A Recombinant Oncolytic Pseudorabies Virus Expressing Interleukin-18, Interferon-Gamma and PH20 Genes Promotes Systemic Antitumor Immunity
Abstract
1. Introduction
2. Materials and Methods
2.1. E. coli Strains and Plasmids
2.2. Cells and Viruses
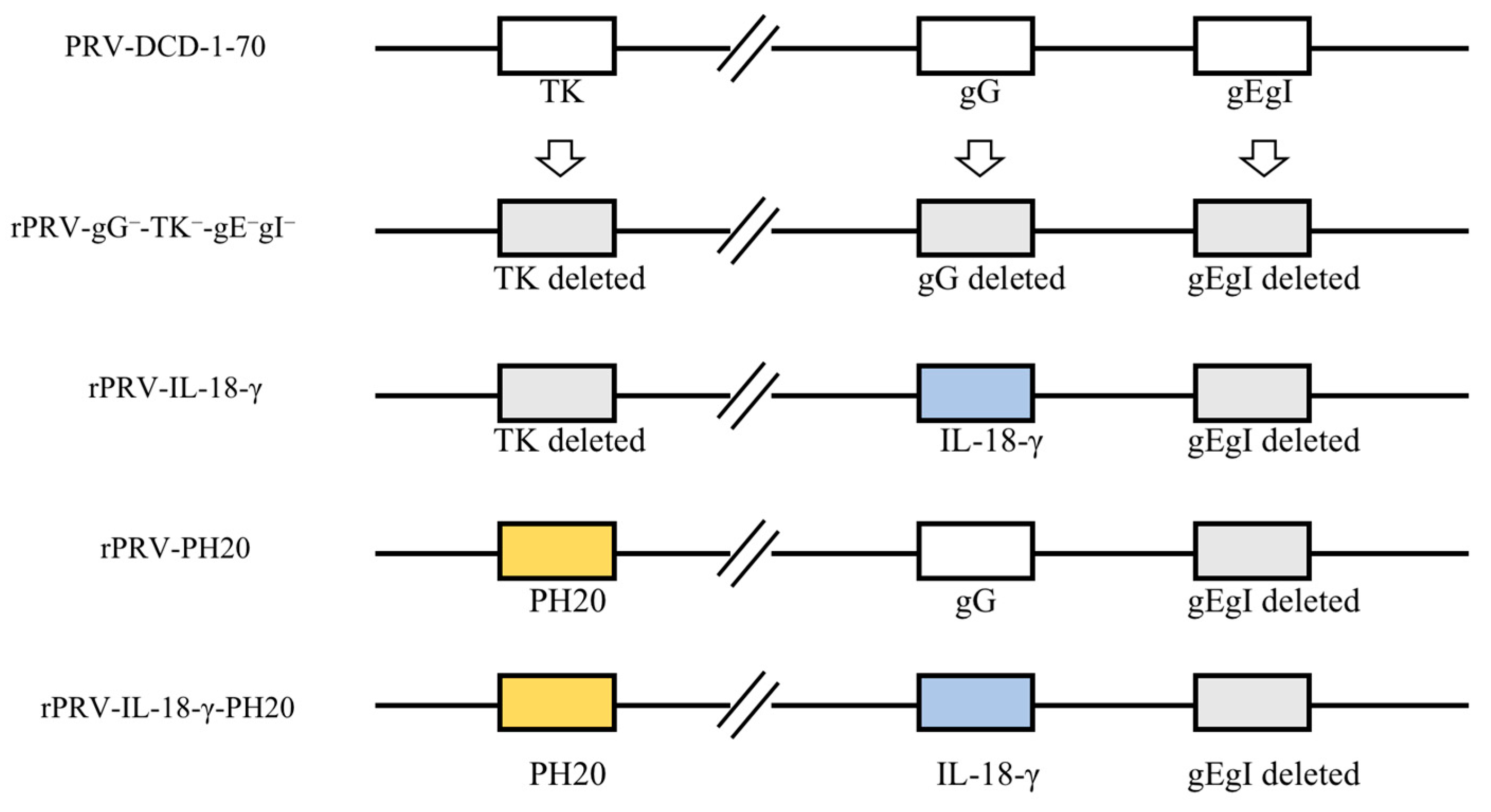
2.3. Construction of PRV Recombinant Virus
2.4. Rescue and Validation of Recombinant Viruses
2.5. Cell Viability Assay
2.6. Animal Experiments
2.7. Histopathological Examination of Tumor
2.8. Immunohistochemical Analysis of Tumor
2.9. Statistical Analysis
3. Result
3.1. Construction of PRV Recombinant Virus Infectious Clones and Virus Rescue
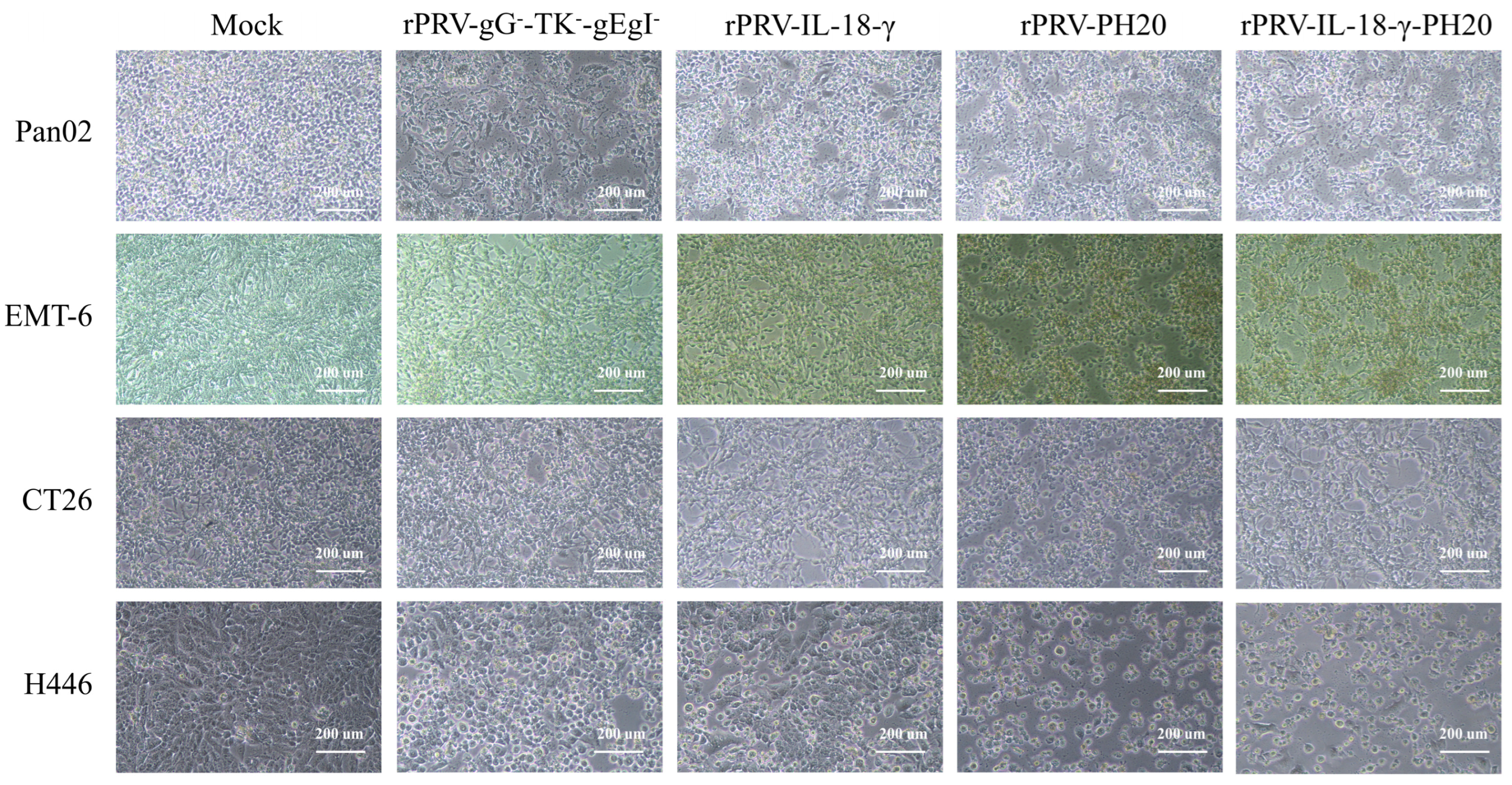
3.2. Pathological Characteristics of PRV Recombinant Virus on Various Tumor Cell Lines
3.3. Oncolytic Spectrum of Recombinant PRV In Vitro
3.4. Antitumor Effect in Pan02 Tumor Models
3.5. Histopathological Examination of Tumor
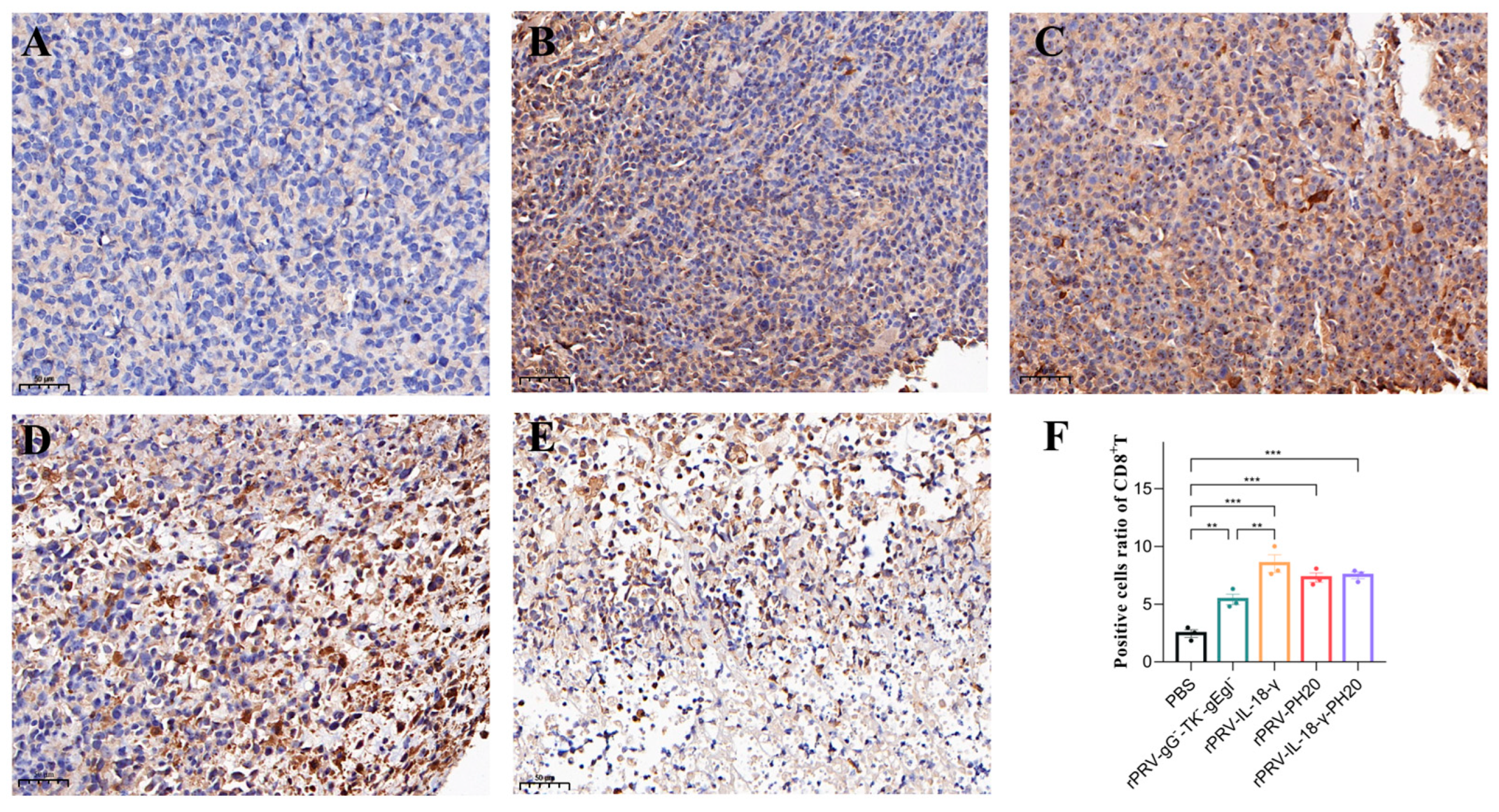
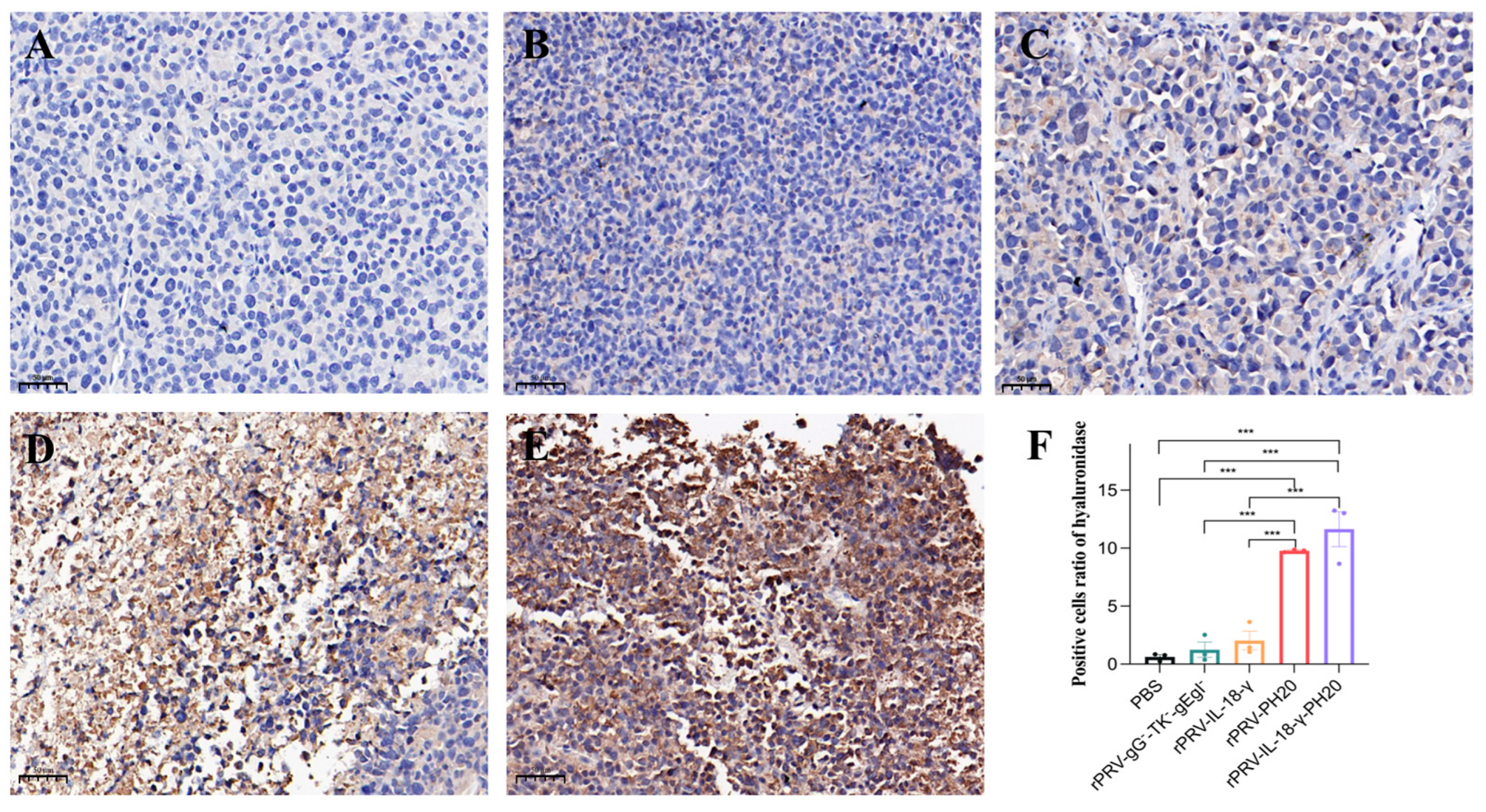
3.6. Immunohistochemical Analysis of Tumor
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hemminki, O.; Dos Santos, J.M.; Hemminki, A. Oncolytic viruses for cancer immunotherapy. J. Hematol. Oncol. 2020, 13, 84. [Google Scholar] [CrossRef]
- Cuddington, B.P.; Verschoor, M.; Ashkar, A.; Mossman, K.L. Enhanced efficacy with azacytidine and oncolytic BHV-1 in a tolerized cotton rat model of breast adenocarcinoma. Mol. Ther. Oncolytics 2015, 2, 15004. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.; Sahu, U.; Mullarkey, M.P.; Kaur, B. Replication and Spread of Oncolytic Herpes Simplex Virus in Solid Tumors. Viruses 2022, 14, 118. [Google Scholar] [CrossRef]
- Uchida, H.; Marzulli, M.; Nakano, K.; Goins, W.F.; Chan, J.; Hong, C.-S.; Mazzacurati, L.; Yoo, J.Y.; Haseley, A.; Nakashima, H.; et al. Effective Treatment of an Orthotopic Xenograft Model of Human Glioblastoma Using an EGFR-retargeted Oncolytic Herpes Simplex Virus. Mol. Ther. 2013, 21, 561–569. [Google Scholar] [CrossRef]
- Chai, C.; Zhang, J.; Zhou, Y.; Yin, H.; Zhang, F.; Diao, Y.; Zan, X.; Ma, Y.; Wang, Y.; Wu, Y.; et al. The Effects of Oncolytic Pseudorabies Virus Vaccine Strain Inhibited the Growth of Colorectal Cancer HCT-8 Cells In Vitro and In Vivo. Animals 2022, 12, 2416. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, H.; Coffman, Z.; Bettencourt, J.; Shipley, T.; Bramblett, D.E. Herpes simplex virus 1 as an oncolytic viral therapy for refractory cancers. Front. Oncol. 2022, 12, 940019. [Google Scholar] [CrossRef] [PubMed]
- Lerma, L.; Alcalá, S.; Piñero, C.; Torres, M.; Martin, B.; Lim, F.; Sainz, B.; Tabarés, E. Expression of the immediate early IE180 protein under the control of the hTERT and CEA tumor-specific promoters in recombinant pseudorabies viruses: Effects of IE180 protein on promoter activity and apoptosis induction. Virology 2016, 488, 9–19. [Google Scholar] [CrossRef]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Gong, J.S.; Yao, Z.Y.; Jiang, J.-Y.; Su, C.; Li, H.; Kang, C.-L.; Liu, L.; Xu, Z.H.; Shi, J.-S.; et al. Insights into the source, mechanism and biotechnological applications of hyaluronidases. Biotechnol. Adv. 2022, 60, 108018. [Google Scholar] [CrossRef]
- Chen, E.; Han, S.; Song, B.; Xu, L.; Yuan, H.; Liang, M.; Sun, Y. Mechanism Investigation of Hyaluronidase-Combined Multistage Nanoparticles for Solid Tumor Penetration and Antitumor Effect. Int. J. Nanomed. 2020, 15, 6311–6324. [Google Scholar] [CrossRef]
- Kiyokawa, J.; Kawamura, Y.; Ghouse, S.M.; Acar, S.; Barcin, E.; Martinez-Quintanilla, J.; Martuza, R.; Alemany, R.; Rabkin, S.D.; Shah, K.; et al. Modification of Extracellular Matrix Enhances Oncolytic Adenovirus Immunotherapy in Glioblastoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Cui, Y.; Zheng, Y.; Li, S.; Lv, J.; Wu, Q.; Long, Y.; Wang, S.; Yao, Y.; Wei, W.; et al. Human Hyaluronidase PH20 Potentiates the Antitumor Activities of Mesothelin-Specific CAR-T Cells Against Gastric Cancer. Front. Immunol. 2021, 12, 660488. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, A.; Giménez-Alejandre, M.; Rojas, J.J.; Moreno, R.; Bazan-Peregrino, M.; Cascalló, M.; Alemany, R. Safety and Efficacy of VCN-01, an Oncolytic Adenovirus Combining Fiber HSG-Binding Domain Replacement with RGD and Hyaluronidase Expression. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Tse, B.W.C.; Russell, P.J.; Lochner, M.; Förster, I.; Power, C.A. IL-18 inhibits growth of murine orthotopic prostate carcinomas via both adaptive and innate immune mechanisms. PLoS ONE 2011, 6, e24241. [Google Scholar] [CrossRef]
- Ihim, S.A.; Abubakar, S.D.; Zian, Z.; Sasaki, T.; Saffarioun, M.; Maleknia, S.; Azizi, G. Interleukin-18 cytokine in immunity, inflammation, and autoimmunity: Biological role in induction, regulation, and treatment. Front. Immunol. 2022, 13, 919973. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Robertson, M.J.; Mier, J.W.; Logan, T.; Atkins, M.; Koon, H.; Koch, K.M.; Kathman, S.; Pandite, L.N.; Oei, C.; Kirby, L.C.; et al. Clinical and biological effects of recombinant human interleukin-18 administered by intravenous infusion to patients with advanced cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 4265–4273. [Google Scholar] [CrossRef]
- Yuan, H.; Zheng, Y.; Yan, X.; Wang, H.; Zhang, Y.; Ma, J.; Fu, J. Direct cloning of a herpesvirus genome for rapid generation of infectious BAC clones. J. Adv. Res. 2023, 43, 97–107. [Google Scholar] [CrossRef]
- Shiau, A.L.; Lin, Y.P.; Shieh, G.S.; Su, C.-H.; Wu, W.-L.; Tsai, Y.-S.; Cheng, C.-W.; Lai, M.-D.; Wu, C.-L. Development of a conditionally replicating pseudorabies virus for HER-2/neu-overexpressing bladder cancer therapy. Mol. Ther. 2007, 15, 131–138. [Google Scholar] [CrossRef]
- Jung, K.H.; Choi, I.-K.; Lee, H.-S.; Yan, H.H.; Son, M.K.; Ahn, H.M.; Hong, J.; Yun, C.-O.; Hong, S.-S. Oncolytic adenovirus expressing relaxin (YDC002) enhances therapeutic efficacy of gemcitabine against pancreatic cancer. Cancer Lett. 2017, 396, 155–166. [Google Scholar] [CrossRef]
- Cheng, J.; Sauthoff, H.; Huang, Y.; Kutler, D.I.; Bajwa, S.; Rom, W.N.; Hay, J.G. Human matrix metalloproteinase-8 gene delivery increases the oncolytic activity of a replicating adenovirus. Mol. Ther. J. Am. Soc. Gene Ther. 2007, 15, 1982–1990. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.-F.; Shang, W.-T.; Lu, G.-H.; Guo, K.-X.; Deng, H.; Zhu, X.-H.; Wang, C.-C.; Tian, J. Decreasing hyaluronic acid combined with drug-loaded nanoprobes improve the delivery and efficacy of chemotherapeutic drugs for pancreatic cancer. Cancer Lett. 2021, 523, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Guedan, S.; Rojas, J.J.; Gros, A.; Mercade, E.; Cascallo, M.; Alemany, R. Hyaluronidase expression by an oncolytic adenovirus enhances its intratumoral spread and suppresses tumor growth. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Farrera-Sal, M.; Moreno, R.; Mato-Berciano, A.; Maliandi, M.V.; Bazan-Peregrino, M.; Alemany, R. Hyaluronidase expression within tumors increases virotherapy efficacy and T cell accumulation. Mol. Ther. Oncolytics 2021, 22, 27–35. [Google Scholar] [CrossRef]
- Alspach, E.; Lussier, D.M.; Schreiber, R.D. Interferon γ and Its Important Roles in Promoting and Inhibiting Spontaneous and Therapeutic Cancer Immunity. Cold Spring Harb. Perspect. Biol. 2019, 11, a28480. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.H.; Qiu, W.H.; Wu, G.S.; Yang, X.-P.; Zou, S.-Q.; Qiu, F.-Z. The immunotherapeutic effect of dendritic cells vaccine modified with interleukin-18 gene and tumor cell lysate on mice with pancreatic carcinoma. World J. Gastroenterol. 2002, 8, 908–912. [Google Scholar] [CrossRef]
- Hoekstra, M.E.; Bornes, L.; Dijkgraaf, F.E.; Philips, D.; Pardiech, I.N.; Toebes, M.; Thommmen, D.S.; van Rheenen, J.; Schumacher, T.N.M. Long-distance modulation of bystander tumor cells by CD8+T cell-secreted IFN-γ. Nat. Cancer 2020, 1, 749. [Google Scholar] [CrossRef]
- Sims, J.E.; Smith, D.E. The IL-1 family: Regulators of immunity. Nat. Rev. Immunol. 2010, 10, 89–102. [Google Scholar] [CrossRef]
- Nakanishi, K.; Yoshimoto, T.; Tsutsui, H.; Okamura, H. Interleukin-18 is a unique cytokine that stimulates both Th1 and Th2 responses depending on its cytokine milieu. Cytokine Growth Factor Rev. 2001, 12, 53–72. [Google Scholar] [CrossRef]
- Iwai, Y.; Hemmi, H.; Mizenina, O.; Kuroda, S.; Suda, K.; Steinman, R.M. An IFN-gamma-IL-18 signaling loop accelerates memory CD8+ T cell proliferation. PLoS ONE 2008, 3, e2404. [Google Scholar] [CrossRef]
- Nakamura, K.; Bald, T.; Smyth, M.J. Cancer-killing, decoy-resistant interleukin-18. Immunol. Cell Biol. 2020, 98, 434–436. [Google Scholar] [CrossRef]
- Zhou, T.; Damsky, W.; Weizman, O.-E.; McGeary, M.K.; Hartmann, K.P.; Rosen, C.E.; Fischer, S.; Jackson, R.; Flavell, R.A.; Wang, J.; et al. IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature 2020, 583, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, Y.; Seng, Z.; Cui, G.; Qiu, J. Bone marrow-derived mesenchymal stem cells co-expressing interleukin-18 and interferon-beta exhibit potent antitumor effect against intracranial glioma in rats. Oncol. Rep. 2015, 34, 1915–1922. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.M.; Horton, K.J.; Coveler, A.L.; Hingorani, S.; Harris, W.P. Targeting the Tumor Stroma: The Biology and Clinical Development of Pegylated Recombinant Human Hyaluronidase (PEGPH20). Curr. Oncol. Rep. 2017, 19, 47. [Google Scholar] [CrossRef] [PubMed]





| Primers | Sequences (5′ to 3′) |
|---|---|
| PH20-homologous arm-F | AGCTCTTAAGGCTAGAGTACTTAATACGACTCACTATAGGGCCACCATGGCTATGGGAGTGCTAAAATTCAAGCA |
| PH20-homologous arm-R | CGTCCCTATACTTTATTCATCATTTTAATGCTAAACGCCTTCAGAAGAAACCAATTCTGCT |
| Primers | Sequences (5′ to 3′) |
|---|---|
| gB-F | tcgacgatgcagttgacggag |
| gB-R | gtgctcttcaaggagaacatcg |
| gG-check-F | gtacgccgggacccatcgccag |
| gG-check-R | gttcagcagccggtccacctg |
| TK-check-F | ttgacttcaaaggccagggtcaag |
| TK-check-R | ggatgcgcgtgtcgttgag |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Sun, J.; Lv, X.; Tang, X.; Zheng, Y.; Ma, J.; Sun, Y. A Recombinant Oncolytic Pseudorabies Virus Expressing Interleukin-18, Interferon-Gamma and PH20 Genes Promotes Systemic Antitumor Immunity. Microorganisms 2023, 11, 1850. https://doi.org/10.3390/microorganisms11071850
Han X, Sun J, Lv X, Tang X, Zheng Y, Ma J, Sun Y. A Recombinant Oncolytic Pseudorabies Virus Expressing Interleukin-18, Interferon-Gamma and PH20 Genes Promotes Systemic Antitumor Immunity. Microorganisms. 2023; 11(7):1850. https://doi.org/10.3390/microorganisms11071850
Chicago/Turabian StyleHan, Xiaohui, Jingshuai Sun, Xiaocheng Lv, Xiaoyu Tang, Yubin Zheng, Jinyun Ma, and Yuan Sun. 2023. "A Recombinant Oncolytic Pseudorabies Virus Expressing Interleukin-18, Interferon-Gamma and PH20 Genes Promotes Systemic Antitumor Immunity" Microorganisms 11, no. 7: 1850. https://doi.org/10.3390/microorganisms11071850
APA StyleHan, X., Sun, J., Lv, X., Tang, X., Zheng, Y., Ma, J., & Sun, Y. (2023). A Recombinant Oncolytic Pseudorabies Virus Expressing Interleukin-18, Interferon-Gamma and PH20 Genes Promotes Systemic Antitumor Immunity. Microorganisms, 11(7), 1850. https://doi.org/10.3390/microorganisms11071850





