Abstract
The first genome sequenced of a eukaryotic organism was for Saccharomyces cerevisiae, as reported in 1996, but it was more than 10 years before any of the zygomycete fungi, which are the early-diverging terrestrial fungi currently placed in the phyla Mucoromycota and Zoopagomycota, were sequenced. The genome for Rhizopus delemar was completed in 2008; currently, more than 1000 zygomycete genomes have been sequenced. Genomic data from these early-diverging terrestrial fungi revealed deep phylogenetic separation of the two major clades—primarily plant—associated saprotrophic and mycorrhizal Mucoromycota versus the primarily mycoparasitic or animal-associated parasites and commensals in the Zoopagomycota. Genomic studies provide many valuable insights into how these fungi evolved in response to the challenges of living on land, including adaptations to sensing light and gravity, development of hyphal growth, and co-existence with the first terrestrial plants. Genome sequence data have facilitated studies of genome architecture, including a history of genome duplications and horizontal gene transfer events, distribution and organization of mating type loci, rDNA genes and transposable elements, methylation processes, and genes useful for various industrial applications. Pathogenicity genes and specialized secondary metabolites have also been detected in soil saprobes and pathogenic fungi. Novel endosymbiotic bacteria and viruses have been discovered during several zygomycete genome projects. Overall, genomic information has helped to resolve a plethora of research questions, from the placement of zygomycetes on the evolutionary tree of life and in natural ecosystems, to the applied biotechnological and medical questions.
1. Introduction
The first “land” fungi retained their flagella for motility and remained adapted to life in water, and several diverging lineages switched to multicellularity while exploiting new environments [1]. A hallmark of the process of adapting to terrestrial life was the loss of the flagellum, which occurred independently several times during fungal evolution, giving rise to fungi with a hyphal growth form [2]. Prominent among early-divergent fungi is a group of lineages historically known as the zygomycetes, which are unified by similarities in reproductive biology (e.g., the formation of sexual zygospores via undifferentiated gametangia) and mostly do not form macroscopic fruiting bodies, making them more difficult to study than Dikarya [3]. Zygomycetes have also been challenging to study because many species are involved in obligate symbiotic interactions and have not been successfully grown in pure culture [4]. However, subsequent molecular studies have shown that the zygomycete fungi are paraphyletic [5]. Today, zygomycetes are recognized as two independent monophyletic groups divided into six subphyla [6]. Terrestrial environments posed novel challenges to early fungi, and the adaptive solutions that these lineages evolved can today be gleaned from the vast library of genes contained within their genomes [7,8]. While these first land colonizers are now extinct, many of their features are preserved in the genomes of extant zygomycetes. One instance, in which whole-genome data have proven useful, is in understanding the origin and evolution of fungi. About 450 million years ago, combined activities of fungi and plants placed the terrestrial world on a trajectory toward its current state, setting the stage for subsequent evolutionary transitions. Primitive mycorrhiza-like relationships and other forms of fungal–plant associations helped facilitate the successful colonization of land by the first terrestrial plants, and the genes responsible for such interactions are still preserved in fungal genomes [9]. Hence, zygomycete genomes have and will continue to play an integral role in our current understanding of the initial transition of organisms to land [10].
The number of sequenced fungal genomes continues to grow exponentially, and genomic data have become widely and freely accessible. While most sequenced genomes are from Ascomycota and Basidiomycota (Dikarya) species, there have been concerted efforts to sequence genomes of early-diverging and underrepresented lineages [11,12,13,14,15]. Since 2014, the number of zygomycete genomes has grown from a dozen [16] to nearly a thousand. However, 70% of these genomes are still in the draft stage, and not all of them are available. While the number of species with sequenced genomes for all organisms is on the rise, the available pool of zygomycete genomes is exceptional due to its diversity, i.e., one-third of ca. 1000 known zygomycete species have already been sequenced (Supplementary Table S1 [17]).
The sequencing efforts have not been uniform among different groups of Mucoromycota and Zoopagomycota; for most species, only a single representative genome is available. While several major clades lack a single published genome (e.g., Eryniopsis and Neozygites in Entomophthoromycotina, and several genera in Zoopagomycotina), numerous isolates of several other species have been sequenced (Figure 1). This discrepancy can be largely explained by practical interests since focal species are sometimes chosen for their relevance to human health or biotechnology [18]. In addition, some taxa are common in culture collections whereas other taxa are challenging to grow in vitro or presumably uncultivable [4,11]. The goal of this review is to (1) provide background into the methods that have been used to sequence zygomycete genomes and which genomes have been sequenced, and (2) consider insights into the genomic architecture, gene content, and evolutionary history gained from these data.
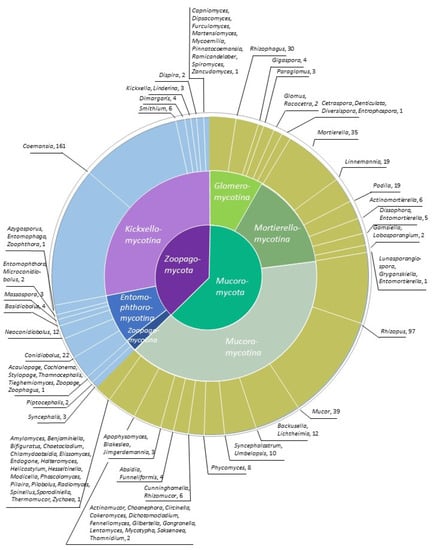
Figure 1.
Distribution of sequenced genomes among genera. Different shades of green and brown—Mucoromycota; purple and blue—Zoopagomycota.
2. Sequencing the Zygomycetes
Axenic cultures remain the preferred and most straightforward method of obtaining high-molecular-weight genomic DNA for sequencing projects [19]. Typically, the easier a species is to grow in culture, the easier it is to obtain pure, unfragmented DNA of high quality. In some cases, however, DNA from a single cell or even nuclei can be extracted and sequenced. This technique has been especially successful for the mycoparasites or fungal parasites of animals (e.g., Piptocephalis, Stylopage, and Zoopage [11,20,21]) or for resolving questions on genetic diversity of the nuclei in Glomeromycotina, such as Rhizophagus [22]. When neither culturing nor single-cell genomics is possible, fungal DNA can be extracted along with that of the host organism. The two genomes can be distinguished bioinformatically (e.g., in the case of Entomopthorales pathogens of insects [23]). In rare cases, high-quality tissue from field-collected fruiting bodies can be used to generate genomes (e.g., Endogone, Jimgerdemannia, and Modicella [12,14,24]).
The diversity of DNA sequencing technologies allows researchers to select the tools that are most appropriate for addressing specific questions but also requires setting minimal reporting standards for each type of sequencing technology. During sequence processing, genomic DNA is cut and sequenced in length spans from 100–300 base pairs (short reads, Illumina) to 1000–60,000 base pairs (long reads, PacBio, or MinION). Illumina short-read sequencing remains the dominant technology used to generate fungal genomes, while long-read technologies such as PacBio and Oxford nanopore are being used with increased frequency, especially for larger genomes (Supplementary Table S1). Illumina sequencing generally provides the greatest sequencing quality and accuracy, although millions of short raw reads (150–300 bp) are produced; as a result, genome assembly can be challenging especially with genomes featuring large amounts of repetitive DNA. Assembly quality might be adjusted by combining the use of short reads from Illumina with long reads from another sequencing platform. The use of longer reads (up to 60,000 bp) facilitates read alignment and/or mapping, often achieving the length of the chromosome [25] and allows precise resolution of repetitive sequences of rDNA [26]. However, PacBio and other long-read technologies typically demand higher quality and quantity of DNA than Illumina. On the other hand, these technologies are changing rapidly as different companies compete for the market share and make efforts to increase the number and length of reads and to maintain and improve sequence accuracy [27].
The majority of zygomycete genomes are assembled de novo, even when assemblies of closely related species are already available. Choice of assembly software usually depends on the data type. For example, AllPaths-LG works best for assembling Illumina short-read sequences, while Falcon is preferred for longer PacBio reads (Supplementary Table S1) [28]. The most widely used genome assemblers for zygomycetes include gsAssembler (Roche Diagnostics, Indianapolis, IN, USA), Newbler [18], Velvet [29], Arachne, Celera [30], HGAP [31], JGI Jazz (especially among earlier assemblies from 2001–2014) [32], Ray [33], CLC Genomics Workbench 20.0 (https://digitalinsights.qiagen.com, accessed on 10 July 2023), MaSuRCA [34], Falcon, AllPaths-LG [35], SOAPdenovo [36], SPAdes [37], SOLiD [38], Canu [39], FinisherSC [40], Flye [41], and MEGAHIT [42] (Figure 2).
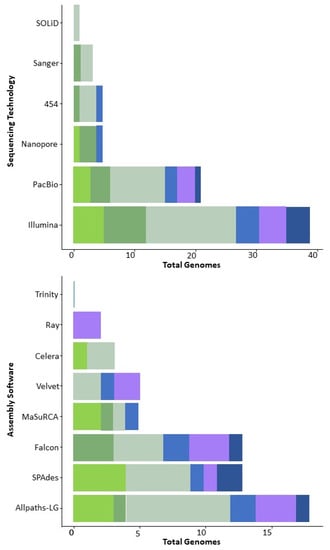
Figure 2.
Approaches used in the generation of zygomycete genomes: sequencing (upper panel) and assembly (lower panel) technologies. Different shades of green and brown—Mucoromycota; purple and blue—Zoopagomycota.
Most zygomycete genome assemblies are similar in size to the average fungal genome: 30–50 Mb (Supplementary Table S1). Notable exceptions include species in Entomophthoromycotina, Glomeromycotina, and Mucoromycotina, whose large genomes compare in size to Arabidopsis thaliana (135 Mb), Caenorhabditis elegans (100 Mb), or Drosophila melanogaster (140 Mb) [43,44]. However, there are still far smaller than the genomes of some of the Basidiomycete fungi that cause rust diseases on plants, which includes the largest fungal genome sequenced to date (Austropuccinia psidii, 1 Gb) [45]. Most zygomycete genomes contain over 10,000 predicted genes. Rhizophagus spp. (Glomeromycotina) are a notable exception; they contain more than double the typical number with 26–30 K predicted genes (Figure 3). The smallest numbers of genes are found in Zoopagomycotina (4–8 K), followed by 6–11 K in Kickxellomycotina, 9–16 K in Entomophthoromycotina, Mucoromycotina and Mortierellomycotina, and 20–30 K in Glomeromycotina (Supplementary Table S1). However, these numbers may change for Entomophthoromycotina and Zoopagomycotina since there are only a few representative genomes available for those lineages.
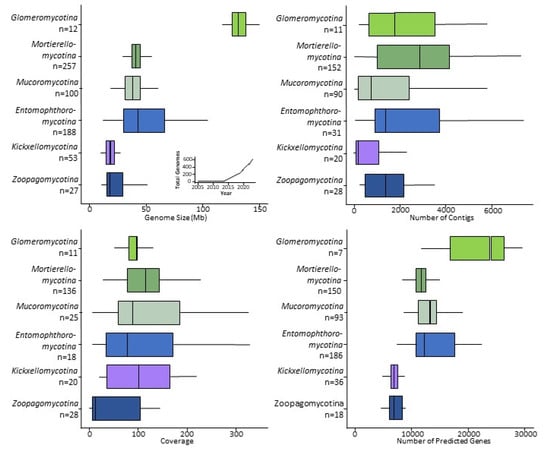
Figure 3.
Genome statistics for zygomycete sequences. Sizes of the genomes and number of the genomes sequenced since 2005 (upper left), number of contigs in assemblies (upper right), estimated sequencing depth coverage relative to the single genome (lower left), and the number of the predicted genes (lower right). Different shades of green and brown—Mucoromycota; purple and blue—Zoopagomycota.
During the assembly process DNA sequences are combined into contigs, which are sometimes merged to form scaffolds (if sufficient evidence exists to support these merges). These scaffolds of assembled DNA sequences should ideally equal the number of chromosomes [46]. In practice, the number of scaffolds rarely matches exactly the number of chromosomes and, for most species, the exact number of chromosomes is unknown [47]. Many genomes of easily culturable Mucoromycota with small genomes have the lowest number of scaffolds (e.g., 12 in Actinomortierella wolfii), while the genomes of species with larger genomes (e.g., taxa in Glomeromycotina and Entomophthoromycotina) are more fragmented such as 210 contigs in Rhizophagus irregularis [26], or 373,021 in Massospora cicadina [48]. These larger assemblies are due to larger overall genomes (e.g., larger number of chromosomes) and because of the difficulties with assembly caused by polyploidy, numerous repeats, and transposons [49]; these aspects are prevalent in many Mucoromycota and Zoopagomycota genomes (Supplementary Table S1). However, the sequenced genomes with hundreds of thousands of scaffolds often reflect low sequence coverage relative to the size of the genome, resulting in highly fragmented assemblies. All these factors contribute to the number of the scaffolds, not reflecting the actual numbers of chromosomes.
Genome quality is also improved by increasing the sequence coverage. Some zygomycete genomes have been sequenced at a depth of 500–700× and many genome projects aim for a mean depth of ca. 100× coverage (Supplement Table S1). High-quality genome assemblies rely on increased sequencing depth, particularly in cases of high intraspecific polymorphism or variable genome architecture, e.g., species of Glomeromycotina and many noncoding regions and transposons, as in the genomes of Entomophthoromycotina species. However, some researchers have used low-coverage genome sequencing (ca. 10× coverage) as a cheap and efficient method to obtain a smaller number of orthologous gene regions for phylogenomic analyses [12,15,24].
3. Genomes and the Evolution of the Zygomycetes
The first genome sequence of a zygomycete fungus was from a clinical strain of Rhizopus delemar that was isolated from a patient with mucormycosis—a highly destructive and lethal infection that is typically seen in immunocompromised hosts. Sequencing of the R. delemar genome was driven from the clinical perspective, but it also provided the first genome of a fungus outside of the Dikarya [18]. Moving forward a decade, a landmark project generated genomic sequences of all species known to cause mucoromycosis in humans. Thirty zygomycete genomes were sequenced and assembled for this project [50]. Another substantial advance in zygomycete genomics was made by the ZyGoLife team, comprising the efforts of 12 mycological labs across the USA and Canada in collaboration with the US Department of Energy Joint Genome Institute (JGI) [14]. The phylogenomic study by Spatafora et al. [6] found that the former Zygomycota phylum was not monophyletic but instead comprised two distinct lineages—the primarily plant-associated saprotrophic and mycorrhizal Mucoromycota versus the primarily mycoparasitic or animal-associated parasites and commensals in the Zoopagomycota. However, further analyses using phylogenomics data to resolve the evolutionary relationships of zygomycetes have yielded slightly conflicting results. Although Zoopagomycota and Mucoromycota are maintained as monophyletic lineages, a recent study by Li et al. [51] found that Zoopagomycota and Mucoromycota formed one larger monophyletic clade in some (but not all) of their analyses. In contrast, other recent phylogenomic studies by Strassert and Monaghan [52] and Galindo et al. [53] recovered a topology that was similar to Spatafora et al. [6], where the Zoopagomycota were resolved as the sister group to all terrestrial fungi. In many of the studies, the position of Glomeromycotina has been problematic, suggesting that more work will be needed to resolve the placement of this clade [51,54].
One of the greatest debates in early-diverging fungal evolution was the ploidy of the dominant life form given the predominantly haploid nature seen in the Dikarya. One outcome of the analysis of genome sequences of zygomycetes species is that diploids are more common than expected [9,55,56,57].
3.1. Mucoromycota
Mucoromycota comprises many critically important plant symbionts, including the arbuscular mycorrhizal fungi (Glomeromycotina) [58], Endogonales (Mucoromycotina) [24], “fine root endophytes” (Mucoromycotina) [59], and putative root endophytic species including Mortierella (Mortierellomycotina) [60], Pygmaeomycetaceae (Mucoromycotina) [61], and Umbelopsis (Mucoromycotina) [62]. On the other hand, many fungi within this group are also noted plant pathogens (e.g., Rhizopus microsporus, which causes rice seedling blight, and various Choanephora, Gilbertella, and Mucor species that cause post-harvest rots) as well as numerous species (Actinomucor, Apophysomyces, Cokeromyces, Cunninghamella, Lichtheimia, Mucor, Rhizomucor, Rhizopus, Saksenea, Syncephalastrum) that can cause the disease mucormycosis in humans and other animals [63].
Most zygomycete genomes are of species within Mucoromycota (Figure 1); as a result, much of what we know about genome evolution among early-diverging fungi comes from this phylum. Genome duplication appears to have played a significant role in their evolution, as first described for Rhizopus delemar and Mucor circinelloides [18,64]. Genome sequencing has also revealed a diversity of endobacteria that may be found as symbionts in the Mucoromycota, e.g., in Rhizopus microsporus and Endogone pisiformis (Mucoromycotina), Linnemannia elongata and Mortierella alpina (Mortierellomycotina), and Rhizophagus irregularis (Glomeromycotina) [65,66,67,68,69,70]. Many plant-associated fungal lineages contain endobacteria, but the potential role of these symbionts in the land colonization process remain to be characterized. “Foreign” DNA in the form of diverse mycoviruses has also been frequently detected during genome sequencing of zygomycetes [30,71]. Genomics enabled the systematic study of genome architecture [72,73,74], genetic manipulations [75], and intra-isolate genomic variations [76]. Genome-wide searches and comparisons have also detected evidence for transposon spread through genomes [77,78,79] and uniquely high 6-methyladenine DNA modifications in zygomycete fungi as compared to Dikarya [80]. Studies on metabolic patterns across the fungal kingdom, the search for the mating gene loci and protein families [81,82,83], and the phylogenies of certain taxonomic groups [84,85] have also been conducted using-whole genome sequencing data. Genome-based research has also provided insights into the trophic modes of zygomycete fungi, including the evolution of mycorrhizae [24,86] and commonalities in the pathogenicity of Mucoralean fungi [50,87].
3.2. Mucoromycotina
Genome sequencing of this subphylum has been the most comprehensive among all the basal fungal lineages although genomes are still unavailable for several genera, including species of Siepmannia, Chlamydoabsidia, and Utharomyces, as well as for two new root-associated species of Pygmaeomyces of the new genus and family Pygmaeomycetaceae [61]. Furthermore, the plant-associated order Endogonales remains highly undersampled despite recent advances [24,88]. Some of its “fine root endophytes” colonize early-diverging plants, and sometimes co-occur with AMF [89,90]. The average genome size in Mucoromycotina species ranges from 35 to 50 Mbp, with species of Umbelopsis at the low end of the spectrum (22–30 Mbp) and Endogonales such as Jimgerdemannia species at the higher end (240 Mbp) (Supplementary Table S1). For some genera, several hundred genomes have already been sequenced. For example, there are >220 genomes available for species of Rhizopus, mostly of R. arrhizus (syn. R. oryzae) and its varieties and synonyms, and of R. microsporus, R. delemar, and R. stolonifer (Supplement Table S1). Rhizopus is increasingly studied as a model organism thanks to its availability in culture collections and to its important role in biotechnology and human health. Genomic information might be helpful for future studies of pathogenicity in Rhizopus species since infections by members of this genus are the leading cause of human mucormycosis [50,84]. Phylogenomic data suggest that species of Rhizopus are part of a well resolved monophyletic group. However, this is in stark contrast to the situation among members of the genus Mucor (>150 genomes), which is incredibly diverse and is likely polyphyletic (Figure 4). The additional Mucoromycotina genomes consist mostly of saprotrophic species that are relevant to human health (species of Lichtheimia and Cunninghamella) and biotechnology (Phycomyces blakesleeanus) [91,92,93]. Genomes of Endogonales (as well as Glomeromycotina) contain effectors to facilitate plant–fungus interactions, which apparently aided in the terrestrialization of Earth by early land plants. Genome information helped to describe and clarify the phylogenetic placement of new lineages and species, such as Bifiguratus [94] and Calcarisporiella [95].
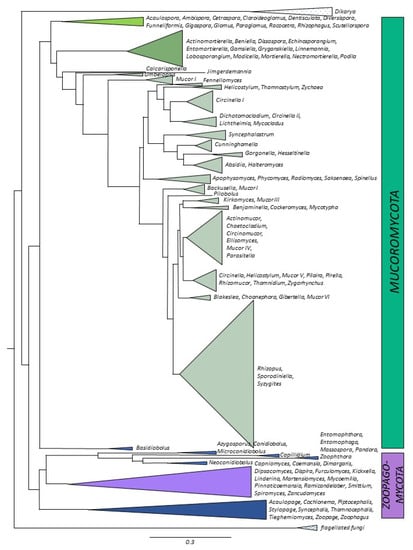
Figure 4.
Distributions of the number of genomes sequenced imposed onto phylogenetic tree. Different shades of green and brown—Mucoromycota, purple and blue—Zoopagomycota.
3.3. Mortierellomycotina
This subphylum contains an estimated 170 species [96] and was recently circumscribed into 13 monophyletic genera using multi-locus and low-coverage genome sequencing [12]. Reference genome sequences of both low quality and high quality are now available for representatives for most genera, including Actinomortierella, Benniella, Dissophora, Entomortierella, Gamsiella, Gryganskiella, Linnemannia, Lobosporangium, Lunasporangiospora, Mortierella, Necromortierella, Podila, and the sporocarpic genus Modicella [12,97]. A high diversity of Mortierellomycotina species can be isolated from soil, but it appears that genera may differ in their ecological roles. For example, Necromortierella species appear to be necrotrophic, whereas Actinomortierella spp. are often associated with millipedes, and Entomortierella spp. are often associated with arthropods [98]. Many genera, such as Linnemannia and Podila, have been recovered as rhizosphere associates and endophytes of plant roots [99,100], and they appear to improve plant growth and impact flower and seed production [101]. Several species such as Mortierella alpina have been sequenced due to their use in industry and biotechnology for lipid production [102].
3.4. Glomeromycotina
Arbuscular mycorrhizal fungi (AMFs) are arguably the most common and oldest symbionts of terrestrial plants [58], credited with the role in the facilitation of plant transition to the terrestrial habitat [103]. AMFs supply their plant hosts with mineral nutrients translocated from the soil in return for plant-assimilated carbon [104]. Yet, there are not many genomic sequences of these organisms available due to historical impediments hampering sequencing projects, including multinucleate spores with different genotypes, obligate biotrophy of AMFs, and the initial uncertainty concerning their genome sizes, with conflicting size estimates in Rhizophagus irregularis ranging from 14.1 [105] to 154.8 Mb [106]. Once these obstacles were surmounted, genomic data confirmed that AMFs have unusually large genomes, ranging from 153 Mb in R. irregularis [26] to 773.1 Mb in Gigaspora margarita [107]. This size enhancement relative to other Mucoromycota is driven by lineage-specific expansions of gene families and massive proliferation of transposable elements [26,86,107]. Genomic data also revealed that AMFs display several other features that differentiate them not only from Mucoromycota but also from other fungi. For example, despite the lack of morphological evidence of sexual reproduction, the R. irregularis genome harbors a candidate mating-type locus with two genes encoding homeodomain-like transcription factors [108]. If these homeodomain-like genes indeed control sex, then regulation of the reproductive processes in AMFs would resemble mating in Dikarya rather than in Mucoromycota, which rely on HMG domain proteins for sex determination [109]. Rhizophagus irregularis differs also from other fungi in the organization of its rRNA operons. Instead of being tandemly arrayed as they are in other fungi, the rRNA operons in Glomeromycotina are dispersed individually across the genome [26]. This important discovery put an end to a long-lasting debate on whether intraindividual rRNA gene variation typical for AMFs is distributed among dissimilar nuclei [110] or contained in each nucleus [111], supporting the latter scenario. Lastly, genome-based inferences of metabolic capacity revealed that AMFs lack the gene encoding fatty acid synthase [112,113,114], which makes them obligately dependent on plant hosts for lipids [115,116,117,118]. As in other fastidious organisms, this information is important for devising cultivation strategies that complement fatty acid auxotrophy of AMF [119,120]. Even though accumulation of genomic data for AMF has lagged behind other Mucoromycota, the insights gathered so far have been critical for resolving several puzzles in the biology of these unique organisms. PacBio HiFi and Hi-C sequencing of all available AMF heterokaryons helped to discover the two sets of coexisting homologous chromosomes in one heterokaryon. Genes required for plant colonization were found in gene-sparse, repeat-reach compartments [121]. Another important question that remains to be addressed is which genomic features of AMFs contribute to superior yield outcomes in agronomic crop species, as AMFs do not universally benefit all plant hosts.
3.5. Zoopagomycota
In contrast to Mucoromycota, most species of Zoopagomycota are parasites or pathogenes of animals and fungi. This includes the insect pathogenic group Entomophthoromycotina (some members of which exhibit behavioral control over their hosts) [122], the Kickxellomycotina that includes a mixture of ecologies (including commensalistic arthropods gut fungi) [15], and Zoopagomycotina, which are parasites of fungi (mainly in Mucoromycota) and microinvertebrates (e.g., amoebae, nematodes, rotifers) [20]. There are also opportunistic human pathogens in this phylum, including species of Basidiobolus and Conidiobolus [123]. Axenic culturing of some fungi in Zoopagomycota is possible [124,125] but extracting high-quality genomic DNA from many species remains difficult. Sometimes there is a need to extract genomic material of the parasite together with the host (often another fungus or insect) and separate their genomes during assembly, which creates additional challenges. Small amounts of available DNA have necessitated the use of single-cell DNA extraction in some cases, especially in species with dramatically reduced vegetative mycelial structures. Much greater success has been achieved for putative saprotrophic fungi such as dung-inhabiting species of Kickxellales (Supplementary Table S1). Genomes and transcriptomes have been used primarily to resolve various evolutionary questions, including the monophyletic origin of Zoopagales [20], evolution of ploidy from a diploid ancestral stage [11], and loss of a large number of pectinases and other plant cell-wall-degrading enzymes in Zoopagomycota [126]. Genome data were also successfully used to resolve evolutionary relationships within particular taxonomic groups, such as Massospora [98], to track other evolutionary events such as horizontal gene transfer events in gut fungi [127], and to reveal the genetic toolbox in insect symbionts [128]. These studies help us to understand the evolutionary trajectories of genome size, namely, a dramatic increase in genome size for the insect pathogens and insect endosymbionts as compared to the genome size in the saprobes and mycoparasites. Genome data have also helped to clarify fundamental differences among Zoopagomycota lineages in their production of secondary metabolites [129], as well as metabolic pathways in general [11]. Mating genetics in this phylum is still not well understood, and that is a question that remains to be addressed with genomic data.
3.6. Kickxellomycotina
This subphylum contains an interesting mix of mycoparasites (Dimargaritales), putative saprotrophs (Kickxellales), and obligate insect endobionts (Asselariales, Barbatosporales, Harpellales, and Orphellales). The Dimargaritales are similar to mycoparasites in the Zoopagales in that they form penetration hyphae (haustoria) and mostly attack similar host species in the Mucoromycota. In contrast to the Zoopagales mycoparasites, species of Dimargaritales appear to be rare in the environment and have mostly been isolated from dung rather than soil. Recent genomic analyses have also highlighted that members of the Dimargaritales that have larger genomes and more predicted secondary metabolite genes compared to their close relatives [15,129]. Furthermore, single-cell genome data have suggested that Dimargaris cristalligena may be nonhaploid and missing certain enzymes from metabolic pathways (e.g., thiamine biosynthesis and biotin metabolism) [11]. Identifying such genomic features of symbiotic species can provide targets for further experimental tests to understand the molecular interactions between hosts and parasites.
On the other hand, the Kickxellales is the only group of Zoopagomycota containing mostly saprotrophic species, and many appear to be rare in the environment, having only been reported in the literature one or a few times. An exception is the genus Coemansia, the species of which are commonly isolated from soil and dung samples [4,130]. Genome sequencing efforts have demonstrated that Kickxellales species have small genomes (less than 40 MB) and variability in their predicted secondary metabolite genes, with Coemansia and Linderina, along with more polyketide synthases than other Kickxellales and D. cristalligena having more nonribosomal peptide synthetases than other Kickxellales [129]. A preliminary rDNA phylogeny showed polyphyly among Kickxellales [130], and a large-scale sequencing project that utilized low-coverage genome data from >100 isolates to test these hypotheses, as well as explore the diversity of Coemansia species, resolved many of these relationships [15].
The gut fungi (Trichomycetes) include arthropod symbionts for which several draft genomes have been available since 2016 [131]. These fungi are distinct from the saprotrophic and coprophilic relatives (i.e., Kickxellales) and belong to four distinct phylogenetic lineages—Asellariales, Barbatosporales, Harpellales, and Orphellales [132]. Asellariales are associated with isopods and spring tails (Collembola) [133]. Species within the three genera (Asellaria, Baltomyces, and Orchesellaria) have been recovered from the both terrestrial and aquatic hosts [133], but their phylogenetic relationships remain unclear, as sequence data have only been obtained from Asellaria [132], and no cultures are available for this group.
Harpellales species are usually found in lower Diptera insect larvae and have coevolved with their hosts for over 200 million years [134]. Species of Orphellales are stonefly nymph symbionts with unusual morphological characters (i.e., asexual and sexual spore shapes, and thalli protruding beyond the anus of insect hosts) [135], which cluster together with other Trichomycetes and the Spiromycetales for a sister clade to Kickxellales [15]. Smittium is a well-studied genus within Harpellales with approximately 100 described species [136]. However, there are only nine whole-genome sequences available for these insect gut-dwelling fungi, and all were made from culturable Harpellales species, with the best assembly—Capniomyces stellatus—having 72 scaffolds [131]. Barbatosporales are monotypic, and the single species (Barbatospora ambicaudata) has only been recorded from black fly larvae collected in the Great Smoky Mountains National Park in Tennessee, obtained in axenic culture [137].
Although we only have a handful of Harpellales genomes, they have already expanded our knowledge and understanding of these enigmatic microbial fungi in multiple ways. For example, a mosquito-like polyubiquitin gene has been identified in Zancudomyces culisetae (Harpellales), which was presumably acquired via a horizontal gene transfer event [127]. Comparative genomics revealed a fungus–insect symbiotic core gene toolbox (FISCoG) that is shared among the insect-associated fungi (e.g., Harpellales, Basidiobolus, and Conidiobolus) and higher-ranked Hypocreales (Ascomycota). However, the insect pathogens (Hypocreales and Entomophthoromycotina members) tend to have genomes enriched in genes that are useful for pathogenic processes such as the platelet-activating factor acetyl-hydrolase coding genes, whereas the gut commensals have genomes enriched in cell adhesion genes for a successful gut-dwelling lifestyle [128]. In addition, Harpellales genomes also facilitated a kingdom-wide study to confirm the production of selenoproteins in early-diverging fungal lineages [138]. As a major group of early-diverging fungi, representing seven of the nine fungal species that utilize selenoproteins, Harpellales may take the advantages of selenocysteine over cysteine for specific oxidoreductase functions.
Unfortunately, no efforts have been successful in obtaining genome information of the Asellariales, Barbatosporales, and Orphellales, which colonize different host types. However, multigene analyses incorporating low-coverage genome data (along with data from Asellariales and Orphellales) suggested an alternate hypothesis for the evolution of fungal–insect associations. Contrary to previous topologies that suggested multiple events [132], the larger dataset showed a possible single origin of gut fungi [15]. Further evolutionary evidence may be identified with the help of their genome sequences to understand how these gut-dwelling microbial fungi interact with various aquatic insect hosts and evaluate the origins of the symbiosis.
3.7. Zoopagomycotina
Zoopagomycotina species are fascinating, partially due to their cryptic ecological niches and ability to parasitize various hosts such as amoebae, nematodes, rotifers, and other fungi. It is still challenging to culture most of the Zoopagomycotina isolates. Thus, it is not surprising to see that the Zoopagomycotina has the lowest number of available genomes, the qualities of which are not comparable to other lineages, especially the well-sequenced Mucoromycotina. Recently developed single-cell genomic techniques have provided an alternative, culture-independent method to obtain their genomic information [11]. On the basis of single-cell or multiple-cell libraries, the completeness of genome assemblies can be as high as 89.6% (Piptocephalis tieghemiana RSA 1565) [20]. The assembled Zoopagomycotina genomes indicate that this clade of fungi is characterized by small genome size, but the numbers of protein-coding genes are not necessarily low. Some of the genomes can encode close to 10,000 genes (e.g., Zoophagus insidians, a predator of rotifers and nematodes (Supplement Table S1). It seems likely that these completely microscopic fungi are capable of more complicated and yet-to-be identified interactions with their animal and/or fungal hosts, but more work will be needed to explore these interactions.
Two additional Zoopagales have remarkably small genomes: a species of Acaulopage (11 Mbp) and a species of Zoopage (14 Mbp). No genomes have yet been published for the following genera: Amoebophilus, Aplectosoma, Bdellospora, Brachymyces, Cystopage, Endocochlus, Euryancale, Helicocephalum, Reticulocephalis, and Sigmoideomyces.
3.8. Entomophthoromycotina
This lineage remains the most insufficiently sequenced fungal group at the subphylum level. Despite many genomes from the genus Conidiobolus, there are only a few other species (all from the key family Entomophthoraceae) for which genomes are available. The genus Conidiobolus is a polyphyletic assemblage of multiple lineages. This group of Conidiobolus-like fungi was recently phylogenetically divided into three new families, using a combined multi-gene and phylogenomic approach, revealing that the ballistic conidia arose prior to their transition to a parasitic lifestyle [85,139]. Difficulties with culturing and, therefore, obtaining enough high-molecular-weight genomic DNA represent a reason why some species only have transcriptomes available. Alternatively, in some cases, the fungal pathogen is sequenced together with its host, and then the reads are separated bioinformatically. Aside from the polyphyletic genus Conidiobolus and entomophthoroid-like Basidiobolus, genome or transcriptome data are available only for a few species of Entomophaga, Entomophthora, Massospora, Pandora, and Zoophthora. No genomes yet exist for Ancylistes, Apterivorax, Batkoa, Erynia, Eryniopsis, Furia, Macrobiotophthora, Meristacrum, Neozygites, Orthomyces, Tabanomyces, Thaxterosporium, Schizangiella, and Strongwellsea. The largest genomes recovered are over 2000 Mbp in Massospora and 650 Mbp in Zoophthora, with possibly similar genome sizes in Entomophthora and Entomophaga (Supplementary Table S1). Phylogenetic analysis recently demonstrated that Basidiobolus, a genus of amphibian gut-dwelling fungi, does not belong to Entomophthorales, where it was traditionally placed, mostly on the basis of its production of forcibly discharged conidia that are morphologically similar to those in other entomophthoralean fungi [140]. The genus was difficult to place with rDNA data [141], but was inferred either as a separate clade sister to the Mucoromycota [51] or as the earliest diverging lineage within Zoopagomycota [128], on the basis of phylogenomic data. The ballistic spore dispersal mechanism, which differs structurally from that found in the Entomophthorales, is likely an example of convergent evolution among fungi that appear to be similar but are distantly related. In addition to being evolutionarily enigmatic, the genomes of B. meristosporus and B. heterosporus are larger than most other basal fungi and contained a much higher proportion of secondary metabolite-encoding genes than other Zoopagomycota species that have been studied thus far. Furthermore, Basidiobolus genomes contain numerous genes that putatively originated from horizontal gene transfer events from bacteria [129].
4. Insights from Genomics
4.1. Adaptation to Terrestrial Life Inferred from Genomes
The transition to a terrestrial environment placed novel constraints on species of early-diverging fungi, especially in terms of their resistance to desiccation and solar radiation. Genomes have been essential to studying which zygomycete genes made this transition possible.
Sensing of and response to light have been explored in the model Mucorales species Phycomyces blakesleeanus. Mutants impaired in photoresponses, especially phototropism, were isolated as reported in the 1970s [142]. However, the first genes related to photoresponses were not reported until 2006 [143]. These were identified using a candidate gene approach in which P. blakesleeanus was assumed to have similar blue light sensors as the Dikarya species [143,144], wherein a two-protein complex was first identified in Neurospora crassa, defining a sensing and signaling system that uses a light-sensing domain coupled to transcription factor roles for signal transmission. With the release of the P. blakesleeanus genome sequence in 2010, identifying a candidate gene for madB as encoding the interacting protein was achieved using BLAST against the genome sequence [145]. The genome sequence was performed along with that of M. circinelloidies and revealed an expansion of this pair of genes required for light responses (3–4 copies of each). Being able to make targeted gene mutations in M. circinelloides of these homologs, as based of genome sequence information, provided a clear example of subfunctionalization after gene duplication [146]. The identification of the mutations in madI and madC mutants in P. blakesleeanus used traditional genetic mapping with molecular markers derived from the comparison of the genomes between two strains. With the regions narrowed, candidate genes could be identified and confirmed by sequencing [64,147]. These genes play conserved roles in photobiology in the Dikarya species, suggesting that this innovation for terrestrial life had already evolved in early-diverging fungal lineages. For instance, phenotypic analysis of the P. blakesleeanus madA and madB mutants indicated that this pair of genes has a conserved function in protecting fungi from harmful ultraviolet light [148].
In addition to responding to light, many Mucorales species can sense gravity. This has been correlated with the formation of vacuolar crystals in P. blakesleeanus [149]. Through purification of such crystals, followed by mass spectrometry of the associated proteins using the mass database derived from the genome sequence, the OCTIN protein was identified. A gravity-insensitive mutant had a mutation within the corresponding gene. Remarkably, phylogenetic comparison revealed that the origin of the gravity-sensing gene OCTIN in the Mucorales was most likely via a horizontal gene transfer from a bacterial species [150]. Other challenges associated with terrestrial life, such as fluctuations in temperature and desiccation, have yet to explored from a genomic standpoint among zygomycetes. Some of the early diverging lineages such as Glomeromycotina and Mortierellomycotina lack ergosterol, what can increase their membrane stiffness [77,112]. Ergosterol has long been considered as one of the regulators of dry–wet cycles [151].
4.2. Endobacteria
Symbiotic associations with highly coevolved endocellular bacteria stand out among the most prominent features differentiating Mucoromycota from other fungi, including the Dikarya [152,153,154]. Thus far, most of these endosymbionts have been identified as representatives of two bacterial families, Burkholderiaceae and Mycoplasmataceae. While some of the Mucoromycota associations with bacteria are mutually beneficial for both partners, in others, the fungi appear to be negatively impacted by their endosymbionts. The application of “omics” tools has been essential in elucidating the molecular underpinnings of the mutualism between Glomeromycotina and their ancient and uncultivable endosymbiont “Candidatus Glomeribacter gigasporarum” (CaGg, Burkholderiaceae) [155]. In this symbiosis, CaGg improves hyphal proliferation in the presymbiotic growth stage of the host fungus [156], which can be attributed to endobacteria-mediated reprograming of the fungal metabolism [157,158]. “Omics” approaches were also critical in understanding the biology and devising a cultivation strategy for Mycoavidus cysteinexigens (Burkholderiaceae) [159], a cysteine auxotroph and ancient antagonistic endosymbiont of Mortierellomycotina [69,160,161]. Comparative genomics also revealed that “Candidatus Mycoavidus necroximicus” produces several nematocidal metabolites that protect the fungal host Mortierella verticillata from feeding by nematodes [162]. Endobacteria have also been shown to regulate and control mating and asexual reproduction in Rhizopus microsporus [163], produce toxins (rhizoxin) that enable pathogenicity on plants, and provide protection from predators [164], as well as potentially protect fungal spores from the human immune system via anti-phagocyte activity [165]. The ease of partner manipulation in the mutualism between R. microsporus and Mortierellaceae (Mucoromycotina) and Mycetohabitans spp. (formerly Burkholderia) [166,167] elevated this association to a model system for studying fungal-bacterial symbioses [163,168].
While the symbioses of Mucoromycota with Burkholderiaceae endobacteria are well characterized for some species, Mucoromycota associations with Mycoplasmataceae, referred to as Mollicutes- or mycoplasma-related endobacteria (MREs) [169,170], are only beginning to be unraveled. MREs have been detected in all three subphyla of Mucoromycota: Glomeromycotina [169], Mortierellomycotina [171], and Mucoromycotina [24,172]. MREs of Glomeromycotina are classified as “Candidatus Moeniiplasma glomeromycotorum” (CaMg) [173]. Genomic features of CaMg have been examined in depth [66,174], including evidence of gene acquisition from fungal hosts. However, the role of CaMg in the biology of Glomeromycotina remains uncertain. Naito et al. [174] speculated that CaMg is an antagonist of Glomeromycotina. If this is confirmed, then CaMg would resemble the lifestyle of the MREs associated with Mortierellomycotina [171].
Moreover, bacterial–fungal interactions seem to be much more common and diverse than previously thought [175], and Mucoromycota representatives are no exception. Telagathoti et al. [176] showed recently that 30% of Mortierellales isolates were associated with different groups of bacteria. Another survey of Mucoromycota cultures from soil indicated diverse Paraburkholderia-related sequences detected with 20% frequency from Mortierella and Umbelopsis strains [154]. The ecological meaning of these interactions remains to be described.
Outside of the Mucoromycota, however, the importance or prevalence of endosymbiotic bacteria remains less clear. Despite bacterial sequences always present in the environmental samples of entomophthoralean fungi (A.P. Gryganskyi, personal observation), nothing is known about their connection to the entomopathogens and their role in pathogenic processes that impact insects. However, six recently constructed metagenome assemblies of four various bacterial species in connection with Massospora cicadina might indicate their symbiotic relationship to this entomopathogenic species [177]. Among Zoopagomycotina, Davis et al. [20] detected 16S sequences with close matches to Mycoavidus cysteinexexigens in the genome of Stylopage hadra (nematode parasitic fungus). A recent genomic assessment of Kickxellomycotina fungi did not reveal any obvious colonization by endosymbiotic bacteria [15], suggesting that these bacterial associations might be less important or prevalent in Zoopagomycota than in Mucoromycota.
4.3. Secretomes, Gene Clusters, and Secondary Metabolites
All fungi explore and exploit their environment by secreting various substances. The secretome composition of basal fungal lineages was shaped by diversification early in their evolution. Characterization of secretome compositions of 132 zygomycete genomes revealed that current trophic mode plays a less significant role as compared to the phylogenetic relationships of the fungi [178]. This is in part because Mucoromycota and Zoopagomycota each had their own unique lineage-specific expansion of secreted digestive enzymes, according to their historical substrate differences and their different evolutionary trajectories [178].
Genomic screenings of available genomes for novel natural compounds also show promising potential [179,180,181]. These screenings discovered the presence of gene clusters harboring polyketide synthases (PKS), non-ribosomal peptides, terpenoids, and L-tryptophan dimethylallyl transferases with transcription factor coding genes in proximity to some of the gene clusters. Since then, NRPS clusters have been detected in some Mortierellomycotina and Mucoromycotina. For example, the mycelium of Mortierella alpina produces diverse novel compounds [182]. Similarly, the transcripts corresponding to PKS and NRPS were recently described in transcriptomes of five Mucor species [183]. Mucor circinelloides was shown to produce carotenoids and terpenoids [180], and the genes associated with terpene biosynthesis were identified recently in Mucor and Rhizopus genomes [74]. Siderophore production has been reported from Glomus [184], Apophysomyces [185], Rhizopus [186], and Basidiobolus [129]. Rhizoferrin from Rhizopus delemar has been further characterized and showed a glycosylation pattern [187] that was classified as a member of the sodium (Na)–iodide symporter gene family [186]. The best-studied compound isolated from early diverging fungi is an antimitotic agent called rhizoxin produced by an endosymbiotic bacterium (Mycetohabitans rhizoxinica) and encoded by a huge NRPS–PKS cluster [166]. The genome of amphibian gut symbiont Basidiobolus is enriched with the genes and gene clusters involved in the production of secondary metabolites through horizontal gene transfer from bacteria [129]. Genomes of mycoparasitic Dimargaritales (Kickxellomycotina) are also enriched for secondary metabolites [15], especially when compared to mycoparasitic Zoopagomycotina (Piptocephalis and Syncephalis) whose genomes appear depauperate in these genes [129]. These groups of mycoparasites are very similar in their morphology and ecology; thus, the reasons underpinning the observed difference in metabolite genes remain unclear.
4.4. Transposable Elements
Transposable elements (TEs) are abundant in zygomycete genomes [79]. Genomes of Entomophtorales, Basidiobolales [129], and Endogonales [24] are particularly rich in TEs, while species of Umbelopsidales [188] and Zoopagomycotina [11] have very few. Genomes of Rhizopus species comprised approximately 40% repetitive sequences with only ~10% potentially active elements [84]. In taxa with more compact genomes such as Umbelopsis isabellina, transposons account for as little as 5% of their genome size. In contrast, Jimgerdemannia flammicorona can have more than 77% of its genome occupied by repetitive elements and transposons [24]. In most of the analyzed genomes, long terminal repeat retrotransposons from the Ty3/Gypsy family dominate the TE landscape [84]. Mucoromycotina, Kickxellomycotina, and Blastocladiomycota possess transposons with a tyrosine recombinase from a gene family which seem to be lost in Dikarya [78]. While the function of mobile elements in zygomycete genomes is not well understood, in Ascomycota, large mobile elements (termed “starships”) may carry a variety of genes (e.g., secondary metabolites, virulence factors, and heavy-metal tolerance) and contribute to the overall variation in genome structure [189,190].
5. Conclusions
There has been renewed research interest in the zygomycete fungi during the last decade, and this is reflected in the rapidly increasing number of genome sequences from this group of fungi. While the quality of the genomes varies according to the sequencing platform and starting materials available, the genomes are sufficient for phylogenomic reconstructions, as well as for the discovery of functional genes and gene clusters of interest. Much of the work has focused on several key topics, including the evolution and phylogeny of the group, adaptation, and transition to a terrestrial habitat (e.g., light and gravity sensors, and reinforcement of cell-wall structure to tolerate a nonaquatic environment), genome duplication, endosymbionts and horizontal gene transfer, genome architecture, pathogenicity, and coevolution with host species. The number of genomes has grown rapidly over the past decade, and we expect that the next decade will bring increased taxon sampling (e.g., new genomes for taxa that have not yet been sampled) and improvements in genome quality due to advances in long-read sequencing technologies and bioinformatics approaches. In the future, we expect that genomic analyses of these fungi will take new directions, including a detailed comparison of the genomes of aquatic vs. terrestrial lineages, structural studies, a comparison of the zygomycete vs. Dikarya genomes, experimental manipulations of host/parasite interactions to understand the molecular mechanisms underpinning pathogenesis, and building links among genomic, proteomic, and metabolomic data.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms11071830/s1: Supplementary Table S1. The genomes of Mucoromycota and Zoopagomycota available in open public sources: NCBI Bioproject, Genome and Nucleotide databases.
Funding
This material is based upon work supported by the National Science Foundation (DEB-1441604 to JWS, DEB-1441715 to JES, DEB-1441677 to TYJ). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. The work (proposals 10.46936/10.25585/60001019 and 10.46936/10.25585/60001062) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. J.E.S. is a paid consultant for Michroma and Sincarne and CIFAR fellow in the program Fungal Kingdom: Threats and Opportunities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Heaton, L.L.M.; Jones, N.S.; Fricker, M.D. A Mechanistic Explanation of the Transition to Simple Multicellularity in Fungi. Nat. Commun. 2020, 11, 2594. [Google Scholar] [CrossRef]
- Nagy, L.G.; Varga, T.; Csernetics, Á.; Virágh, M. Fungi Took a Unique Evolutionary Route to Multicellularity: Seven Key Challenges for Fungal Multicellular Life. Fungal Biol. Rev. 2020, 34, 151–169. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Guarro, J.; Gené, J.; Figueras, M.J. Atlas of Clinical Fungi, 2nd ed.; Centraalbureau voor Schimmelcultures (CBS): Utrecht, The Netherlands, 2000; ISBN 90-70351-43-9. [Google Scholar]
- Benny, G.L.; Smith, M.E.; Kirk, P.M.; Tretter, E.D.; White, M.M. Challenges and Future Perspectives in the Systematics of Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina. In Biology of Microfungi; Li, D.-W., Ed.; Fungal Biology; Springer International Publishing: Cham, Switzerland, 2016; pp. 65–126. ISBN 978-3-319-29137-6. [Google Scholar]
- James, T.Y.; Letcher, P.M.; Longcore, J.E.; Mozley-Standridge, S.E.; Porter, D.; Powell, M.J.; Griffith, G.W.; Vilgalys, R. A Molecular Phylogeny of the Flagellated Fungi (Chytridiomycota) and Description of a New Phylum (Blastocladiomycota). Mycologia 2006, 98, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A Phylum-Level Phylogenetic Classification of Zygomycete Fungi Based on Genome-Scale Data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.G.; Kovács, G.M.; Krizsán, K. Complex Multicellularity in Fungi: Evolutionary Convergence, Single Origin, or Both? Biol. Rev. Camb. Philos. Soc. 2018, 93, 1778–1794. [Google Scholar] [CrossRef]
- Kiss, E.; Hegedüs, B.; Virágh, M.; Varga, T.; Merényi, Z.; Kószó, T.; Bálint, B.; Prasanna, A.N.; Krizsán, K.; Kocsubé, S.; et al. Comparative Genomics Reveals the Origin of Fungal Hyphae and Multicellularity. Nat. Commun. 2019, 10, 4080. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E. Fungal Genomes and Insights into the Evolution of the Kingdom. Microbiol. Spectr. 2017, 5, 1–25. [Google Scholar] [CrossRef]
- Berbee, M.L.; James, T.Y.; Strullu-Derrien, C. Early Diverging Fungi: Diversity and Impact at the Dawn of Terrestrial Life. Annu. Rev. Microbiol. 2017, 71, 41–60. [Google Scholar] [CrossRef]
- Ahrendt, S.R.; Quandt, C.A.; Ciobanu, D.; Clum, A.; Salamov, A.; Andreopoulos, B.; Cheng, J.-F.; Woyke, T.; Pelin, A.; Henrissat, B.; et al. Leveraging Single-Cell Genomics to Expand the Fungal Tree of Life. Nat. Microbiol. 2018, 3, 1417–1428. [Google Scholar] [CrossRef]
- Vandepol, N.; Liber, J.; Desirò, A.; Na, H.; Kennedy, M.; Barry, K.; Grigoriev, I.V.; Miller, A.N.; O’Donnell, K.; Stajich, J.E.; et al. Resolving the Mortierellaceae Phylogeny through Synthesis of Multi-Gene Phylogenetics and Phylogenomics. Fungal Divers. 2020, 104, 267–289. [Google Scholar] [CrossRef]
- Chang, M.C.; Hur, J.; Park, D. Interpreting the COVID-19 Test Results: A Guide for Physiatrists. Am. J. Phys. Med. Rehabil. 2020, 99, 583–585. [Google Scholar] [CrossRef] [PubMed]
- Spatafora, J.W.; Aime, M.C.; Grigoriev, I.V.; Stajich, J.E.; Blackwell, M. The Fungal Tree of Life: From Molecular Systematics to Genome-Scale Phylogenies. Microbiol. Spectrum. 2017, 5, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, N.K.; Stajich, J.E.; Benny, G.L.; Barry, K.; Mondo, S.; LaButti, K.; Lipzen, A.; Daum, C.; Grigoriev, I.V.; Ho, H.-M.; et al. Mycoparasites, Gut Dwellers, and Saprotrophs: Phylogenomic Reconstructions and Comparative Analyses of Kickxellomycotina Fungi. Genome Biol. Evol. 2023, 15, evac185. [Google Scholar] [CrossRef]
- Gryganskyi, A.P.; Muszewska, A. Whole Genome Sequencing and the Zygomycota. Fungal Genom. Biol. 2014, 4, e116. [Google Scholar] [CrossRef]
- Stephens, Z.D.; Lee, S.Y.; Faghri, F.; Campbell, R.H.; Zhai, C.; Efron, M.J.; Iyer, R.; Schatz, M.C.; Sinha, S.; Robinson, G.E. Big Data: Astronomical or Genomical? PLoS Biol. 2015, 13, e1002195. [Google Scholar] [CrossRef]
- Ma, L.-J.; Ibrahim, A.S.; Skory, C.; Grabherr, M.G.; Burger, G.; Butler, M.; Elias, M.; Idnurm, A.; Lang, B.F.; Sone, T.; et al. Genomic Analysis of the Basal Lineage Fungus Rhizopus oryzae Reveals a Whole-Genome Duplication. PLoS Genet. 2009, 5, e1000549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yang, Y.; Castlebury, L.A.; Cerniglia, C.E. A Method for the Large Scale Isolation of High Transformation Efficiency Fungal Genomic DNA. FEMS Microbiol. Lett. 1996, 145, 261–265. [Google Scholar] [CrossRef]
- Davis, W.J.; Amses, K.R.; Benny, G.L.; Carter-House, D.; Chang, Y.; Grigoriev, I.; Smith, M.E.; Spatafora, J.W.; Stajich, J.E.; James, T.Y. Genome-Scale Phylogenetics Reveals a Monophyletic Zoopagales (Zoopagomycota, Fungi). Mol. Phylogenetics Evol. 2019, 133, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Montoliu-Nerin, M.; Sánchez-García, M.; Bergin, C.; Grabherr, M.; Ellis, B.; Kutschera, V.E.; Kierczak, M.; Johannesson, H.; Rosling, A. Building de Novo Reference Genome Assemblies of Complex Eukaryotic Microorganisms from Single Nuclei. Sci. Rep. 2020, 10, 1303. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Limpens, E.; Zhang, Z.; Ivanov, S.; Saunders, D.G.O.; Mu, D.; Pang, E.; Cao, H.; Cha, H.; Lin, T.; et al. Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus. PLoS Genet. 2014, 10, e1004078. [Google Scholar] [CrossRef]
- Małagocka, J.; Grell, M.N.; Lange, L.; Eilenberg, J.; Jensen, A.B. Transcriptome of an Entomophthoralean Fungus (Pandora formicae) Shows Molecular Machinery Adjusted for Successful Host Exploitation and Transmission. J. Invertebr. Pathol. 2015, 128, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Desirò, A.; Na, H.; Sandor, L.; Lipzen, A.; Clum, A.; Barry, K.; Grigoriev, I.V.; Martin, F.M.; Stajich, J.E.; et al. Phylogenomics of Endogonaceae and Evolution of Mycorrhizas within Mucoromycota. New Phytol. 2019, 222, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, M.O.; Russ, C.; Ross, M.G.; Gabriel, S.B.; Nusbaum, C.; DePristo, M.A. Pacific Biosciences Sequencing Technology for Genotyping and Variation Discovery in Human Data. BMC Genom. 2012, 13, 375. [Google Scholar] [CrossRef]
- Maeda, T.; Kobayashi, Y.; Kameoka, H.; Okuma, N.; Takeda, N.; Yamaguchi, K.; Bino, T.; Shigenobu, S.; Kawaguchi, M. Evidence of Non-Tandemly Repeated RDNAs and Their Intragenomic Heterogeneity in Rhizophagus irregularis. Commun. Biol. 2018, 1, 87. [Google Scholar] [CrossRef]
- Segerman, B. The Most Frequently Used Sequencing Technologies and Assembly Methods in Different Time Segments of the Bacterial Surveillance and RefSeq Genome Databases. Front. Cell. Infect. Microbiol. 2020, 10, 527102. [Google Scholar] [CrossRef] [PubMed]
- El-Metwally, S.; Hamza, T.; Zakaria, M.; Helmy, M. Next-Generation Sequence Assembly: Four Stages of Data Processing and Computational Challenges. PLoS Comput. Biol. 2013, 9, e1003345. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Myers, J.M.; Bonds, A.E.; Clemons, R.A.; Thapa, N.A.; Simmons, D.R.; Carter-House, D.; Ortanez, J.; Liu, P.; Miralles-Durán, A.; Desirò, A.; et al. Survey of Early-Diverging Lineages of Fungi Reveals Abundant and Diverse Mycoviruses. mBio 2020, 11, e02027-20. [Google Scholar] [CrossRef]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, Finished Microbial Genome Assemblies from Long-Read SMRT Sequencing Data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Shapiro, H. Outline of the Assembly Process: JAZZ, the JGI In-House Assembler. Available online: https://escholarship.org/uc/item/0g8792vf (accessed on 13 March 2023).
- Boisvert, S.; Raymond, F.; Godzaridis, É.; Laviolette, F.; Corbeil, J. Ray Meta: Scalable de Novo Metagenome Assembly and Profiling. Genome Biol. 2012, 13, R122. [Google Scholar] [CrossRef]
- Zimin, A.V.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.L.; Yorke, J.A. The MaSuRCA Genome Assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef]
- Butler, J.; MacCallum, I.; Kleber, M.; Shlyakhter, I.A.; Belmonte, M.K.; Lander, E.S.; Nusbaum, C.; Jaffe, D.B. ALLPATHS: De Novo Assembly of Whole-Genome Shotgun Microreads. Genome Res. 2008, 18, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Miller, J.R.; Koren, S.; Sutton, G. Assembly Algorithms for Next-Generation Sequencing Data. Genomics 2010, 95, 315–327. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Lam, K.-K.; LaButti, K.; Khalak, A.; Tse, D. FinisherSC: A Repeat-Aware Tool for Upgrading de Novo Assembly Using Long Reads. Bioinformatics 2015, 31, 3207–3209. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.R. CHAPTER 1—Genome Size Evolution in Animals. In The Evolution of the Genome; Gregory, T.R., Ed.; Academic Press: Burlington, NJ, USA, 2005; pp. 3–87. ISBN 978-0-12-301463-4. [Google Scholar]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef]
- Tavares, S.; Ramos, A.P.; Pires, A.S.; Azinheira, H.G.; Caldeirinha, P.; Link, T.; Abranches, R.; do Céu Silva, M.; Voegele, R.T.; Loureiro, J.; et al. Genome Size Analyses of Pucciniales Reveal the Largest Fungal Genomes. Front. Plant Sci. 2014, 5, 422. [Google Scholar] [CrossRef]
- Salazar, A.N.; Gorter de Vries, A.R.; van den Broek, M.; Brouwers, N.; de la Torre Cortès, P.; Kuijpers, N.G.A.; Daran, J.-M.G.; Abeel, T. Chromosome Level Assembly and Comparative Genome Analysis Confirm Lager-Brewing Yeasts Originated from a Single Hybridization. BMC Genom. 2019, 20, 916. [Google Scholar] [CrossRef]
- Wieloch, W. Chromosome Visualisation in Filamentous Fungi. J. Microbiol. Methods 2006, 67, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Boyce, G.R.; Gluck-Thaler, E.; Slot, J.C.; Stajich, J.E.; Davis, W.J.; James, T.Y.; Cooley, J.R.; Panaccione, D.G.; Eilenberg, J.; De Fine Licht, H.H.; et al. Psychoactive Plant- and Mushroom-Associated Alkaloids from Two Behavior Modifying Cicada Pathogens. Fungal Ecol. 2019, 41, 147–164. [Google Scholar] [CrossRef]
- Vinson, J.P.; Jaffe, D.B.; O’Neill, K.; Karlsson, E.K.; Stange-Thomann, N.; Anderson, S.; Mesirov, J.P.; Satoh, N.; Satou, Y.; Nusbaum, C.; et al. Assembly of Polymorphic Genomes: Algorithms and Application to Ciona savignyi. Genome Res. 2005, 15, 1127–1135. [Google Scholar] [CrossRef]
- Chibucos, M.C.; Soliman, S.; Gebremariam, T.; Lee, H.; Daugherty, S.; Orvis, J.; Shetty, A.C.; Crabtree, J.; Hazen, T.H.; Etienne, K.A.; et al. An Integrated Genomic and Transcriptomic Survey of Mucormycosis-Causing Fungi. Nat. Commun. 2016, 7, 12218. [Google Scholar] [CrossRef]
- Li, Y.; Steenwyk, J.L.; Chang, Y.; Wang, Y.; James, T.Y.; Stajich, J.E.; Spatafora, J.W.; Groenewald, M.; Dunn, C.W.; Hittinger, C.T.; et al. A Genome-Scale Phylogeny of the Kingdom Fungi. Curr. Biol. 2021, 31, 1653–1665.e5. [Google Scholar] [CrossRef]
- Strassert, J.F.H.; Monaghan, M.T. Phylogenomic Insights into the Early Diversification of Fungi. Curr. Biol. 2022, 32, 3628–3635.e3. [Google Scholar] [CrossRef]
- Galindo, L.J.; López-García, P.; Torruella, G.; Karpov, S.; Moreira, D. Phylogenomics of a New Fungal Phylum Reveals Multiple Waves of Reductive Evolution across Holomycota. Nat. Commun. 2021, 12, 4973. [Google Scholar] [CrossRef] [PubMed]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal Evolution: Diversity, Taxonomy and Phylogeny of the Fungi. Biol. Rev. Camb. Philos. Soc. 2019, 94, 2101–2137. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Ganley, A.R.D.; Gabaldón, T.; Cox, M.P. The Case of the Missing Ancient Fungal Polyploids. Am. Nat. 2016, 188, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.N.; Salgado, J.F.M.; Natsopoulou, M.E.; Kristensen, T.; Stajich, J.E.; De Fine Licht, H.H. Diploidy within a Haploid Genus of Entomopathogenic Fungi. Genome Biol. Evol. 2021, 13, evab158. [Google Scholar] [CrossRef]
- Amses, K.R.; Simmons, D.R.; Longcore, J.E.; Mondo, S.J.; Seto, K.; Jerônimo, G.H.; Bonds, A.E.; Quandt, C.A.; Davis, W.J.; Chang, Y.; et al. Diploid-Dominant Life Cycles Characterize the Early Evolution of Fungi. Proc. Natl. Acad. Sci. USA 2022, 119, e2116841119. [Google Scholar] [CrossRef]
- Genre, A.; Lanfranco, L.; Perotto, S.; Bonfante, P. Unique and Common Traits in Mycorrhizal Symbioses. Nat. Rev. Microbiol. 2020, 18, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Hoysted, G.A.; Jacob, A.S.; Kowal, J.; Giesemann, P.; Bidartondo, M.I.; Duckett, J.G.; Gebauer, G.; Rimington, W.R.; Schornack, S.; Pressel, S.; et al. Mucoromycotina Fine Root Endophyte Fungi Form Nutritional Mutualisms with Vascular Plants. Plant Physiol. 2019, 181, 565–577. [Google Scholar] [CrossRef]
- Wani, Z.A.; Kumar, A.; Sultan, P.; Bindu, K.; Riyaz-Ul-Hassan, S.; Ashraf, N. Mortierella alpina CS10E4, an Oleaginous Fungal Endophyte of Crocus sativus L. Enhances Apocarotenoid Biosynthesis and Stress Tolerance in the Host Plant. Sci. Rep. 2017, 7, 8598. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.; Luo, J.; Khiste, S.; Scalera, A.; Sajjad, S.; Zhang, N. Pygmaeomycetaceae, a New Root-Associated Family in Mucoromycotina from the Pygmy Pine Plains. Mycologia 2021, 113, 134–145. [Google Scholar] [CrossRef]
- Liu, S.; Liu, M.; Liao, Q.-G.; Lü, F.-B.; Zhao, X.-L. Effects of Inoculated Mycorrhizal Fungi and Non-Mycorrhizal Beneficial Micro-Organisms on Plant Traits, Nutrient Uptake and Root-Associated Fungal Community Composition of the Cymbidium hybridum in Greenhouse. J. Appl. Microbiol. 2021, 131, 413–424. [Google Scholar] [CrossRef]
- Steinbrink, J.M.; Miceli, M.H. Clinical Review of Mucormycosis. Infect. Dis. Clin. N. Am. 2021, 35, 435–452. [Google Scholar] [CrossRef]
- Corrochano, L.M.; Kuo, A.; Marcet-Houben, M.; Polaino, S.; Salamov, A.; Villalobos-Escobedo, J.M.; Grimwood, J.; Álvarez, M.I.; Avalos, J.; Bauer, D.; et al. Expansion of Signal Transduction Pathways in Fungi by Extensive Genome Duplication. Curr. Biol. 2016, 26, 1577–1584. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Hertweck, C. Pathogenic Fungus Harbours Endosymbiotic Bacteria for Toxin Production. Nature 2005, 437, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Torres-Cortés, G.; Ghignone, S.; Bonfante, P.; Schüßler, A. Mosaic Genome of Endobacteria in Arbuscular Mycorrhizal Fungi: Transkingdom Gene Transfer in an Ancient Mycoplasma-Fungus Association. Proc. Natl. Acad. Sci. USA 2015, 112, 7785–7790. [Google Scholar] [CrossRef]
- Mondo, S.J.; Salvioli, A.; Bonfante, P.; Morton, J.B.; Pawlowska, T.E. Nondegenerative Evolution in Ancient Heritable Bacterial Endosymbionts of Fungi. Mol. Biol. Evol. 2016, 33, 2216–2231. [Google Scholar] [CrossRef] [PubMed]
- Bonfante, P.; Desirò, A. Who Lives in a Fungus? The Diversity, Origins and Functions of Fungal Endobacteria Living in Mucoromycota. ISME J. 2017, 11, 1727–1735. [Google Scholar] [CrossRef] [PubMed]
- Uehling, J.; Gryganskyi, A.; Hameed, K.; Tschaplinski, T.; Misztal, P.K.; Wu, S.; Desirò, A.; Vande Pol, N.; Du, Z.; Zienkiewicz, A.; et al. Comparative Genomics of Mortierella elongata and Its Bacterial Endosymbiont Mycoavidus cysteinexigens. Environ. Microbiol. 2017, 19, 2964–2983. [Google Scholar] [CrossRef] [PubMed]
- Lastovetsky, O.A.; Ahn, E.; Mondo, S.J.; Toomer, K.H.; Zhang, A.; Johnson, L.M.; Pawlowska, T.E. Distribution and Population Structure of Endobacteria in Arbuscular Mycorrhizal Fungi at North Atlantic Dunes. ISME J. 2018, 12, 3001–3013. [Google Scholar] [CrossRef]
- Espino-Vázquez, A.N.; Bermúdez-Barrientos, J.R.; Cabrera-Rangel, J.F.; Córdova-López, G.; Cardoso-Martínez, F.; Martínez-Vázquez, A.; Camarena-Pozos, D.A.; Mondo, S.J.; Pawlowska, T.E.; Abreu-Goodger, C.; et al. Narnaviruses: Novel Players in Fungal-Bacterial Symbioses. ISME J. 2020, 14, 1743–1754. [Google Scholar] [CrossRef] [PubMed]
- Bever, J.D.; Kang, H.-J.; Kaonongbua, W.; Wang, M. Genomic Organization and Mechanisms of Inheritance in Arbuscular Mycorrhizal Fungi: Contrasting the Evidence and Implications of Current Theories. In Mycorrhiza; Varma, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 135–148. ISBN 978-3-540-78824-9. [Google Scholar]
- Martin, F.; Gianinazzi-Pearson, V.; Hijri, M.; Lammers, P.; Requena, N.; Sanders, I.R.; Shachar-Hill, Y.; Shapiro, H.; Tuskan, G.A.; Young, J.P.W. The Long Hard Road to a Completed Glomus intraradices Genome. New Phytol. 2008, 180, 747–750. [Google Scholar] [CrossRef]
- Lebreton, A.; Corre, E.; Jany, J.-L.; Brillet-Guéguen, L.; Pèrez-Arques, C.; Garre, V.; Monsoor, M.; Debuchy, R.; Le Meur, C.; Coton, E.; et al. Comparative Genomics Applied to Mucor Species with Different Lifestyles. BMC Genom. 2020, 21, 135. [Google Scholar] [CrossRef]
- Vellanki, S.; Navarro-Mendoza, M.I.; Garcia, A.E.; Murcia, L.; Perez-Arques, C.; Garre, V.; Nicolas, F.E.; Lee, S.C. Mucor circinelloides: Growth, Maintenance and Genetic Manipulation. Curr. Protoc. Microbiol. 2018, 49, e53. [Google Scholar] [CrossRef]
- Boon, E.; Zimmerman, E.; Lang, B.F.; Hijri, M. Intra-Isolate Genome Variation in Arbuscular Mycorrhizal Fungi Persists in the Transcriptome. J. Evol. Biol. 2010, 23, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Muszewska, A.; Okrasińska, A.; Steczkiewicz, K.; Drgas, O.; Orłowska, M.; Perlińska-Lenart, U.; Aleksandrzak-Piekarczyk, T.; Szatraj, K.; Zielenkiewicz, U.; Piłsyk, S.; et al. Metabolic Potential, Ecology and Presence of Associated Bacteria Is Reflected in Genomic Diversity of Mucoromycotina. Front. Microbiol. 2021, 12, 636986. [Google Scholar] [CrossRef] [PubMed]
- Muszewska, A.; Steczkiewicz, K.; Ginalski, K. DIRS and Ngaro Retrotransposons in Fungi. PLoS ONE 2013, 8, e76319. [Google Scholar] [CrossRef] [PubMed]
- Muszewska, A.; Steczkiewicz, K.; Stepniewska-Dziubinska, M.; Ginalski, K. Cut-and-Paste Transposons in Fungi with Diverse Lifestyles. Genome Biol. Evol. 2017, 9, 3463–3477. [Google Scholar] [CrossRef]
- Mondo, S.J.; Dannebaum, R.O.; Kuo, R.C.; Louie, K.B.; Bewick, A.J.; LaButti, K.; Haridas, S.; Kuo, A.; Salamov, A.; Ahrendt, S.R.; et al. Widespread Adenine N6-Methylation of Active Genes in Fungi. Nat. Genet. 2017, 49, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Corradi, N.; Sanders, I.R. Evolution of the P-Type II ATPase Gene Family in the Fungi and Presence of Structural Genomic Changes among Isolates of Glomus intraradices. BMC Evol. Biol. 2006, 6, 21. [Google Scholar] [CrossRef]
- Halary, S.; Malik, S.-B.; Lildhar, L.; Slamovits, C.H.; Hijri, M.; Corradi, N. Conserved Meiotic Machinery in Glomus spp., a Putatively Ancient Asexual Fungal Lineage. Genome Biol. Evol. 2011, 3, 950–958. [Google Scholar] [CrossRef]
- Halary, S.; Daubois, L.; Terrat, Y.; Ellenberger, S.; Wöstemeyer, J.; Hijri, M. Mating Type Gene Homologues and Putative Sex Pheromone-Sensing Pathway in Arbuscular Mycorrhizal Fungi, a Presumably Asexual Plant Root Symbiont. PLoS ONE 2013, 8, e80729. [Google Scholar] [CrossRef]
- Gryganskyi, A.P.; Golan, J.; Dolatabadi, S.; Mondo, S.; Robb, S.; Idnurm, A.; Muszewska, A.; Steczkiewicz, K.; Masonjones, S.; Liao, H.-L.; et al. Phylogenetic and Phylogenomic Definition of Rhizopus Species. G3 (Bethesda) 2018, 8, 2007–2018. [Google Scholar] [CrossRef]
- Gryganskyi, A.P.; Nie, Y.; Hajek, A.E.; Hodge, K.T.; Liu, X.-Y.; Aadland, K.; Voigt, K.; Anishchenko, I.M.; Kutovenko, V.B.; Kava, L.; et al. The Early Terrestrial Fungal Lineage of Conidiobolus-Transition from Saprotroph to Parasitic Lifestyle. J. Fungi 2022, 8, 789. [Google Scholar] [CrossRef]
- Tisserant, E.; Malbreil, M.; Kuo, A.; Kohler, A.; Symeonidi, A.; Balestrini, R.; Charron, P.; Duensing, N.; Frei dit Frey, N.; Gianinazzi-Pearson, V.; et al. Genome of an Arbuscular Mycorrhizal Fungus Provides Insight into the Oldest Plant Symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, 20117–20122. [Google Scholar] [CrossRef]
- Wang, Y.; Chang, Y.; Ortañez, J.; Peña, J.F.; Carter-House, D.; Reynolds, N.K.; Smith, M.E.; Benny, G.; Mondo, S.J.; Salamov, A.; et al. Divergent Evolution of Early Terrestrial Fungi Reveals the Evolution of Mucormycosis Pathogenicity Factors. Genome Biol. Evol. 2023, 15, evad046. [Google Scholar] [CrossRef]
- Desirò, A.; Rimington, W.R.; Jacob, A.; Pol, N.V.; Smith, M.E.; Trappe, J.M.; Bidartondo, M.I.; Bonito, G. Multigene Phylogeny of Endogonales, an Early Diverging Lineage of Fungi Associated with Plants. IMA Fungus 2017, 8, 245–257. [Google Scholar] [CrossRef]
- Ogura-Tsujita, Y.; Yamamoto, K.; Hirayama, Y.; Ebihara, A.; Morita, N.; Imaichi, R. Fern Gametophytes of Angiopteris lygodiifolia and Osmunda japonica Harbor Diverse Mucoromycotina Fungi. J. Plant Res. 2019, 132, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Rimington, W.R.; Pressel, S.; Duckett, J.G.; Field, K.J.; Bidartondo, M.I. Evolution and Networks in Ancient and Widespread Symbioses between Mucoromycotina and Liverworts. Mycorrhiza 2019, 29, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Polaino, S.; Shakya, V.P.S.; Idnurm, A. A New Genetic Linkage Map of the Zygomycete Fungus Phycomyces blakesleeanus. PLoS ONE 2013, 8, e58931. [Google Scholar] [CrossRef]
- Linde, J.; Schwartze, V.; Binder, U.; Lass-Flörl, C.; Voigt, K.; Horn, F. De Novo Whole-Genome Sequence and Genome Annotation of Lichtheimia ramosa. Genome Announc. 2014, 2, e00888-14. [Google Scholar] [CrossRef] [PubMed]
- Schwartze, V.U.; Winter, S.; Shelest, E.; Marcet-Houben, M.; Horn, F.; Wehner, S.; Linde, J.; Valiante, V.; Sammeth, M.; Riege, K.; et al. Gene Expansion Shapes Genome Architecture in the Human Pathogen Lichtheimia corymbifera: An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina). PLoS Genet. 2014, 10, e1004496. [Google Scholar] [CrossRef]
- Torres-Cruz, T.J.; Billingsley Tobias, T.L.; Almatruk, M.; Hesse, C.N.; Kuske, C.R.; Desirò, A.; Benucci, G.M.N.; Bonito, G.; Stajich, J.E.; Dunlap, C.; et al. Bifiguratus adelaidae, Gen. et Sp. Nov., a New Member of Mucoromycotina in Endophytic and Soil-Dwelling Habitats. Mycologia 2017, 109, 363–378. [Google Scholar] [CrossRef]
- Hirose, D.; Degawa, Y.; Inaba, S.; Tokumasu, S. The Anamorphic Genus Calcarisporiella Is a New Member of the Mucoromycotina. Mycoscience 2012, 53, 256–260. [Google Scholar] [CrossRef]
- Nagy, L.G.; Petkovits, T.; Kovács, G.M.; Voigt, K.; Vágvölgyi, C.; Papp, T. Where Is the Unseen Fungal Diversity Hidden? A Study of Mortierella Reveals a Large Contribution of Reference Collections to the Identification of Fungal Environmental Sequences. New Phytol. 2011, 191, 789–794. [Google Scholar] [CrossRef]
- Smith, M.E.; Gryganskyi, A.; Bonito, G.; Nouhra, E.; Moreno-Arroyo, B.; Benny, G. Phylogenetic Analysis of the Genus Modicella Reveals an Independent Evolutionary Origin of Sporocarp-Forming Fungi in the Mortierellales. Fungal Genet. Biol. 2013, 61, 61–68. [Google Scholar] [CrossRef]
- Macias, A.M.; Geiser, D.M.; Stajich, J.E.; Łukasik, P.; Veloso, C.; Bublitz, D.C.; Berger, M.C.; Boyce, G.R.; Hodge, K.; Kasson, M.T. Evolutionary Relationships among Massospora Spp. (Entomophthorales), Obligate Pathogens of Cicadas. Mycologia 2020, 112, 1060–1074. [Google Scholar] [CrossRef] [PubMed]
- Bonito, G.; Hameed, K.; Ventura, R.; Krishnan, J.; Schadt, C.W.; Vilgalys, R. Isolating a Functionally Relevant Guild of Fungi from the Root Microbiome of Populus. Fungal Ecol. 2016, 22, 35–42. [Google Scholar] [CrossRef]
- Vandepol, N.; Liber, J.; Yocca, A.; Matlock, J.; Edger, P.; Bonito, G. Linnemannia elongata (Mortierellaceae) Stimulates Arabidopsis thaliana Aerial Growth and Responses to Auxin, Ethylene, and Reactive Oxygen Species. PLoS ONE 2022, 17, e0261908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Bonito, G.; Hsu, C.-M.; Hameed, K.; Vilgalys, R.; Liao, H.-L. Mortierella elongata Increases Plant Biomass among Non-Leguminous Crop Species. Agronomy 2020, 10, 754. [Google Scholar] [CrossRef]
- Wurlitzer, J.M.; Stanišić, A.; Wasmuth, I.; Jungmann, S.; Fischer, D.; Kries, H.; Gressler, M. Bacterial-like Nonribosomal Peptide Synthetases Produce Cyclopeptides in the Zygomycetous Fungus Mortierella alpina. Appl. Environ. Microbiol. 2021, 87, e02051-20. [Google Scholar] [CrossRef]
- Remy, W.; Taylor, T.N.; Hass, H.; Kerp, H. Four Hundred-Million-Year-Old Vesicular Arbuscular Mycorrhizae. Proc. Natl. Acad. Sci. USA 1994, 91, 11841–11843. [Google Scholar] [CrossRef]
- Smith, S.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: Cambridge, MA, USA, 2008; ISBN 978-0-12-370526-6. [Google Scholar]
- Hijri, M.; Sanders, I.R. The Arbuscular Mycorrhizal Fungus Glomus intraradices Is Haploid and Has a Small Genome Size in the Lower Limit of Eukaryotes. Fungal Genet. Biol. 2004, 41, 253–261. [Google Scholar] [CrossRef]
- Sędzielewska, K.A.; Fuchs, J.; Temsch, E.M.; Baronian, K.; Watzke, R.; Kunze, G. Estimation of the Glomus intraradices Nuclear DNA Content. New Phytol. 2011, 192, 794–797. [Google Scholar] [CrossRef] [PubMed]
- Venice, F.; Ghignone, S.; Salvioli di Fossalunga, A.; Amselem, J.; Novero, M.; Xianan, X.; Sędzielewska Toro, K.; Morin, E.; Lipzen, A.; Grigoriev, I.V.; et al. At the Nexus of Three Kingdoms: The Genome of the Mycorrhizal Fungus Gigaspora margarita Provides Insights into Plant, Endobacterial and Fungal Interactions. Environ. Microbiol. 2020, 22, 122–141. [Google Scholar] [CrossRef]
- Ropars, J.; Toro, K.S.; Noel, J.; Pelin, A.; Charron, P.; Farinelli, L.; Marton, T.; Krüger, M.; Fuchs, J.; Brachmann, A.; et al. Evidence for the Sexual Origin of Heterokaryosis in Arbuscular Mycorrhizal Fungi. Nat. Microbiol. 2016, 1, 16033. [Google Scholar] [CrossRef]
- Idnurm, A.; Walton, F.J.; Floyd, A.; Heitman, J. Identification of the Sex Genes in an Early Diverged Fungus. Nature 2008, 451, 193–196. [Google Scholar] [CrossRef]
- Kuhn, G.; Hijri, M.; Sanders, I.R. Evidence for the Evolution of Multiple Genomes in Arbuscular Mycorrhizal Fungi. Nature 2001, 414, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Pawlowska, T.E.; Taylor, J.W. Organization of Genetic Variation in Individuals of Arbuscular Mycorrhizal Fungi. Nature 2004, 427, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Wewer, V.; Brands, M.; Dörmann, P. Fatty Acid Synthesis and Lipid Metabolism in the Obligate Biotrophic Fungus Rhizophagus irregularis during Mycorrhization of Lotus japonicus. Plant J. 2014, 79, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; San Clemente, H.; Roy, S.; Bécard, G.; Zhao, B.; Roux, C. A Survey of the Gene Repertoire of Gigaspora rosea Unravels Conserved Features among Glomeromycota for Obligate Biotrophy. Front. Microbiol. 2016, 7, 233. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Maeda, T.; Yamaguchi, K.; Kameoka, H.; Tanaka, S.; Ezawa, T.; Shigenobu, S.; Kawaguchi, M. The Genome of Rhizophagus clarus HR1 Reveals a Common Genetic Basis for Auxotrophy among Arbuscular Mycorrhizal Fungi. BMC Genom. 2018, 19, 465. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Brands, M.; Wewer, V.; Dörmann, P.; Harrison, M.J. Arbuscular Mycorrhiza-Specific Enzymes FatM and RAM2 Fine-Tune Lipid Biosynthesis to Promote Development of Arbuscular Mycorrhiza. New Phytol. 2017, 214, 1631–1645. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, W.; Xie, Q.; Liu, N.; Liu, L.; Wang, D.; Zhang, X.; Yang, C.; Chen, X.; Tang, D.; et al. Plants Transfer Lipids to Sustain Colonization by Mutualistic Mycorrhizal and Parasitic Fungi. Science 2017, 356, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Keymer, A.; Pimprikar, P.; Wewer, V.; Huber, C.; Brands, M.; Bucerius, S.L.; Delaux, P.-M.; Klingl, V.; von Röpenack-Lahaye, E.; Wang, T.L.; et al. Lipid Transfer from Plants to Arbuscular Mycorrhiza Fungi. eLife 2017, 6, e29107. [Google Scholar] [CrossRef] [PubMed]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty Acids in Arbuscular Mycorrhizal Fungi Are Synthesized by the Host Plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef] [PubMed]
- Kameoka, H.; Tsutsui, I.; Saito, K.; Kikuchi, Y.; Handa, Y.; Ezawa, T.; Hayashi, H.; Kawaguchi, M.; Akiyama, K. Stimulation of Asymbiotic Sporulation in Arbuscular Mycorrhizal Fungi by Fatty Acids. Nat. Microbiol. 2019, 4, 1654–1660. [Google Scholar] [CrossRef]
- Sugiura, Y.; Akiyama, R.; Tanaka, S.; Yano, K.; Kameoka, H.; Marui, S.; Saito, M.; Kawaguchi, M.; Akiyama, K.; Saito, K. Myristate Can Be Used as a Carbon and Energy Source for the Asymbiotic Growth of Arbuscular Mycorrhizal Fungi. Proc. Natl. Acad. Sci. USA 2020, 117, 25779–25788. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Yildirir, G.; Rizzi, Y.; Malar, C.M.; Sorwar, E.; Chen, E.C.; Iwasaki, W.; Brauer, E.K.; Bosnich, W.; Gutjahr, C.; et al. Resolving the Haplotypes of Arbuscular Mycorrhizal Fungi Highlights the Role of Two Nuclear Populations in Host Interactions. bioRxiv 2023. [Google Scholar] [CrossRef]
- Hughes, D.P.; Araújo, J.P.M.; Loreto, R.G.; Quevillon, L.; de Bekker, C.; Evans, H.C. From So Simple a Beginning: The Evolution of Behavioral Manipulation by Fungi. Adv. Genet. 2016, 94, 437–469. [Google Scholar] [CrossRef] [PubMed]
- Vilela, R.; Mendoza, L. Human Pathogenic Entomophthorales. Clin. Microbiol. Rev. 2018, 31, e00014-18. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, K.L.; Benny, G.L.; Ho, H.-M.; Smith, M.E. Phylogenetic Systematics of Syncephalis (Zoopagales, Zoopagomycotina), a Genus of Ubiquitous Mycoparasites. Mycologia 2017, 109, 333–349. [Google Scholar] [CrossRef]
- Lovett, B.; Macias, A.; Stajich, J.E.; Cooley, J.; Eilenberg, J.; de Fine Licht, H.H.; Kasson, M.T. Behavioral Betrayal: How Select Fungal Parasites Enlist Living Insects to Do Their Bidding. PLoS Pathog. 2020, 16, e1008598. [Google Scholar] [CrossRef]
- Chang, Y.; Wang, S.; Sekimoto, S.; Aerts, A.L.; Choi, C.; Clum, A.; LaButti, K.M.; Lindquist, E.A.; Yee Ngan, C.; Ohm, R.A.; et al. Phylogenomic Analyses Indicate That Early Fungi Evolved Digesting Cell Walls of Algal Ancestors of Land Plants. Genome Biol. Evol. 2015, 7, 1590–1601. [Google Scholar] [CrossRef]
- Wang, Y.; White, M.M.; Kvist, S.; Moncalvo, J.-M. Genome-Wide Survey of Gut Fungi (Harpellales) Reveals the First Horizontally Transferred Ubiquitin Gene from a Mosquito Host. Mol. Biol. Evol. 2016, 33, 2544–2554. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Stata, M.; Wang, W.; Stajich, J.E.; White, M.M.; Moncalvo, J.-M. Comparative Genomics Reveals the Core Gene Toolbox for the Fungus-Insect Symbiosis. mBio 2018, 9, e00636-18. [Google Scholar] [CrossRef]
- Tabima, J.F.; Trautman, I.A.; Chang, Y.; Wang, Y.; Mondo, S.; Kuo, A.; Salamov, A.; Grigoriev, I.V.; Stajich, J.E.; Spatafora, J.W. Phylogenomic Analyses of Non-Dikarya Fungi Supports Horizontal Gene Transfer Driving Diversification of Secondary Metabolism in the Amphibian Gastrointestinal Symbiont, Basidiobolus. G3 (Bethesda) 2020, 10, 3417–3433. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.-C.; Ho, H.-M.; Reynolds, N.; Smith, M.E.; Benny, G.L.; Chien, C.-Y.; Tsai, J.-L. Preliminary Phylogeny of Coemansia (Kickxellales), with Descriptions of Four New Species from Taiwan. Mycologia 2017, 109, 815–831. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; White, M.M.; Moncalvo, J.-M. Draft Genome Sequence of Capniomyces stellatus, the Obligate Gut Fungal Symbiont of Stonefly. Genome Announc. 2016, 4, e00761-16. [Google Scholar] [CrossRef]
- Tretter, E.D.; Johnson, E.M.; Benny, G.L.; Lichtwardt, R.W.; Wang, Y.; Kandel, P.; Novak, S.J.; Smith, J.F.; White, M.M. An Eight-Gene Molecular Phylogeny of the Kickxellomycotina, Including the First Phylogenetic Placement of Asellariales. Mycologia 2014, 106, 912–935. [Google Scholar] [CrossRef] [PubMed]
- Lichtward, R.W. The Trichomycetes, Fungal Associates of Arthropods, 2nd ed.; Springer Science & Business Media: New York, NY, USA; Berlin/Heidelberg, Germany; Tokio, Japan, 2012. [Google Scholar]
- Wang, Y.; White, M.M.; Moncalvo, J.-M. Diversification of the Gut Fungi Smittium and Allies (Harpellales) Co-Occurred with the Origin of Complete Metamorphosis of Their Symbiotic Insect Hosts (Lower Diptera). Mol. Phylogenet. Evol. 2019, 139, 106550. [Google Scholar] [CrossRef]
- White, M.M.; Guàrdia Valle, L.; Lichtwardt, R.W.; Siri, A.; Strongman, D.B.; William, R.T.; Gause, W.J.; Tretter, E.D. New Species and Emendations of Orphella: Taxonomic and Phylogenetic Reassessment of the Genus to Establish the Orphellales, for Stonefly Gut Fungi with a Twist. Mycologia 2018, 110, 147–178. [Google Scholar] [CrossRef]
- Wang, Y.; Tretter, E.D.; Lichtwardt, R.W.; White, M.M. Overview of 75 Years of Smittium Research, Establishing a New Genus for Smittium culisetae, and Prospects for Future Revisions of the “Smittium” Clade. Mycologia 2013, 105, 90–111. [Google Scholar] [CrossRef]
- White, M.M.; Siri, A.; Lichtwardt, R.W. Trichomycete Insect Symbionts in Great Smoky Mountains National Park and Vicinity. Mycologia 2006, 98, 333–352. [Google Scholar] [CrossRef]
- Mariotti, M.; Salinas, G.; Gabaldón, T.; Gladyshev, V.N. Utilization of Selenocysteine in Early-Branching Fungal Phyla. Nat. Microbiol. 2019, 4, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Yu, D.-S.; Wang, C.-F.; Liu, X.-Y.; Huang, B. A Taxonomic Revision of the Genus Conidiobolus (Ancylistaceae, Entomophthorales): Four Clades Including Three New Genera. MycoKeys 2020, 66, 55–81. [Google Scholar] [CrossRef] [PubMed]
- Humber, R.A. Synopsis of a Revised Classification for the Entomophthorales (Zygomycotina). Mycotaxon 1989, 34, 441–460. [Google Scholar]
- Nagahama, T.; Sato, H.; Shimazu, M.; Sugiyama, J. Phylogenetic Divergence of the Entomophthoralean Fungi: Evidence from Nuclear 18S Ribosomal RNA Gene Sequences. Mycologia 1995, 87, 203–209. [Google Scholar] [CrossRef]
- Bergman, K.; Eslava, A.P.; Cerdá-Olmedo, E. Mutants of Phycomyces with Abnormal Phototropism. Mol. Gen. Genet. 1973, 123, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Rodríguez-Romero, J.; Corrochano, L.M.; Sanz, C.; Iturriaga, E.A.; Eslava, A.P.; Heitman, J. The Phycomyces MadA Gene Encodes a Blue-Light Photoreceptor for Phototropism and Other Light Responses. Proc. Natl. Acad. Sci. USA 2006, 103, 4546–4551. [Google Scholar] [CrossRef]
- Liu, Y.; He, Q.; Cheng, P. Photoreception in Neurospora: A Tale of Two White Collar Proteins. Cell. Mol. Life Sci. 2003, 60, 2131–2138. [Google Scholar] [CrossRef]
- Sanz, C.; Rodríguez-Romero, J.; Idnurm, A.; Christie, J.M.; Heitman, J.; Corrochano, L.M.; Eslava, A.P. Phycomyces MadB Interacts with MadA to Form the Primary Photoreceptor Complex for Fungal Phototropism. Proc. Natl. Acad. Sci. USA 2009, 106, 7095–7100. [Google Scholar] [CrossRef]
- Silva, F.; Torres-Martínez, S.; Garre, V. Distinct White Collar-1 Genes Control Specific Light Responses in Mucor circinelloides. Mol. Microbiol. 2006, 61, 1023–1037. [Google Scholar] [CrossRef]
- Polaino, S.; Villalobos-Escobedo, J.M.; Shakya, V.P.S.; Miralles-Durán, A.; Chaudhary, S.; Sanz, C.; Shahriari, M.; Luque, E.M.; Eslava, A.P.; Corrochano, L.M.; et al. A Ras GTPase Associated Protein Is Involved in the Phototropic and Circadian Photobiology Responses in Fungi. Sci. Rep. 2017, 7, 44790. [Google Scholar] [CrossRef]
- Verma, S.; Idnurm, A. The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect Cryptococcus neoformans from UV Damage. PLoS Genet. 2013, 9, e1003769. [Google Scholar] [CrossRef]
- Schimek, C.; Eibel, P.; Horie, T.; Galland, P.; Ootaki, T. Protein Crystals in Phycomyces Sporangiophores Are Involved in Graviperception. Adv. Space Res. 1999, 24, 687–696. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Greig, J.; Khan, A.; Goh, C.; Jedd, G. Evolutionary Novelty in Gravity Sensing through Horizontal Gene Transfer and High-Order Protein Assembly. PLoS Biol. 2018, 16, e2004920. [Google Scholar] [CrossRef]
- Dupont, S.; Lemetais, G.; Ferreira, T.; Cayot, P.; Gervais, P.; Beney, L. Ergosterol Biosynthesis: A Fungal Pathway for Life on Land? Evolution 2012, 66, 2961–2968. [Google Scholar] [CrossRef]
- Pawlowska, T.E.; Gaspar, M.L.; Lastovetsky, O.A.; Mondo, S.J.; Real-Ramirez, I.; Shakya, E.; Bonfante, P. Biology of Fungi and Their Bacterial Endosymbionts. Annu. Rev. Phytopathol. 2018, 56, 289–309. [Google Scholar] [CrossRef] [PubMed]
- Bonfante, P.; Venice, F.; Lanfranco, L. The Mycobiota: Fungi Take Their Place between Plants and Bacteria. Curr. Opin. Microbiol. 2019, 49, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Okrasińska, A.; Bokus, A.; Duk, K.; Gęsiorska, A.; Sokołowska, B.; Miłobędzka, A.; Wrzosek, M.; Pawłowska, J. New Endohyphal Relationships between Mucoromycota and Burkholderiaceae Representatives. Appl. Environ. Microbiol. 2021, 87, e02707-20. [Google Scholar] [CrossRef] [PubMed]
- Bianciotto, V.; Lumini, E.; Bonfante, P.; Vandamme, P. “Candidatus Glomeribacter Gigasporarum” Gen. Nov., Sp. Nov., an Endosymbiont of Arbuscular Mycorrhizal Fungi. Int. J. Syst. Evol. Microbiol. 2003, 53, 121–124. [Google Scholar] [CrossRef]
- Lumini, E.; Bianciotto, V.; Jargeat, P.; Novero, M.; Salvioli, A.; Faccio, A.; Bécard, G.; Bonfante, P. Presymbiotic Growth and Sporal Morphology Are Affected in the Arbuscular Mycorrhizal Fungus Gigaspora margarita Cured of Its Endobacteria. Cell. Microbiol. 2007, 9, 1716–1729. [Google Scholar] [CrossRef]
- Salvioli, A.; Ghignone, S.; Novero, M.; Navazio, L.; Venice, F.; Bagnaresi, P.; Bonfante, P. Symbiosis with an Endobacterium Increases the Fitness of a Mycorrhizal Fungus, Raising Its Bioenergetic Potential. ISME J. 2016, 10, 130–144. [Google Scholar] [CrossRef]
- Dearth, S.P.; Castro, H.F.; Venice, F.; Tague, E.D.; Novero, M.; Bonfante, P.; Campagna, S.R. Metabolome Changes Are Induced in the Arbuscular Mycorrhizal Fungus Gigaspora margarita by Germination and by Its Bacterial Endosymbiont. Mycorrhiza 2018, 28, 421–433. [Google Scholar] [CrossRef]
- Ohshima, S.; Sato, Y.; Fujimura, R.; Takashima, Y.; Hamada, M.; Nishizawa, T.; Narisawa, K.; Ohta, H. Mycoavidus cysteinexigens Gen. Nov., Sp. Nov., an Endohyphal Bacterium Isolated from a Soil Isolate of the Fungus Mortierella elongata. Int. J. Syst. Evol. Microbiol. 2016, 66, 2052–2057. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yao, Q.; Dearth, S.P.; Entler, M.R.; Castro Gonzalez, H.F.; Uehling, J.K.; Vilgalys, R.J.; Hurst, G.B.; Campagna, S.R.; Labbé, J.L.; et al. Integrated Proteomics and Metabolomics Suggests Symbiotic Metabolism and Multimodal Regulation in a Fungal-Endobacterial System. Environ. Microbiol. 2017, 19, 1041–1053. [Google Scholar] [CrossRef]
- Takashima, Y.; Seto, K.; Degawa, Y.; Guo, Y.; Nishizawa, T.; Ohta, H.; Narisawa, K. Prevalence and Intra-Family Phylogenetic Divergence of Burkholderiaceae-Related Endobacteria Associated with Species of Mortierella. Microbes Environ. 2018, 33, 417–427. [Google Scholar] [CrossRef]
- Büttner, H.; Niehs, S.P.; Vandelannoote, K.; Cseresnyés, Z.; Dose, B.; Richter, I.; Gerst, R.; Figge, M.T.; Stinear, T.P.; Pidot, S.J.; et al. Bacterial Endosymbionts Protect Beneficial Soil Fungus from Nematode Attack. Proc. Natl. Acad. Sci. USA 2021, 118, e2110669118. [Google Scholar] [CrossRef]
- Mondo, S.J.; Lastovetsky, O.A.; Gaspar, M.L.; Schwardt, N.H.; Barber, C.C.; Riley, R.; Sun, H.; Grigoriev, I.V.; Pawlowska, T.E. Bacterial Endosymbionts Influence Host Sexuality and Reveal Reproductive Genes of Early Divergent Fungi. Nat. Commun. 2017, 8, 1843. [Google Scholar] [CrossRef]
- Richter, I.; Radosa, S.; Cseresnyés, Z.; Ferling, I.; Büttner, H.; Niehs, S.P.; Gerst, R.; Scherlach, K.; Figge, M.T.; Hillmann, F.; et al. Toxin-Producing Endosymbionts Shield Pathogenic Fungus against Micropredators. mBio 2022, 13, e01440-22. [Google Scholar] [CrossRef] [PubMed]
- Itabangi, H.; Sephton-Clark, P.C.S.; Tamayo, D.P.; Zhou, X.; Starling, G.P.; Mahamoud, Z.; Insua, I.; Probert, M.; Correia, J.; Moynihan, P.J.; et al. A Bacterial Endosymbiont of the Fungus Rhizopus microsporus Drives Phagocyte Evasion and Opportunistic Virulence. Curr. Biol. 2022, 32, 1115–1130.e6. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Hertweck, C. A Gene Cluster Encoding Rhizoxin Biosynthesis in “Burkholderia rhizoxina”, the Bacterial Endosymbiont of the Fungus Rhizopus microsporus. Chembiochem 2007, 8, 41–45. [Google Scholar] [CrossRef]
- Estrada-de Los Santos, P.; Palmer, M.; Chávez-Ramírez, B.; Beukes, C.; Steenkamp, E.T.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E.; et al. Whole Genome Analyses Suggests That Burkholderia sensu lato Contains Two Additional Novel Genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the Evolution of Diazotrophy and Nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef] [PubMed]
- Lastovetsky, O.A.; Krasnovsky, L.D.; Qin, X.; Gaspar, M.L.; Gryganskyi, A.P.; Huntemann, M.; Clum, A.; Pillay, M.; Palaniappan, K.; Varghese, N.; et al. Molecular Dialogues between Early Divergent Fungi and Bacteria in an Antagonism versus a Mutualism. mBio 2020, 11, e02088-20. [Google Scholar] [CrossRef]
- Naumann, M.; Schüßler, A.; Bonfante, P. The Obligate Endobacteria of Arbuscular Mycorrhizal Fungi Are Ancient Heritable Components Related to the Mollicutes. ISME J. 2010, 4, 862–871. [Google Scholar] [CrossRef]
- Toomer, K.H.; Chen, X.; Naito, M.; Mondo, S.J.; den Bakker, H.C.; VanKuren, N.W.; Lekberg, Y.; Morton, J.B.; Pawlowska, T.E. Molecular Evolution Patterns Reveal Life History Features of Mycoplasma-Related Endobacteria Associated with Arbuscular Mycorrhizal Fungi. Mol. Ecol. 2015, 24, 3485–3500. [Google Scholar] [CrossRef]
- Desirò, A.; Hao, Z.; Liber, J.A.; Benucci, G.M.N.; Lowry, D.; Roberson, R.; Bonito, G. Mycoplasma-Related Endobacteria within Mortierellomycotina Fungi: Diversity, Distribution and Functional Insights into Their Lifestyle. ISME J. 2018, 12, 1743–1757. [Google Scholar] [CrossRef]
- Desirò, A.; Faccio, A.; Kaech, A.; Bidartondo, M.I.; Bonfante, P. Endogone, One of the Oldest Plant-Associated Fungi, Host Unique Mollicutes-Related Endobacteria. New Phytol. 2015, 205, 1464–1472. [Google Scholar] [CrossRef]
- Naito, M.; Desirò, A.; González, J.B.; Tao, G.; Morton, J.B.; Bonfante, P.; Pawlowska, T.E. “Candidatus Moeniiplasma Glomeromycotorum”, an Endobacterium of Arbuscular Mycorrhizal Fungi. Int. J. Syst. Evol. Microbiol. 2017, 67, 1177–1184. [Google Scholar] [CrossRef]
- Naito, M.; Morton, J.B.; Pawlowska, T.E. Minimal Genomes of Mycoplasma-Related Endobacteria Are Plastic and Contain Host-Derived Genes for Sustained Life within Glomeromycota. Proc. Natl. Acad. Sci. USA 2015, 112, 7791–7796. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.J.; House, G.L.; Morales, D.P.; Kelliher, J.M.; Gallegos-Graves, L.V.; LeBrun, E.S.; Davenport, K.W.; Palmieri, F.; Lohberger, A.; Bregnard, D.; et al. Widespread Bacterial Diversity within the Bacteriome of Fungi. Commun. Biol. 2021, 4, 1168. [Google Scholar] [CrossRef] [PubMed]
- Telagathoti, A.; Probst, M.; Peintner, U. Habitat, Snow-Cover and Soil PH, Affect the Distribution and Diversity of Mortierellaceae Species and Their Associations to Bacteria. Front. Microbiol. 2021, 12, 669784. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, C.L.; Lovett, B.; Kasson, M.T.; Stajich, J.E. Metagenome-Assembled Genomes of Bacteria Associated with Massospora cicadina Fungal Plugs from Infected Brood VIII Periodical Cicadas. Microbiol. Resour. Announc. 2022, 11, e00413-22. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Wang, Y.; Mondo, S.; Ahrendt, S.; Andreopoulos, W.; Barry, K.; Beard, J.; Benny, G.L.; Blankenship, S.; Bonito, G.; et al. Evolution of Zygomycete Secretomes and the Origins of Terrestrial Fungal Ecologies. iScience 2022, 25, 104840. [Google Scholar] [CrossRef] [PubMed]
- Shelest, E.; Voigt, K. 2 Genomics to Study Basal Lineage Fungal Biology: Phylogenomics Suggests a Common Origin. In Fungal Genomics; The Mycota; Springer: Berlin/Heidelberg, Germany, 2014; Volume XIII, pp. 31–60. ISBN 978-3-642-45217-8. [Google Scholar]
- Voigt, K.; Wolf, T.; Ochsenreiter, K.; Nagy, G.; Kaerger, K.; Shelest, E.; Papp, T. 15 Genetic and Metabolic Aspects of Primary and Secondary Metabolism of the Zygomycetes. In Biochemistry and Molecular Biology; The Mycota; Springer: Berlin/Heidelberg, Germany, 2016; Volume III, pp. 361–385. ISBN 978-3-319-27788-2. [Google Scholar]
- Koczyk, G.; Pawłowska, J.; Muszewska, A. Terpenoid Biosynthesis Dominates among Secondary Metabolite Clusters in MUcoromycotina Genomes. J. Fungi 2021, 7, 285. [Google Scholar] [CrossRef]
- Baldeweg, F.; Warncke, P.; Fischer, D.; Gressler, M. Fungal Biosurfactants from Mortierella alpina. Org. Lett. 2019, 21, 1444–1448. [Google Scholar] [CrossRef]
- Lebreton, A.; Meslet-Cladière, L.; Morin-Sardin, S.; Coton, E.; Jany, J.-L.; Barbier, G.; Corre, E. Comparative Analysis of Five Mucor Species Transcriptomes. Genomics 2019, 111, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Winkelmann, G. A Search for Glomuferrin: A Potential Siderophore of Arbuscular Mycorrhizal Fungi of the Genus Glomus. Biometals 2017, 30, 559–564. [Google Scholar] [CrossRef]
- Prakash, H.; Rudramurthy, S.M.; Gandham, P.S.; Ghosh, A.K.; Kumar, M.M.; Badapanda, C.; Chakrabarti, A. Apophysomyces variabilis: Draft Genome Sequence and Comparison of Predictive Virulence Determinants with Other Medically Important Mucorales. BMC Genom. 2017, 18, 736. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.S.; Grieve, C.L.; Murugathasan, I.; Bennet, A.J.; Czekster, C.M.; Liu, H.; Naismith, J.; Moore, M.M. The Rhizoferrin Biosynthetic Gene in the Fungal Pathogen Rhizopus delemar Is a Novel Member of the NIS Gene Family. Int. J. Biochem. Cell Biol. 2017, 89, 136–146. [Google Scholar] [CrossRef]
- Škríba, A.; Patil, R.H.; Hubáček, P.; Dobiáš, R.; Palyzová, A.; Marešová, H.; Pluháček, T.; Havlíček, V. Rhizoferrin Glycosylation in Rhizopus microsporus. J. Fungi 2020, 6, 89. [Google Scholar] [CrossRef]
- Takeda, I.; Tamano, K.; Yamane, N.; Ishii, T.; Miura, A.; Umemura, M.; Terai, G.; Baker, S.E.; Koike, H.; Machida, M. Genome Sequence of the Mucoromycotina Fungus Umbelopsis isabellina, an Effective Producer of Lipids. Genome Announc. 2014, 2, e00071-14. [Google Scholar] [CrossRef]
- Gluck-Thaler, E.; Ralston, T.; Konkel, Z.; Ocampos, C.G.; Ganeshan, V.D.; Dorrance, A.E.; Niblack, T.L.; Wood, C.W.; Slot, J.C.; Lopez-Nicora, H.D.; et al. Giant Starship Elements Mobilize Accessory Genes in Fungal Genomes. Mol. Biol. Evol. 2022, 39, msac109. [Google Scholar] [CrossRef]
- Urquhart, A.S.; Chong, N.F.; Yang, Y.; Idnurm, A. A Large Transposable Element Mediates Metal Resistance in the Fungus Paecilomyces variotii. Curr. Biol. 2022, 32, 937–950.e5. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).