review of probiotic intervention. J. Anim. Physiol. Anim. Nutr. 2013, 97, 405–443. [Google Scholar] [CrossRef] [PubMed]
- Prol-García, J.M.; Gómez, M.; Lorenzo Sánchez, L.; Pintado, J. Phaeobacter grown in biofilters: A new strategy for the control of Vibrionaceae in aquaculture. Aquac. Res. 2014, 45, 1012–1025. [Google Scholar] [CrossRef]
- Verma, G.; Gupta, A. Probiotics Application in Aquaculture: Improving Nutrition and Health. J. Anim. Feed. Sci. Technol. 2015, 3, 53–64. [Google Scholar]
- Bjornsdottir, R.; Johannsdottir, J.; Coe, J.; Smaradottir, H.; Agustsson, T.; Sigurgisladottir, S.; Gudmundsdottir, K.B. Survival and quality of halibut larvae (Hippoglossus hippoglossus L.) in intensive farming: Possible impact of the intestinal bacterial community. Aquaculture 2009, 286, 53–63. [Google Scholar] [CrossRef]
- Lazado, C.C.; Caipang, C.M.; Estante, E.G. Prospects of host-associated microorganisms in fish and penaeids as probiotics with immunomodulatory functions. Fish Shellfish Immunol. 2015, 45, 2–12. [Google Scholar] [CrossRef]
- Conceicao, L.; Yufera, M.; Makridis, P.; Morais, S.; Dinis, M. Live feeds for early stages of fish rearing. Aquac. Res. 2010, 41, 613–640. [Google Scholar] [CrossRef]
- Loufi, K.; Sfakianakis, D.G.; Makridis, P. The effect of feeding with the copepod Acartia tonsa during the first days of larval rearing on skeleton ontogeny and skeletal deformities in greater amberjack (Seriola dumerili). In Proceedings of the Aquaculture Europe 2022, Rimini, Italy, 27–30 September 2022; pp. 742–743. Available online: https://eposters.blob.core.windows.net/eas-eposters/AE2022AbstractBook.pdf (accessed on 2 May 2023).
- Dittman, K.K.; Rasmussen, B.B.; Melchiorsen, J.; Sonnenschein, E.C.; Lone, G.; Bentzon-Tilia, M. Changes in the microbiome of mariculture feed organisms after treatment with a potentially probiotic strain of Phaeobacter inhibens. Appl. Environm. Microbiol. 2020, 86, e0049920. [Google Scholar] [CrossRef]
- Rangel, F.; Enes, P.; Gasko, L.; Gai, F.; Hausmann, B.; Berry, D.; Oliva-Teles, A.; Serra, C.R.; Pereira, F.C. Differential modulation of the European sea bass gut microbiota by distinct insect meals. Front. Microbiol. 2022, 13, 831034. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Kokou, F.; Sasson, G.; Mizrahi, I.; Cnaani, A. Antibiotic effect and microbiome persistence vary along the European seabass gut. Sci. Rep. 2020, 10, 10003. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, M.G.; Tiedje, M.J.; Cole, R.J. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- McLaren, M.R. Silva SSU Taxonomic Training Data Formatted for DADA2 (Silva Version 138). Available online: https://zenodo.org/record/3986799 (accessed on 15 April 2021).
- Pinheiro, J.C.; Bates, D.M. Unconstrained parametrizations for variance-covariance matrices. Stat. Comput. 1996, 6, 289–296. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Jensen, S.; Bergh, Ò.; Enger, Ò.; Hjeltnes, B. Use of PCR-RFLP for genotyping 16S rRNA and characterizing bacteria cultured from halibut fry. Can. J. Microbiol. 2002, 48, 379–386. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics, 1st ed.; Stackebrandt, E.M., Goodfellow, M., Eds.; John Wiley and Sons: Chichester, UK, 1991; pp. 177–203. [Google Scholar]
- Makridis, P.; Martins, S.; Tsalavouta, M.; Dionisio, L.C.; Kotoulas, G.; Magoulas, A.; Dinis, M.T. Antimicrobial activity in bacteria isolated from Senegalese sole, Solea senegalensis, fed with natural prey. Aquac. Res. 2005, 36, 1619–1627. [Google Scholar] [CrossRef]
- Castillo, D.; D’Alvise, P.; Kalatzis, P.G.; Kokkari, C.; Middelboe, M.; Gram, l.; Liu, S.; Katharios, P. Draft genome sequences of Vibrio alginolyticus strains V1 and V2, opportunistic marine pathogens. Genome Announc. 2015, 3, e00729-15. [Google Scholar] [CrossRef] [PubMed]
- Castillo Bermúdez, D.E.; D’Alvise, P.; Middelboe, M.; Gram, L.; Liu, S.; Kalatzis, P.; Kokkari, C.; Katharios, P. Draft genome sequences of the fish pathogen Vibrio harveyi strains VH2 and VH5. Genome Announc. 2015, 3, e01062-15. [Google Scholar] [CrossRef]
- Smyrli, M.; Prapas, A.; Rigos, G.; Kokkari, C.; Pavlidis, M.; Katharios, P. Aeromonas veronii infection associated with high morbility and mortality in farmed European Seabass Dicentrarchus labrax in the Aegean Sea, Greece. Fish Pathol. 2017, 52, 68–81. [Google Scholar] [CrossRef]
- Neu, T.A.; Allen, E.E.; Roy, K. Defining and quantifying the core microbiome: Challenges and prospects. Proc. Natl. Acad. Sci. USA 2021, 118, e2104429118. [Google Scholar] [CrossRef] [PubMed]
- Nijssen, J.E.; Reinders, J.M.; Krystallis, A.; Tacken, G. Developing an Internationalization Strategy Using Diffusion Modeling: The Case of Greater Amberjack. Fishes 2019, 4, 12. [Google Scholar] [CrossRef]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The gut microbiota of marine fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef]
- Stephens, W.Z.; Burns, A.R.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J. 2016. The composition of the Zebrafish intestinal microbial community varies across development. ISME J. 2016, 10, 644–654. [Google Scholar] [CrossRef]
- Li, X.; Zhou, L.; Yu, Y.; Ni, J.; Xu, W.; Yan, Q. Composition of Gut Microbiota in the Gibel Carp (Carassius auratus gibelio) Varies with Host Development. Microb. Ecol. 2016, 74, 239–249. [Google Scholar] [CrossRef]
- Yan, Q.; Li, J.; Yu, Y.; Wang, J.; He, Z.; Van Nostrand, J.D.; Kempher, M.L.; Wu, L.; Wang, Y.; Liao, L.; et al. Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ. Microbiol. 2016, 18, 4739–4754. [Google Scholar] [CrossRef]
- Burns, A.R.; Stephens, W.Z.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J.M. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2015, 10, 655–664. [Google Scholar] [CrossRef]
- Vadstein, O.; Attramadal, J.K.; Bakke, I.; Forberg, T.; Olsen, Y.; Verdegem, M.; Giatsis, C.; Skjermo, J.; Aasen, I.M.; Gatesoupe, F.J.; et al. Managing the microbial community of marine fish larvae: A holistic perspective for larviculture. Front. Microbiol. 2018, 9, 1820. [Google Scholar] [CrossRef]
- Kormas, K.A.; Meziti, A.; Mente, E.; Frentzos, A. Dietary differences are reflected on the gut prokaryotic community structure of wild and commercially reared sea bream (Sparus aurata). Microbiol. Open 2014, 3, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Bruhn, J.B.; Gram, R.; Belas, R. Production of antibacterial compounds and biofilm formation by Roseobacter species are influenced by culture conditions. Appl. Environ. Microb. 2007, 73, 442–450. [Google Scholar] [CrossRef]
- Cardona, E.; Gueguen, Y.; Magré, K.; Lorgeoux, B.; Piquemal, D.; Pierrat, F.; Noguier, F.; Saulnier, D. Bacterial community characterization of water and intestine of the shrimp Litopenaeus stylirostris in a biofloc system. BMC Microbiol. 2016, 16, 157–166. [Google Scholar] [CrossRef]
- Fjellheim, A.J.; Klinkenberg, G.; Skjermo, J.; Aasen, I.M.; Vadstein, O. Selection of candidate probionts by two different screening strategies from Atlantic cod (Gadus morhua L.) larvae. Vet. Microbiol 2010, 144, 153–159. [Google Scholar] [CrossRef]
- Copetti, F.; Gregoracci, G.B.; Vadstein, O.; Schveitzer, R. Management of biofloc concentrations as an ecological strategy for microbial control in intensive shrimp culture. Aquaculture 2021, 543, 736969. [Google Scholar] [CrossRef]
- Pujalte, M.J.; Lucena, T.; Rodrigo-Torres, L.; Arahal, D.R. Comparative genomics of Thalassobius including the description of Thalassobius activus sp. nov., and Thalassobius autumnalis sp. nov. Front. Microbiol. 2018, 8, 2645. [Google Scholar] [CrossRef]
- Makridis, P.; Fjellheim, A.J.; Skjermo, J.; Vadstein, O. Colonization of the gut in first feeding turbot by bacterial strains added to the water or bioencapsulated in rotifers. Aquacult. Int. 2000, 8, 367–380. [Google Scholar] [CrossRef]
- van der Meeren, T.; Olsen, R.E.; Hamre, K.; Fyhn, H.J. Biochemical composition of copepods for evaluation of feed quality in production of juvenile marine fish. Aquaculture 2008, 274, 375–397. [Google Scholar] [CrossRef]
- Arahal, D.R.; Macian, M.C.; Garay, E.; Pujalte, M.J. Thalassobius mediterraneus gen. nov., sp. nov., and reclassification of Ruegeria gelatinovorans as Thalassobius gelatinovorus comb. nov. Int. J. Syst. Evol. Micr. 2005, 55, 2371–2376. [Google Scholar] [CrossRef] [PubMed]
- Hjelm, M.; Bergh, Ø.; Riaza, A.; Nielsen, J.; Melchiorsen, J.; Jensen, S.; Duncan, H.; Ahrens, P.; Birkbeck, H.; Gram, L. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. System. Appl. Microbiol. 2004, 27, 360–371. [Google Scholar]
- Sampaio, A.; Silva, V.; Poeta, P.; Aonofriesei, F. Vibrio spp.: Life Strategies, Ecology, and Risks in a changing Environment. Diversity 2022, 14, 97. [Google Scholar] [CrossRef]
- Sugita, H.; Okano, R.; Suzuki, Y.; Iwai, D.; Mizukami, M.; Akiyama, N.; Matsuura, S. Antibacterial abilities of intestinal bacteria from larval and juvenile Japanese flounder against fish pathogens. Fish Sci. 2002, 68, 1004–1011. [Google Scholar] [CrossRef]
- Leyton, Y.; Sayes, C.; Mejias, C.; Abarca, M.; Wilson, R.; Riquelme, C. Increased larval survival of Seriola lalandi using Pseudoalteromonas sp. as probiotics. Rev. Biol. Mar. Oceanog. 2017, 52, 95–101. [Google Scholar] [CrossRef]
- Nayak, S. Role of gastrointestinal microbiota in fish. Aquac. Res. 2010, 41, 1553–1573. [Google Scholar] [CrossRef]
- Picchietti, S.; Fausto, A.M.; Randelli, E.; Carnevali, O.; Taddei, A.R.; Buonocore, F.; Scapigliati, G.; Abelli, L. Early treatment with Lactobacillus delbrueckii strain induces an increase in intestinal T-cells and granulocytes and modulates immune-related genes of larval Dicentrarchus labrax (L.). Fish Shellfish Immun. 2009, 26, 368–376. [Google Scholar] [CrossRef]
- Ringø, E.; Løvmo, L.; Kristiansen, M.; Bakken, Y.; Salinas, I.; Myklebust, R.; Olsen, R.E.; Mayhew, T.M. Lactic acid bacteria vs. pathogens in the gastrointestinal tract of fish: A review. Aquacult. Res. 2009, 41, 451–467. [Google Scholar] [CrossRef]
- D’Alvise, P.W.; Lillebø, S.; Prol-Garcia, M.J.; Wergeland, H.I.; Nielsen, K.F.; Bergh, Ø.; Gram, L. Phaeobacter gallaeciensis reduces Vibrio anguillarum in cultures of microalgae and rotifers, and prevents Vibriosis in cod larvae. PLoS ONE 2012, 7, e43996. [Google Scholar] [CrossRef]
- D’Alvise, P.W.; Lillebø, S.; Wergeland, H.I.; Nielsen, K.F.; Gram, L.; Bergh, Ø. Protection of cod larvae from vibriosis by Phaeobacter spp.: A comparison of strains and introduction times. Aquaculture 2013, 384–387, 82–86. [Google Scholar] [CrossRef]
- Ruiz-Ponte, C.; Cilia, V.; Lambert, C.; Nicolas, J.L. Roseobacter gallaeciencis sp. nov., a new marine bacterium isolated from the rearings and collectors of the scallop Pecten maximus. Int. J. Syst. Bacteriol. 1998, 48, 537–542. [Google Scholar] [CrossRef]
- Porsby, C.H.; Nielsen, K.F.; Gram, L. Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environm. Microbiol. 2008, 74, 7356–7364. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Sun, Y.; Ma, R.; Li, J.; Huang, K. Probiotic Psychrobacter sp. improved the autochthonous microbial diversity along the gastrointestinal tract of grouper Epinephelus coioides. J. Aquac. Res. Dev. 2011, S1, 001. [Google Scholar] [CrossRef]
- Wanka, Μ.Κ.; Damerau, T.; Costas, B.; Krueger, A.; Carsten Schulz, C.; Wuertz, S. Isolation and characterization of native probiotics for fish farming. BMC Microbiol. 2018, 18, 119. [Google Scholar] [CrossRef] [PubMed]

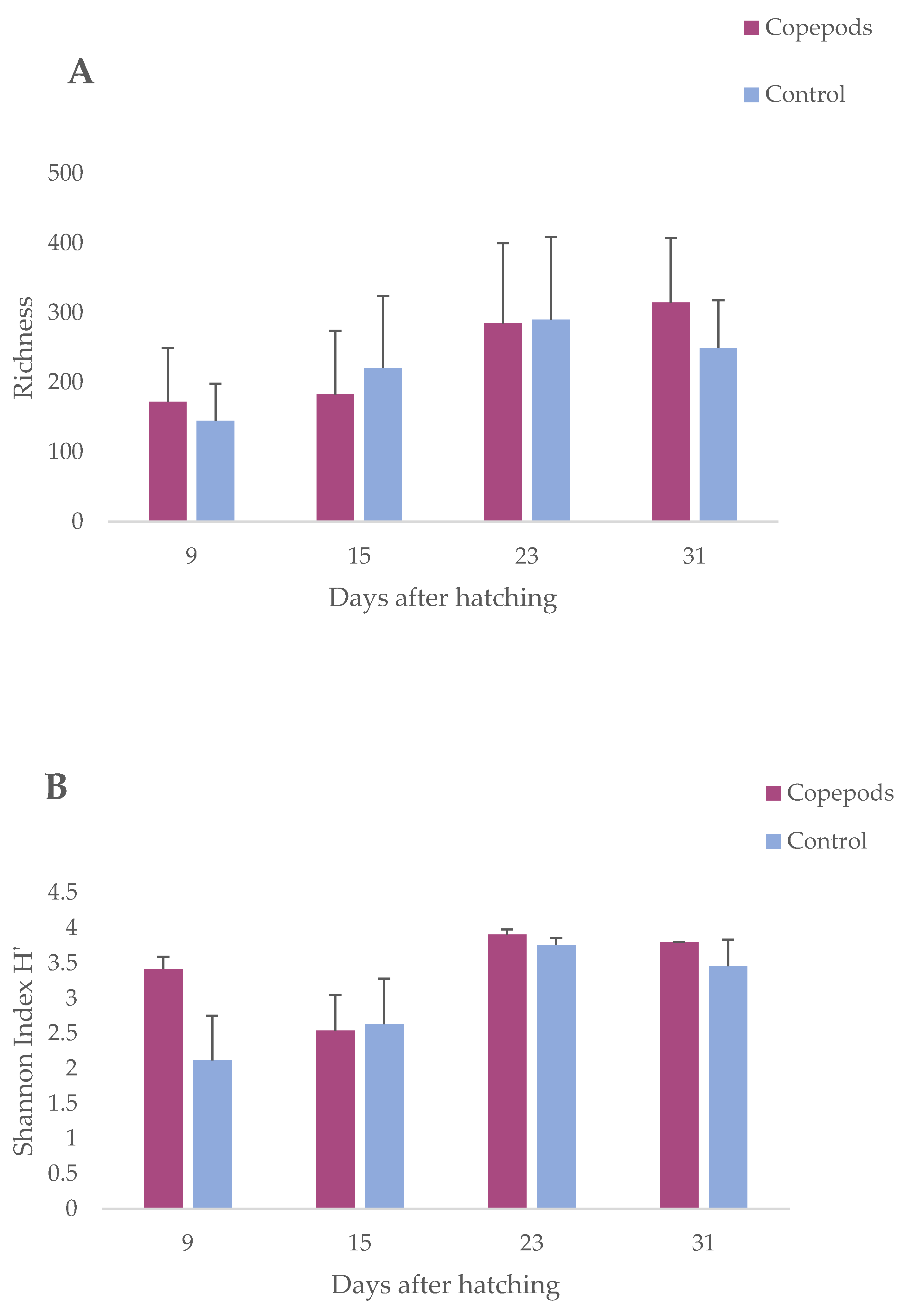

| Richness | |||
|---|---|---|---|
| AIC = 165.4049, BIC = 168.3143, logLik = −76.70243 | |||
| den DF | F | p value | |
| Age | 11 | 10.39 | 0.008 |
| Diet | 11 | 0.72 | 0.41 |
| Age Diet interaction | 11 | 1.51 | 0.24 |
| Diversity (Shannon Index H’) | |||
| AIC = 50.44739, BIC = 53.35683, logLik = −19.2237 | |||
| den DF | F | p-value | |
| Age | 11 | 9.82 | 0.009 |
| Diet | 11 | 1.27 | 0.28 |
| Age Diet interaction | 11 | 2.32 | 0.15 |
| Sum of Square | Degree of Freedom | Mean Square | F | p | |
|---|---|---|---|---|---|
| Diet | 0.18 | 1 | 0.18 | 0.9 | 0.54 |
| Age | 1.80 | 3 | 0.60 | 2.97 | 0.0001 |
| Interaction | 0.53 | 3 | 0.18 | 0.87 | 0.66 |
| Residual | 1.62 | 8 | 0.20 | ||
| Total | 4.14 | 15 |
| Bacterial Strain | Accession Number | Identity (Pct %) | E Value | Bit Score |
|---|---|---|---|---|
| Phaeobacter gallaeciensis JL2886 1 | CP015124.1 | 95 | 0 | 2111 |
| Phaeobacter sp. 1 | KT185144.1 | 96 | 0 | 2159 |
| Ruegeria sp. InS-296 1 | MF359524.1 | 96 | 0 | 2130 |
| Ruegeria sp. InS-119 1 | MF359371.1 | 97 | 0 | 2200 |
| Ruegeria sp. InS-264 1 | MF070517.1 | 99 | 0 | 2350 |
| Nautella italica (Phaeobacter italicus) 1 | HQ908722.1 | 97 | 0 | 2196 |
| Roseobacter sp. 1 | KY770280.1 | 97 | 0 | 2233 |
| Rhodobacteraceae bacterium ZJ3003 1 | KP301108.1 | 98 | 0 | 2281 |
| Rhodobacter sp. 1–5 2 | AF513400.1 | 91 | 0 | 1661 |
| Pathogenic Strain | |||||||
|---|---|---|---|---|---|---|---|
| Bacterial Strain | Aeromonas veronii | Vibrio alginolyticus | Vibrio harveyi | Vibrio anguillarum | Gram | Oxidase | Catalase |
| Vibrio parahaemolyticus strain VP1 | - | ++ | ++++ | ++++ | - | + | + |
| Vibrio alginolyticus strain ZJ-T | ++++ | ++++ | ++++ | ++++ | - | + | + |
| Vibrio alginolyticus strain Xmb025 | +++ | +++ | ++++ | +++ | - | + | + |
| Vibrio alginolyticus strain Xmb019 | +++ | ++++ | +++ | +++ | - | + | + |
| Uncultured Vibrio sp. clone HH101352 | ++++ | +++ | ++++ | ++++ | - | + | + |
| Uncultured Vibrio sp. clone HH101334 | ++++ | +++ | - | +++ | - | + | + |
| Uncultured Vibrio sp. clone C0A05-1 | +++ | +++ | +++ | +++ | - | + | + |
| Uncultured Vibrio sp. clone HH101375 | +++ | ++++ | - | - | - | + | + |
| Vibrio sp. PSMJVIT3 | ++++ | +++ | +++ | +++ | - | + | + |
| Vibrio sp. strain JLT194 | ++++ | ++++ | +++ | ++++ | - | + | + |
| Vibrio sp. CN87 | +++ | ++++ | ++++ | ++++ | - | + | + |
| Pathogenic Strain | ||||
|---|---|---|---|---|
| Bacterial Strain | Aeromonas veronii | Vibrio harveyi | Vibrio anguillarum | Vibrio alginolyticus |
| Phaeobacter gallaeciensis JL2886 1 | + | + | - | - |
| Phaeobacter sp. 1 | + | + | ++ | + |
| Ruegeria sp.InS-296 1 | ++ | ++ | ++ | ++ |
| Rhodobacter sp. 1–5 2 | + | + | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paralika, V.; Kokou, F.; Karapanagiotis, S.; Makridis, P. Characterization of Host-Associated Microbiota and Isolation of Antagonistic Bacteria from Greater Amberjack (Seriola dumerili, Risso, 1810) Larvae. Microorganisms 2023, 11, 1889. https://doi.org/10.3390/microorganisms11081889
Paralika V, Kokou F, Karapanagiotis S, Makridis P. Characterization of Host-Associated Microbiota and Isolation of Antagonistic Bacteria from Greater Amberjack (Seriola dumerili, Risso, 1810) Larvae. Microorganisms. 2023; 11(8):1889. https://doi.org/10.3390/microorganisms11081889
Chicago/Turabian StyleParalika, Vasiliki, Fotini Kokou, Stelios Karapanagiotis, and Pavlos Makridis. 2023. "Characterization of Host-Associated Microbiota and Isolation of Antagonistic Bacteria from Greater Amberjack (Seriola dumerili, Risso, 1810) Larvae" Microorganisms 11, no. 8: 1889. https://doi.org/10.3390/microorganisms11081889
APA StyleParalika, V., Kokou, F., Karapanagiotis, S., & Makridis, P. (2023). Characterization of Host-Associated Microbiota and Isolation of Antagonistic Bacteria from Greater Amberjack (Seriola dumerili, Risso, 1810) Larvae. Microorganisms, 11(8), 1889. https://doi.org/10.3390/microorganisms11081889







