Species Identification and Mycotoxigenic Potential of Aspergillus Section Flavi Isolated from Maize Marketed in the Metropolitan Region of Asunción, Paraguay
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Composed Sample Preparation
2.3. Mycological Analysis of Z. mays var. indurata and Z. mays var. amylacea
2.4. Morphological Identification of Aspergillus Section Flavi
2.5. MALDI-TOF MS Profiling
2.6. Molecular Identification of Aspergillus Section Flavi
2.7. Phylogenetic Analysis and Haplotype Network
2.8. Molecular Characterization of Toxigenic Potential of Aspergillus flavus Isolates
2.9. Aflatoxins and Cyclopiazonic Acid Detections by MALDI-TOF MS
2.10. Mycotoxin Quantification in Maize
2.11. Data Analysis
3. Results
3.1. Mycobiota and Frequency of Aspergillus Section Flavi
3.2. Morphological Identification of Aspergillus Section Flavi
3.3. MALDI-TOF MS Profiling
3.4. Molecular Identification and Phylogenetic Analysis
3.5. Haplotype Network
3.6. Molecular Characterization of the Toxigenic Potential of Aspergillus flavus Isolates
3.7. Aflatoxins and Cyclopiazonic Acid Detections by MALDI-TOF MS
3.8. Mycotoxin Quantification in Maize
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. El Maíz en los Trópicos: Mejoramiento y Producción; Paliwal, R.L., Granados, G., Lafitte, H.R., Violic, A.D., Marathée, J.P., Eds.; Organización de las Naciones Unidas para la Agricultura y la Alimentación-FAO: Roma, Italy, 2001; ISBN 9253044578. [Google Scholar]
- Angulo, P. El maíz en Paraguay. BASE Investig. Soc. 2017, 12, 1–4. [Google Scholar]
- Noldin, O.; Vilaró, M.; Suárez, R.; Abadie, T. Colección Núcleo de Paraguay. In Desarrollo de Colecciones Núcleo de Maíz en el Cono Sur de América Latina: Argentina, Bolivia, Brasil, Chile, Paraguay y Uruguay; Venturini, R.S., Ed.; Serie Documentos; REGENSUR, PROCISUR/IICA: Montevideo, Uruguay, 2005; pp. 67–76. ISBN 92-90-39-698-9. [Google Scholar]
- Salhuana, W.; Machado, V. Races of Maize in Paraguay; Salhuana, W., Machado, V., Eds.; United States Department of Agriculture, Agricultural Research Service and The Maize Program of the Paraguayan Ministry of Agriculture and Livestock: Washington, DC, USA, 1999.
- Noldin, O.; Machado, V. Evaluación Preliminar de la variabilidad de razas tradicionales de maíz (Zea mays L.) en Paraguay. In Avances de Investigación en Recursos Genéticos en el Cono Sur II; Clausen, A., Condón, F., Berretta, A., Eds.; PROCISUR, IICA: Montevideo, Uruguay, 2007; pp. 189–193. ISBN 9789290398264. [Google Scholar]
- Comidas Tipicas del Paraguay. Available online: http://comidastipicasparaguay.blogspot.com/ (accessed on 20 May 2019).
- HOY/Con el Jopara se Ahuyenta Hoy al Karai Octubre. Available online: https://www.hoy.com.py/nacionales/hoy-se-come-jopara-para-ahuyentan-al-karai-octubre (accessed on 20 May 2019).
- USAID/Paraguay. Maíz: Análisis de la cadena de valor. U.S. Agency Int. Dev. 2011, 9–11. Available online: https://2017-2020.usaid.gov/sites/default/files/documents/1862/maiz.pdf (accessed on 20 May 2019).
- Inter-American Institute for Cooperation on Agriculture. Family Farming in the Americas-Family Farming in the Americas: Guiding Principles and Concepts of IICA’s Technical Cooperation; Inter-American Institute for Cooperation on Agriculture: Turrialba, Costa Rica, 2017; Volume 37. [Google Scholar]
- Guerreño, J.O.; Talavera, C.A.L.; Villalba, J.D.G. Guía Técnica Cultivo de Maíz; JICA: Tokyo, Japan, 2019; Volume 48.
- Delgado, V.; Cabral, I. Guía Técnica de Rubros Agropecuarios; DEAG: Berlin, Germany, 2010; pp. 56–57. [Google Scholar]
- Taniwaki, M.H.; Pitt, J.I.; Copetti, M.V.; Teixeira, A.A.; Iamanaka, B.T. Understanding mycotoxin contamination across the food chain in Brazil: Challenges and opportunities. Toxins 2019, 11, 411. [Google Scholar] [CrossRef]
- Mannaa, M.; Kim, K.D. Influence of temperature and water activity on deleterious fungi and mycotoxin production during grain storage. Mycobiology 2017, 45, 240–254. [Google Scholar] [CrossRef]
- Geisen, R.; Touhami, N.; Schmidt-Heydt, M. Mycotoxins as adaptation factors to food related environments. Curr. Opin. Food Sci. 2017, 17, 1–8. [Google Scholar] [CrossRef]
- Neme, K.; Mohammed, A. Mycotoxin occurrence in grains and the role of postharvest management as a mitigation strategies. A review. Food Control 2017, 78, 412–425. [Google Scholar] [CrossRef]
- Chang, S.; Carneiro-Leão, M.P.; de Oliveira, B.F.; Souza-Motta, C.; Lima, N.; Santos, C.; de Oliveira, N.T. Polyphasic approach including MALDI-TOF MS/MS analysis for identification and characterisation of Fusarium verticillioides in Brazilian corn kernels. Toxins 2016, 8, 54. [Google Scholar] [CrossRef]
- Costa, J.; Rodríguez, R.; Garcia-Cela, E.; Medina, A.; Magan, N.; Lima, N.; Battilani, P.; Santos, C. Overview of fungi and mycotoxin contamination in Capsicum pepper and in its derivatives. Toxins 2019, 11, 27. [Google Scholar] [CrossRef]
- Costa, J.; Rodríguez, R.; Santos, C.; Soares, C.; Lima, N.; Santos, C. Mycobiota in Chilean chilli Capsicum annuum L. used for production of Merkén. Int. J. Food Microbiol. 2020, 334, 108833. [Google Scholar] [CrossRef]
- Caceres, I.; Al Khoury, A.; El Khoury, R.; Lorber, S.; Oswald, I.P.; Khoury, E.; Atoui, A.; Puel, O.; Bailly, J. Aflatoxin Biosynthesis and Genetic Regulation: A Review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer (IARC) Aflatoxins. A Review of Human Carcinogens. Part F: Chemical Agents and Related Occupations; IARC: Lyon, France, 2012; Volume 100, pp. 225–244. ISBN 978 92 832 1323 9.
- Kumar, P.; Mahato, D.K.; Kamle, M.; Mohanta, T.K.; Kang, S.G. Aflatoxins: A Global Concern for Food Safety, Human Health and Their Management. Front. Microbiol. 2017, 7, 2170. [Google Scholar] [CrossRef]
- Arrua Alvarenga, A.A.; Moura-Mendes, J.; Fernández Rios, D. Aflatoxins, a real risk. Rep. Científicos La FACEN 2013, 4, 68–81. [Google Scholar]
- Frisvad, J.C. Taxonomy, chemodiversity and chemoconsistency of Aspergillus, Penicillium and Talaromyces species. Front. Microbiol. 2014, 5, 773. [Google Scholar] [CrossRef]
- Perrone, G.; Gallo, A. Aspergillus species and their Associated Mycotoxins. In Mycotoxigenic Fungi: Methods and Protocols, Methods in Molecular Biology; Moretti, A., Susca, A., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1542, pp. 33–49. ISBN 978-1-4939-6705-6. [Google Scholar]
- Lima, N.; Santos, C. MALDI-TOF MS for identification of food spoilage filamentous fungi. Curr. Opin. Food Sci. 2017, 13, 26–30. [Google Scholar] [CrossRef]
- Lima, M.S.; De Lucas, R.C.; Lima, N.; De Lourdes Teixeira De Moraes Polizeli, M.; Santos, C. Fungal community ecology using MALDI-TOF MS demands curated mass spectral databases. Front. Microbiol. 2019, 10, 2017–2020. [Google Scholar] [CrossRef]
- Rodriguez, R.; Santos, C.; Simões, M.F.; Soares, C.; Santos, C.; Lima, N. Polyphasic, Including MALDI-TOF MS, Evaluation of Freeze-Drying Long-Term Preservation on Aspergillus (Section Nigri) strains. Microorganisms 2019, 7, 291. [Google Scholar] [CrossRef]
- Klich, M.A. Identification of Common Aspergillus Species; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2002; p. 116. [Google Scholar]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.-B.; Hubka, V.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification, and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Nováková, A.; Chen, A.J.; Arzanlou, M.; Larsen, T.O.; Sklenář, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef]
- Makhlouf, J.; Carvajal-campos, A.; Querin, A.; Tadrist, S.; Puel, O.; Lorber, S.; Oswald, I.P.; Hamze, M.; Bailly, J.; Bailly, S. Morphologic, molecular and metabolic characterization of Aspergillus section Flavi in spices marketed in Lebanon. Sci. Rep. 2019, 9, 5263. [Google Scholar] [CrossRef]
- Sepúlveda, C.O.; Piontelli, E.L.; Sepúlveda, O.C.; Piontelli, L.E. Poblaciones de Aspergillus en semillas de maíz y soja de importación Argentina: Enfasis en la sección Flavi. Boletín Micológico 2005, 20, 41–55. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Wiley-Blackwell: Ames, IA, USA, 2006; ISBN 978-0-8138-1919-8. [Google Scholar]
- Castellari, C.C.; Cendoya, M.G.; Marcos, F.J.; Barrera, V.; Pacin, M. Factores extrínsecos e intrínsecos asociados a poblaciones fúngicas micotoxigénicas de granos de maíz (Zea mays L.) almacenados en silos bolsa en Argentina. Rev. Argent. Microbiol. 2015, 47, 350–359. [Google Scholar] [CrossRef]
- Pildain, M.B.; Cabral, D.; Vaamonde, G. Poblaciones de Aspergillus flavus en maní cultivado en diferentes zonas agroecológicas de la Argentina, caracterización morfológica y toxigénica. Rev. Investig. Agropecu. 2005, 34, 3–19. [Google Scholar]
- Paziani, M.H.; Carvalho, L.T.; Souza, M.; Melhem, C.; Teresa, M.; Almeida, G.; Emilia, M.; Bonifácio, N.; Martinez, R.; Santos, C.; et al. First Comprehensive Report of Clinical Fusarium Strains Isolated in the State of São Paulo (Brazil) and Identified by MALDI-TOF MS and Molecular Biology. Microorganisms 2020, 8, 66. [Google Scholar] [CrossRef]
- Rodrigues, P.; Venâncio, A.; Lima, N. A polyphasic approach to the identification of aflatoxigenic and non-aflatoxigenic strains of Aspergillus section Flavi isolated from Portuguese almonds. Int. J. Food Microbiol. 2009, 129, 187–193. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, S.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Hong, S.-B.; Go, S.-J.; Shin, H.-D.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2008, 97, 1316–1329. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Criseo, G.; Bagnara, A.; Bisignano, G. Differentiation of aflatoxin-producing and non-producing strains of Aspergillus flavus group. Lett. Appl. Microbiol. 2001, 33, 291–295. [Google Scholar] [CrossRef]
- Catharino, R.R.; Marques, L.A.; Santos, L.S.; Baptista, A.S.; Glória, E.M.; Calori-Domingues, M.A.; Facco, E.M.P.; Eberlin, M.N. Aflatoxin screening by MALDI-TOF mass spectrometry. Anal. Chem. 2005, 77, 8155–8157. [Google Scholar] [CrossRef]
- Romer Labs AgraQuant® Total Aflatoxin Assay. COKAQ1100. Romer Labs Methods. 2009, 1–4. Available online: https://www.romerlabs.com/es/shop/agraquant-r-total-aflatoxin-elisa-test/ (accessed on 1 December 2022).
- Romer Labs AgraQuant® Total Fumonisin Assay COKAQ3000. Romer Labs Methods 2015, 1–4. Available online: https://www.romerlabs.com/es/shop/agraquant-r-fumonisin-elisa-test/ (accessed on 1 December 2022).
- Romer Labs AgraQuant® Zearalenone Assay. COKAQ5000. Romer Labs Methods 2009, 1–4. Available online: romerlabs.com/us_en/shop/agraquant-r-zearalenone-elisa-test/ (accessed on 1 December 2022).
- Romer Labs AgraQuant® T-2 Toxin Assay. COKAQ6000. Romer Labs Methods 2004, 2–5. Available online: https://www.romerlabs.com/es/shop/agraquant-r-t-2-toxin-elisa-test/ (accessed on 1 December 2022).
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Gasperini, A.M.; Garcia-Cela, E.; Sulyok, M.; Medina, A.; Magan, N. Fungal diversity and metabolomic profiles in GM and isogenic non-GM maize cultivars from Brazil. Mycotoxin Res. 2021, 37, 39–48. [Google Scholar] [CrossRef]
- Wit, M.; Warzecha, R.; Mirzwa-Mróz, E.; Jabońska, E.; Ochodzki, P.; Waśkiewicz, A.; Wakulin’ski, W. Susceptibility of flint and dent maize ears to Fusarium species. Phytopathologia 2011, 60, 35–45. [Google Scholar]
- Quéro, L.; Courault, P.; Cellière, B.; Lorber, S.; Jany, J.-L.; Puel, O.; Girard, V.; Vasseur, V.; Nodet, P.; Mounier, J. Application of MALDI-TOF MS to species complex differentiation and strain typing of food related fungi: Case studies with Aspergillus section Flavi species and Penicillium roqueforti isolates. Food Microbiol. 2020, 86, 103311. [Google Scholar] [CrossRef]
- Rodrigues, P.; Santos, C.; Venâncio, A.; Lima, N. Species identification of Aspergillus section Flavi isolates from Portuguese almonds using phenotypic, including MALDI-TOF ICMS, and molecular approaches. J. Appl. Microbiol. 2011, 111, 877–892. [Google Scholar] [CrossRef]
- Da Silva, F.C.; Chalfoun, S.M.; Batista, L.R.; Lima, N. Use of a polyphasic approach including MALDI-TOF MS for identification of Aspergillus section Flavi strains isolated from food commodities in Brazil. Ann. Microbiol. 2015, 65, 2119–2129. [Google Scholar] [CrossRef]
- Hedayati, M.T.; Taghizadeh-Armaki, M.; Zarrinfar, H.; Hoseinnejad, A.; Ansari, S.; Abastabar, M.; Er, H.; Özhak, B.; Öğünç, D.; Ilkit, M.; et al. Discrimination of Aspergillus flavus from Aspergillus oryzae by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass-spectrometry. Mycoses 2019, 62, 1182–1188. [Google Scholar] [CrossRef]
- Quéro, L.; Girard, V.; Pawtowski, A.; Tréguer, S.; Weill, A.; Arend, S.; Cellière, B.; Polsinelli, S.; Valérie, M.; van Belkum, A.; et al. Development and application of MALDI-TOF MS for identification of food spoilage fungi. Food Microbiol. 2019, 81, 76–88. [Google Scholar] [CrossRef]
- Tam, E.W.T.; Chen, J.H.K.; Lau, E.C.L.; Ngan, A.H.Y.; Fung, K.S.C.; Lee, K.C.; Lam, C.W.; Yuen, K.Y.; Lau, S.K.P.; Woo, P.C.Y. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: Characterization by internal transcribed spacer, β-tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser desorption ionization-ti. J. Clin. Microbiol. 2014, 52, 1153–1160. [Google Scholar] [CrossRef]
- Santos, C.; Paterson, R.R.M.; Venâncio, A.; Lima, N. Filamentous fungal characterizations by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Appl. Microbiol. 2010, 108, 375–385. [Google Scholar] [CrossRef]
- Nakamura, S.; Sato, H.; Tanaka, R.; Kusuya, Y.; Takahashi, H.; Yaguchi, T. Ribosomal subunit protein typing using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) for the identification and discrimination of Aspergillus species. BMC Microbiol. 2017, 17, 100. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Islam, M.; Callicott, K.A.; Mutegi, C.; Bandyopadhyay, R.; Cotty, P.J. Aspergillus flavus resident in Kenya: High genetic diversity in an ancient population primarily shaped by clonal reproduction and mutation-driven evolution. Fungal Ecol. 2018, 35, 20–33. [Google Scholar] [CrossRef]
- Reeve, M.A.; Buddie, A.G.; Pollard, K.M.; Varia, S.; Seier, M.K.; Offord, L.C.; Cock, M.J.W. A highly-simplified and inexpensive MALDI-TOF mass spectrometry sample-preparation method with broad applicability to microorganisms, plants, and insects. J. Biol. Methods 2018, 5, 103. [Google Scholar] [CrossRef]
- Hleba, L.; Císarová, M.; Shariati, M.A.; Tancinová, D. Detection of mycotoxins using MALDI-TOF Mass Spectrometry. J. Microbiol. Biotechnol. Food Sci. 2017, 7, 181–185. [Google Scholar] [CrossRef]
- Vaamonde, G.; Patriarca, A.; Fernández Pinto, V.; Comerio, R.; Degrossi, C. Variability of aflatoxin and cyclopiazonic acid production by Aspergillus section flavi from different substrates in Argentina. Int. J. Food Microbiol. 2003, 88, 79–84. [Google Scholar] [CrossRef]
- Razzaghi-Abyaneh, M.; Shams-Ghahfarokhi, M.; Allameh, A.; Kazeroon-Shiri, A.; Ranjbar-Bahadori, S.; Mirzahoseini, H.; Rezaee, M.B. A survey on distribution of Aspergillus section Flavi in corn field soils in Iran: Population patterns based on aflatoxins, cyclopiazonic acid and sclerotia production. Mycopathologia 2006, 161, 183–192. [Google Scholar] [CrossRef]
- Camiletti, B.X.; Torrico, A.K.; Fernanda Maurino, M.; Cristos, D.; Magnoli, C.; Lucini, E.I.; de la Paz Giménez Pecci, M. Fungal screening and aflatoxin production by Aspergillus section Flavi isolated from pre-harvest maize ears grown in two Argentine regions. Crop Prot. 2017, 92, 41–48. [Google Scholar] [CrossRef]
- Probst, C.; Callicott, K.A.; Cotty, P.J. Deadly strains of Kenyan Aspergillus are distinct from other aflatoxin producers. Eur. J. Plant Pathol. 2012, 132, 419–429. [Google Scholar] [CrossRef]
- Rao, B.L.; Husain, A. Presence of cyclopiazonic acid in kodo millet (Paspalum scrobiculatum) causing “kodua poisoning” in man and its production by associated fungi. Mycopathologia 1985, 180, 177–180. [Google Scholar] [CrossRef]
- Antony, M.; Shukla, Y.; Janardhanan, K.K. Potential risk of acute hepatotoxicity of kodo poisoning due to exposure to cyclopiazonic acid. J. Ethnopharmacol. 2003, 87, 211–214. [Google Scholar] [CrossRef]
- Olarte, R.A.; Horn, B.W.; Dorner, J.W.; Monacell, J.T.; Singh, R.; Stone, E.A.; Carbone, I. Effect of sexual recombination on population diversity in aflatoxin production by Aspergillus flavus and evidence for cryptic heterokaryosis. Mol. Ecol. 2012, 21, 1453–1476. [Google Scholar] [CrossRef]
- Donner, M.; Atehnkeng, J.; Sikora, R.A.; Bandyopadhyay, R.; Cotty, P.J. Molecular characterization of atoxigenic strains for biological control of aflatoxins in Nigeria. Food Addit. Contam. Part A 2010, 27, 576–590. [Google Scholar] [CrossRef]
- Cotty, P.J.; Antilla, L.; Wakelyn, P.J. Competitive Exclusion of Aflatoxin Producers: Farmer-driven Research and Development. In Biological Control. A Global Perspective; Vincent, C., Goettel, M.S., Lazarovits, C., Eds.; CAB International: Wallingford, UK, 2007; pp. 241–253. [Google Scholar]
- Cotty, P.J.; Probst, C.; Jaime-garcia, R. Etiology and Management of Aflatoxin Contamination. In Mycotoxins: Detection Methods, Management, Public Health and Agricultural Trade; Leslie, J.F., Bandyopadhyay, R., Visconti, A., Eds.; CABI: Wallingford, UK, 2008; pp. 287–299. ISBN 9781845930820. [Google Scholar]
- Tran-Dinh, N.; Pitt, J.I.; Markwell, P.J. Selection of non-toxigenic strains of Aspergillus flavus for biocontrol of aflatoxins in maize in Thailand. Biocontrol Sci. Technol. 2014, 24, 652–661. [Google Scholar] [CrossRef]
- Santos-Ciscon, B.A.; Van Diepeningen, A.; Machado, C.; Eleutéria, I.; Waalwijk, C. Aspergillus species from Brazilian dry beans and their toxigenic potential. Int. J. Food Microbiol. 2019, 292, 91–100. [Google Scholar] [CrossRef]
- DINAC/Direccional de Aeronáutica Civil. Boletín de Perspectivas Climáticas Enero-Marzo 2020-Paraguay; DINAC/Direccional de Aeronáutica Civil: Asunción, Paraguay, 2020. [Google Scholar]
- Rocha, L.O.; Reis, G.M.; Braghini, R.; Kobashigawa, E.; Araújo, J.; Corrêa, B. Characterization of aflatoxigenic and non-aflatoxigenic strains of Aspergillus section Flavi isolated from corn grains of different geographic origins in Brazil. Eur. J. Plant Pathol. 2012, 132, 353–366. [Google Scholar] [CrossRef]
- Perrone, G.; Susca, A.; Cozzi, G.; Ehrlich, K.; Varga, J.; Frisvad, J.C.; Meijer, M.; Noonim, P.; Mahakarnchanakul, W.; Samson, R.A. Biodiversity of Aspergillus species in some important agricultural products. Stud. Mycol. 2007, 59, 53–66. [Google Scholar] [CrossRef]
- Probst, C.; Bandyopadhyay, R.; Cotty, P.J. Diversity of aflatoxin-producing fungi and their impact on food safety in sub-Saharan Africa. Int. J. Food Microbiol. 2014, 174, 113–122. [Google Scholar] [CrossRef]
- Nesci, A.; Etcheverry, M. Aspergillus section Flavi populations from field maize in Argentina. Lett. Appl. Microbiol. 2002, 34, 343–348. [Google Scholar] [CrossRef]
- Bayman, P.; Cotty, P.J. Genetic diversity in Aspergillus flavus: Association with aflatoxin production and morphology. Can. J. Bot. 1993, 71, 23–31. [Google Scholar] [CrossRef]
- Mauro, A.; Battilani, P.; Callicott, K.A.; Giorni, P.; Pietri, A.; Cotty, P.J. Structure of an Aspergillus flavus population from maize kernels in northern Italy. Int. J. Food Microbiol. 2013, 162, 1–7. [Google Scholar] [CrossRef]
- Astoreca, A.L.; Dalcero, A.M.; Pinto, V.F.; Vaamonde, G. A survey on distribution and toxigenicity of Aspergillus section Flavi in poultry feeds. Int. J. Food Microbiol. 2011, 146, 38–43. [Google Scholar] [CrossRef]
- Probst, C.; Schulthess, F.; Cotty, P.J. Impact of Aspergillus section Flavi community structure on the development of lethal levels of aflatoxins in Kenyan maize (Zea mays). J. Appl. Microbiol. 2010, 108, 600–610. [Google Scholar] [CrossRef]
- Barros, G.; Torres, A.; Chulze, S. Aspergillus flavus population isolated from soil of Argentina’s peanut-growing region. Sclerotia production and toxigenic profile. J. Sci. Food Agric. 2005, 85, 2349–2353. [Google Scholar] [CrossRef]
- Medina, A.; Rodriguez, A.; Magan, N. Effect of climate change on Aspergillus flavus and aflatoxin B1 production. Front. Microbiol. 2014, 5, 348. [Google Scholar] [CrossRef]
- Monda, E.; Masanga, J.; Alakonya, A. Variation in Occurrence and Aflatoxigenicity of Aspergillus flavus from Two Climatically Varied Regions in Kenya. Toxins 2020, 12, 34. [Google Scholar] [CrossRef]
- Medina, A.; Akbar, A.; Baazeem, A.; Rodriguez, A.; Magan, N. Climate change, food security and mycotoxins: Do we know enough? Fungal Biol. Rev. 2017, 31, 143–154. [Google Scholar] [CrossRef]
- Geiser, D.M.; Dorner, J.W.; Horn, B.W.; Taylor, J.W. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet. Biol. 2000, 31, 169–179. [Google Scholar] [CrossRef]
- Gallo, A.; Stea, G.; Battilani, P.; Logrieco, A.F.; Perrone, G. Molecular characterization of an Aspergillus flavus population isolated from maize during the first outbreak of aflatoxin contamination in Italy. Phytopathol. Mediterr. 2012, 51, 198–206. [Google Scholar]
- Oloo, R.D.; Okoth, S.; Wachira, P.; Mutiga, S.; Ochieng, P.; Kago, L.; Nganga, F.; Entfellner, J.B.D.; Ghimire, S. Genetic profiling of Aspergillus isolates with varying aflatoxin production potential from different maize-growing regions of kenya. Toxins 2019, 11, 467. [Google Scholar] [CrossRef]
- Faria, C.B.; dos Santos, F.C.; de Castro, F.F.; Sutil, A.R.; Sergio, L.M.; Silva, M.V.; Machinski Junior, M.; Barbosa-Tessmann, I.P. Occurrence of toxigenic Aspergillus flavus in commercial Bulgur wheat. Food Sci. Technol. 2017, 37, 103–111. [Google Scholar] [CrossRef]
- Sbardelotto Di Domenico, A.; Christ, D.; Hashimoto, E.H.; Busso, C.; Coelho, S.R.M. Evaluation of quality attributes and the incidence of Fusarium sp. and Aspergillus sp. in different types of maize storage. J. Stored Prod. Res. 2014, 61, 59–64. [Google Scholar] [CrossRef]
- Official Journal of the European Union European Commission Regulation (EC) No 1881/2006 of 19 December 2006 Amending Regulation (EC) 1881/2006 Setting Maximum Levels for Certain Contaminants in Foodstuffs. 2006, p. 24. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2006:364:0005:0024:EN:PDF (accessed on 10 May 2020).
- MERCOSUR/GMC/RES. No 25/02 Reglamento Técnico Mercosur Sobre Límites Máximos de Aflatoxinas Admisibles en Leche, Maní y Maíz; MERCOSUR/GMC/RES: Montevideo, Uruguay, 2002. [Google Scholar]
- Munkvold, G.P.; Arias, S.; Taschl, I.; Gruber-Dorninger, C. Mycotoxins in corn: Occurrence, impacts, and management. In Corn: Chemistry and Technology; Serna-Saldivar, S.O., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 235–287. ISBN 9780128119716. [Google Scholar]
- Food and Agriculture Organization (FAO). Estudio Sobre las Particularidades Socioculturales Relacionadas a la Alimentación Escolar y Producción de la Agricultura Familiar; Baranda, D., Ed.; Organización de las Naciones Unidas para la Agricultura y la Alimentación-FAO: Asunción, Paraguay, 2013. [Google Scholar]
- Altomare, C.; Logrieco, A.F.; Gallo, A. Mycotoxins and Mycotoxigenic Fungi: Risk and Management. A Challenge for Future Global Food Safety and Security; Elsevier Ltd.: Amsterdam, The Netherlands, 2021; Volume 1, ISBN 9780128199909. [Google Scholar]
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public Health Impacts of Foodborne Mycotoxins. Annu. Rev. Food Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef]
- Mudili, V.; Siddaih, C.N.; Nagesh, M.; Garapati, P.; Naveen Kumar, K.; Murali, H.S.; Yli Mattila, T.; Batra, H.V. Mould incidence and mycotoxin contamination in freshly harvested maize kernels originated from India. J. Sci. Food Agric. 2014, 94, 2674–2683. [Google Scholar] [CrossRef]
- Hove, M.; Van Poucke, C.; Njumbe-Ediage, E.; Nyanga, L.K.; De Saeger, S. Review on the natural co-occurrence of AFB1 and FB1 in maize and the combined toxicity of AFB1 and FB1. Food Control 2016, 59, 675–682. [Google Scholar] [CrossRef]
- IARC. Effects of aflatoxins and fumonisins on child growth. In Mycotoxin Control in Low- and Middle-Income Countries; Wild, C.P., Miller, J.D., Groopman, J.D., Eds.; International Agency for Research on Cancer: Lyon, France, 2015; ISBN 978-92-832-2510-2. [Google Scholar]
- Kamle, M.; Mahato, D.K.; Devi, S.; Lee, K.E.; Kang, S.G.; Kumar, P. Fumonisins: Impact on Agriculture, Food, and Human Health and their Management Strategies. Toxins 2019, 11, 328. [Google Scholar] [CrossRef]
- Bhat, R.; Rai, R.V.; Karim, A.A. Mycotoxins in Food and Feed: Present Status and Future Concerns. Compr. Rev. Food Sci. Food Saf. 2010, 9, 57–81. [Google Scholar] [CrossRef]
- Magan, N. Fungi of Extreme Environments. In Fungal Ecology; Dix, N.J., Webster, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 322–340. [Google Scholar]
- Chulze, S.N. Strategies to reduce mycotoxin levels in maize during storage: A review. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2010, 27, 651–657. [Google Scholar] [CrossRef]
- Katsurayama, A.M.; Martins, L.M.; Iamanaka, B.T.; Fungaro, M.H.P.; Silva, J.J.; Frisvad, J.C.; Pitt, J.I.; Taniwaki, M.H. Occurrence of Aspergillus section Flavi and aflatoxins in Brazilian rice: From field to market. Int. J. Food Microbiol. 2018, 266, 213–221. [Google Scholar] [CrossRef]
- Varga, J.; Frisvad, J.C.; Samson, R.A. A reappraisal of fungi producing aflatoxins. World Mycotoxin J. 2009, 2, 263–277. [Google Scholar] [CrossRef]
- Giorni, P.; Magan, N.; Pietri, A.; Battilani, P. Growth and aflatoxin production of an Italian strain of Aspergillus flavus: Influence of ecological factors and nutritional substrates. World Mycotoxin J. 2011, 4, 425–432. [Google Scholar] [CrossRef]
- Etcheverry, M.; Nesci, A.; Barros, G.; Torres, A.; Chulze, S. Occurrence of Aspergillus section Flavi and aflatoxin B1 in corn genotypes and corn meal in Argentina. Mycopathologia 2000, 147, 37–41. [Google Scholar] [CrossRef]
- Norlia, M.; Jinap, S.; Nor-khaizura, M.A.R.; Son, R.; Chin, C.K. Polyphasic approach to the identification and characterization of aflatoxigenic strains of Aspergillus section Flavi isolated from peanuts and peanut-based products marketed in Malaysia. Int. J. Food Microbiol. 2018, 282, 9–15. [Google Scholar] [CrossRef]
- Baquião, A.C.; De Oliveira, M.M.M.; Reis, T.A.; Zorzete, P.; Diniz Atayde, D.; Correa, B. Polyphasic approach to the identification of Aspergillus section Flavi isolated from Brazil nuts. Food Chem. 2013, 139, 1127–1132. [Google Scholar] [CrossRef]
- Barros, G.G.; Chiotta, M.L.; Reynoso, M.M.; Torres, A.M.; Chulze, S.N. Molecular characterization of Aspergillus section Flavi isolates collected from peanut fields in Argentina using AFLPs. J. Appl. Microbiol. 2007, 103, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.M.; Sant’Ana, A.S.; Fungaro, M.H.P.; Silva, J.J.; Nascimento, M.d.S.D.; Frisvad, J.C.; Taniwaki, M.H. The biodiversity of Aspergillus section Flavi and aflatoxins in the Brazilian peanut production chain. Food Res. Int. 2017, 94, 101–107. [Google Scholar] [CrossRef] [PubMed]
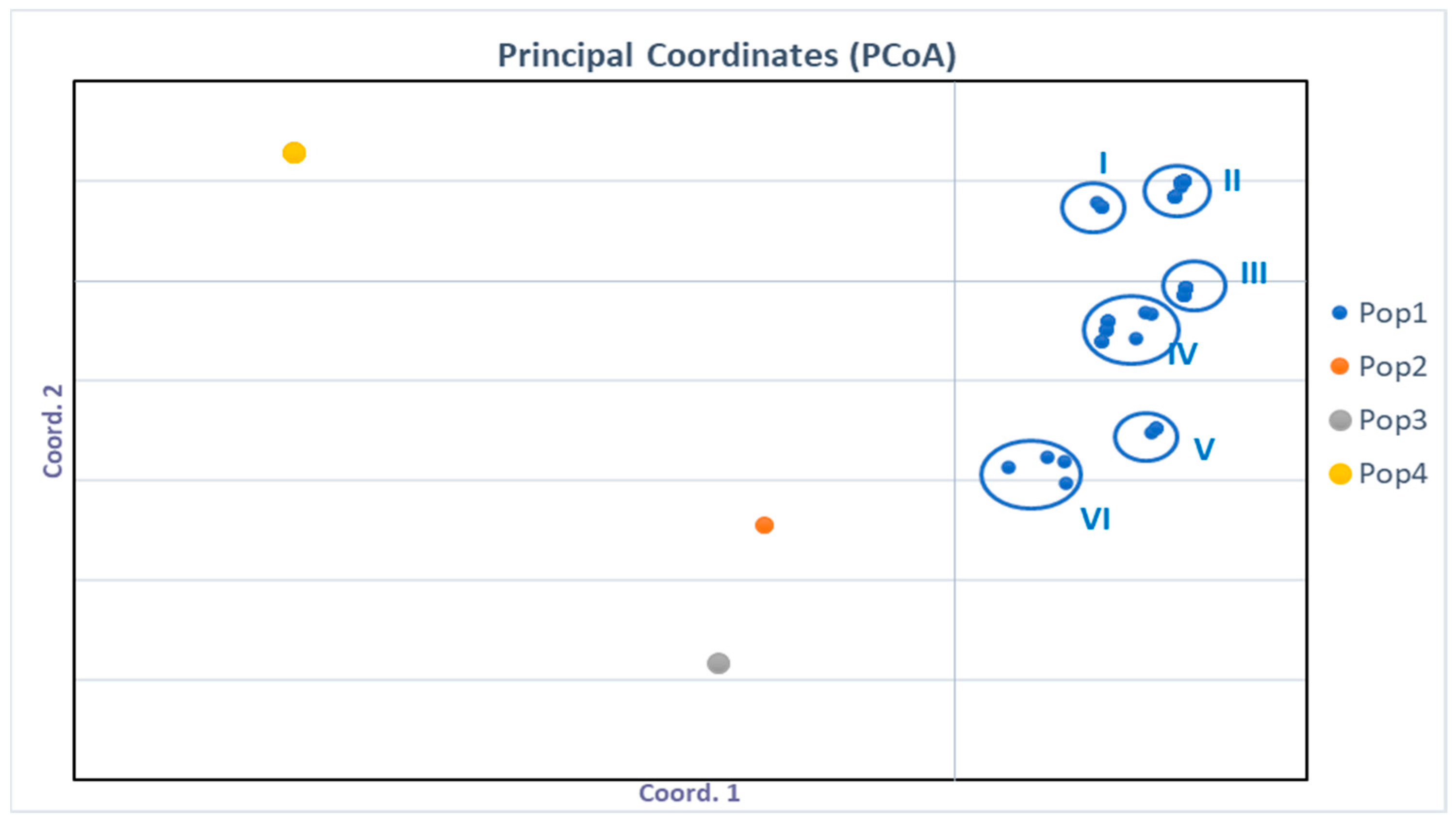
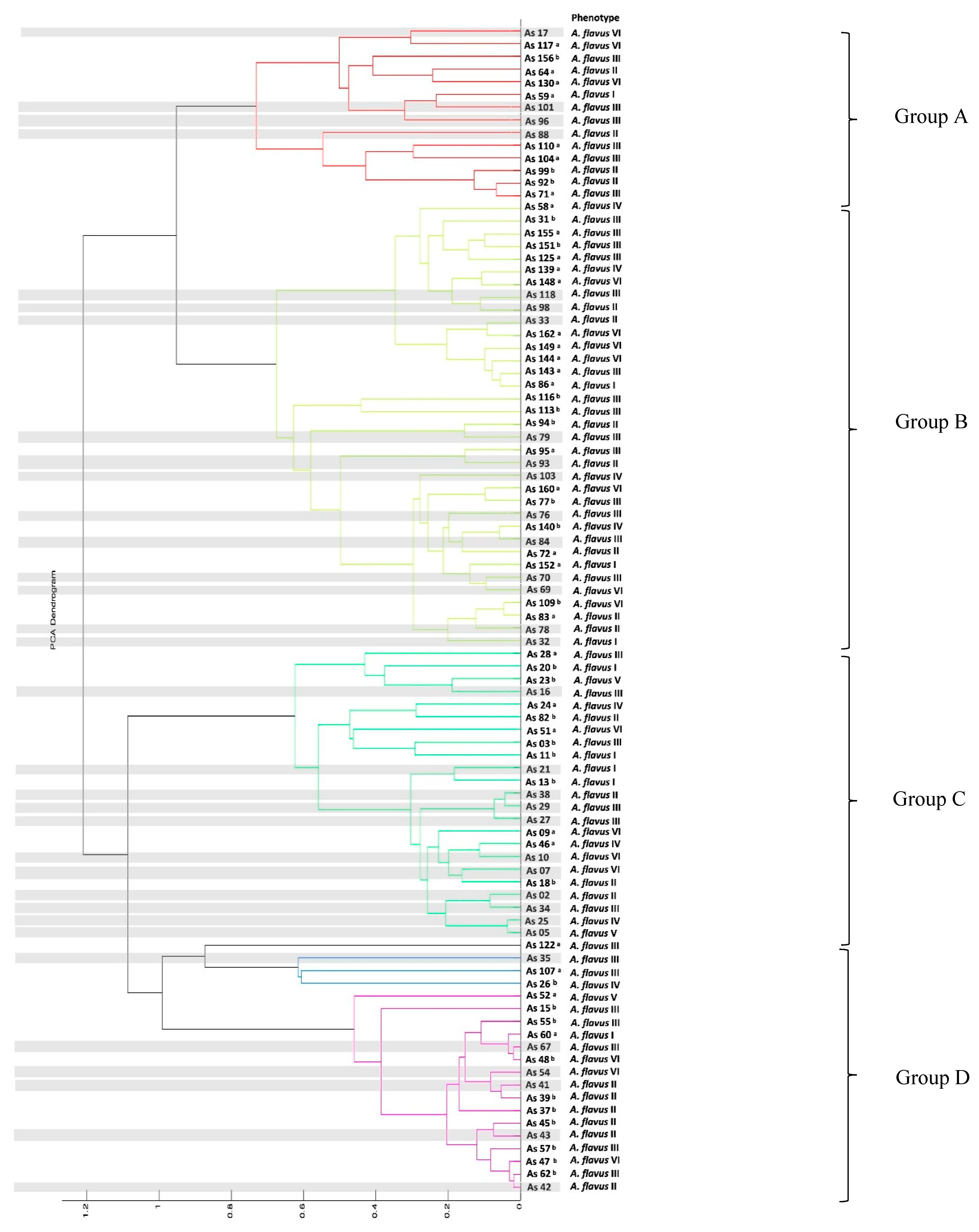


| Maize | Sampling Points (SP) | Frequency (%) | |||
|---|---|---|---|---|---|
| Aspergillus spp. | Fusarium spp. | Penicillium spp. | Sterile Mycelia | ||
| Z. mays var. amylacea | SP1 | 100 ± 0 c | 4± 5 a | 0 ± 0 a | 0 ± 0 a |
| SP2 | 60 ± 5 b | 16 ± 6 b | 10 ± 3 b | 2 ± 1 ab | |
| SP3 | 20 ± 13 d | 45 ± 6 d | 0 ± 0 a | 5 ± 7 ab | |
| SP4 | 58 ± 9 b | 28 ± 8 c | 3 ± 4 a | 2 ± 2 ab | |
| SP5 | 60 ± 4 b | 23 ± 8 bc | 3 ± 3 a | 9 ± 5 b | |
| Z. mays var. indurata | SP1 | 100 ± 0 c | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a |
| SP2 | 11 ± 8 a | 0 ± 0 a | 0 ± 0 a | 4 ± 5 ab | |
| SP3 | 22 ± 19 a | 0 ± 0 a | 1 ± 1 a | 0 ± 0 a | |
| Chemotype | Mycotoxins | N° Isolates (Frequency %) | Producers of Sclerotia (Frequency %) |
|---|---|---|---|
| I | AFB, CPA | 1 (1%) | 1/1 (100%) |
| II | AFB, AFG, CPA | 2 (2%) | 0 |
| III | AFB | 17 (19%) | 16/17 (94%) |
| IV | CPA | 0 | 0 |
| V | Non-producer | 13 (14%) | 12/13 (92%) |
| VI | AFG | 34 (37%) | 25/34 (73%) |
| VII | AFB, AFG | 24 (26%) | 21/24 (87%) |
| VIII | AFG, CPA | 1 (1%) | 0 |
| Maize | Sampling Points (SPs) | Mycotoxins Level (µg.kg−1) ± SD | |||
|---|---|---|---|---|---|
| Total Aflatoxin a | T2 b | ZEA c | FUM d | ||
| SP1 | 20.75 ± 0.6 *A | <LOD | 24.07 ± 0.9 A | 1782.81 ± 271.0 #A | |
| SP2 | <LOD | <LOD | < LOD | <LOD | |
| Z. mays var. | SP3 | <LOD | <LOD | 25.22 ± 3.0 A | 347.98 ± 58.1 B |
| amylacea | SP4 | 1.67 ± 0.2 CD | <LOD | 30.09 ± 3.0 AB | <LOD |
| SP5 | 6.65 ± 1.5 B | <LOD | 21.64 ± 1.6 A | 329.65 ± 36.8 B | |
| Z. mays var. | SP1 | <LOD | <LOD | 22.69 ± 2.0 A | <LOD |
| indurata | SP2 | <LOD | <LOD | 34.88 ± 1.9 B | <LOD |
| SP3 | 2.30 ± 0.7 D | <LOD | 247 ± 6.7 #C | <LOD | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moura-Mendes, J.; Cazal-Martínez, C.C.; Rojas, C.; Ferreira, F.; Pérez-Estigarribia, P.; Dias, N.; Godoy, P.; Costa, J.; Santos, C.; Arrua, A. Species Identification and Mycotoxigenic Potential of Aspergillus Section Flavi Isolated from Maize Marketed in the Metropolitan Region of Asunción, Paraguay. Microorganisms 2023, 11, 1879. https://doi.org/10.3390/microorganisms11081879
Moura-Mendes J, Cazal-Martínez CC, Rojas C, Ferreira F, Pérez-Estigarribia P, Dias N, Godoy P, Costa J, Santos C, Arrua A. Species Identification and Mycotoxigenic Potential of Aspergillus Section Flavi Isolated from Maize Marketed in the Metropolitan Region of Asunción, Paraguay. Microorganisms. 2023; 11(8):1879. https://doi.org/10.3390/microorganisms11081879
Chicago/Turabian StyleMoura-Mendes, Juliana, Cinthia C. Cazal-Martínez, Cinthia Rojas, Francisco Ferreira, Pastor Pérez-Estigarribia, Nathalia Dias, Patrício Godoy, Jéssica Costa, Cledir Santos, and Andrea Arrua. 2023. "Species Identification and Mycotoxigenic Potential of Aspergillus Section Flavi Isolated from Maize Marketed in the Metropolitan Region of Asunción, Paraguay" Microorganisms 11, no. 8: 1879. https://doi.org/10.3390/microorganisms11081879
APA StyleMoura-Mendes, J., Cazal-Martínez, C. C., Rojas, C., Ferreira, F., Pérez-Estigarribia, P., Dias, N., Godoy, P., Costa, J., Santos, C., & Arrua, A. (2023). Species Identification and Mycotoxigenic Potential of Aspergillus Section Flavi Isolated from Maize Marketed in the Metropolitan Region of Asunción, Paraguay. Microorganisms, 11(8), 1879. https://doi.org/10.3390/microorganisms11081879








