Morphometrical Identification and Phylogenetic Analysis of Rhinonyssidae (Acari: Mesostigmata) Parasitizing Avian Hosts: New Molecular Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphometrical Study and Identification
2.2. Molecular Study
Sequence Alignments and Phylogenetic Analyses
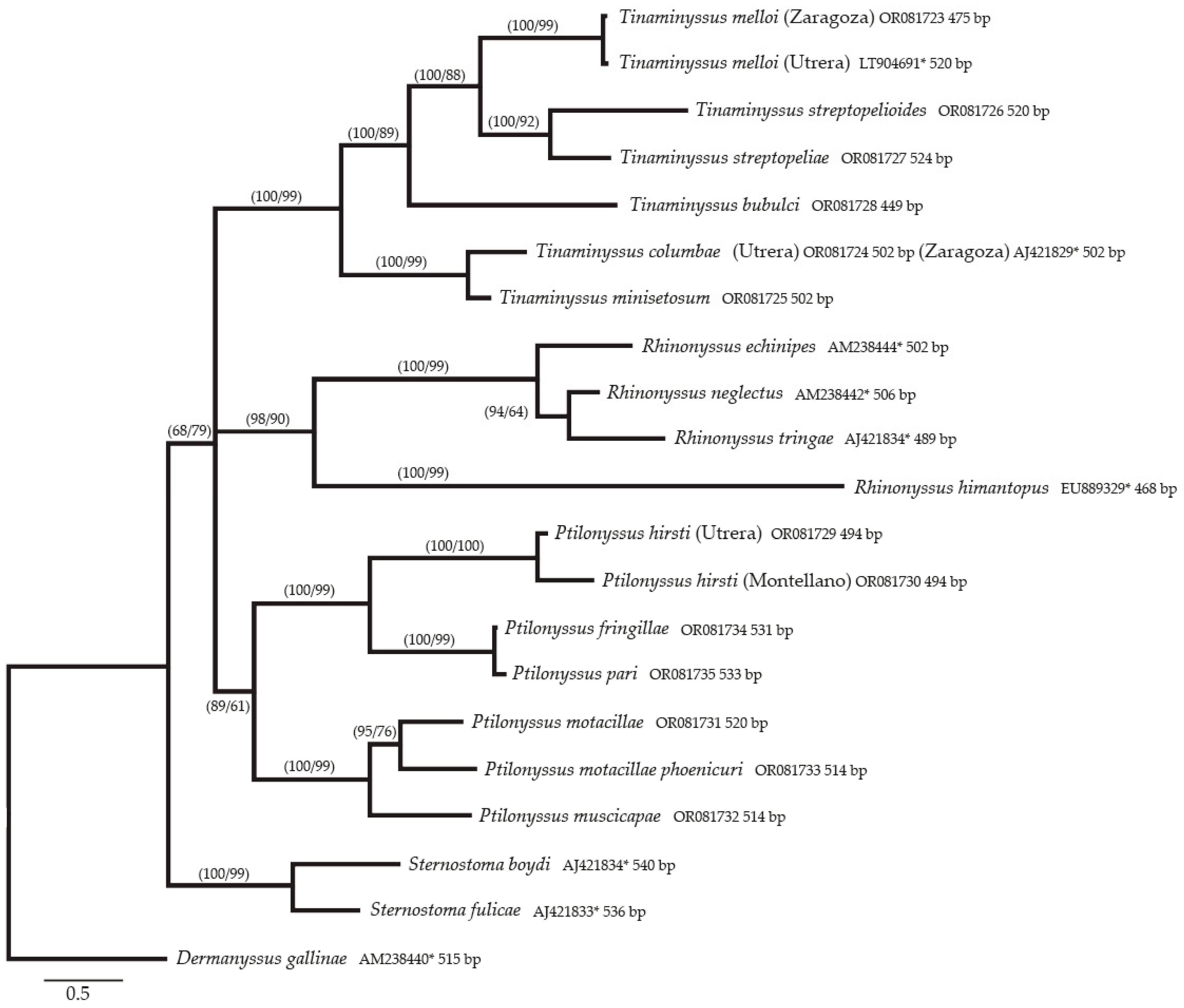
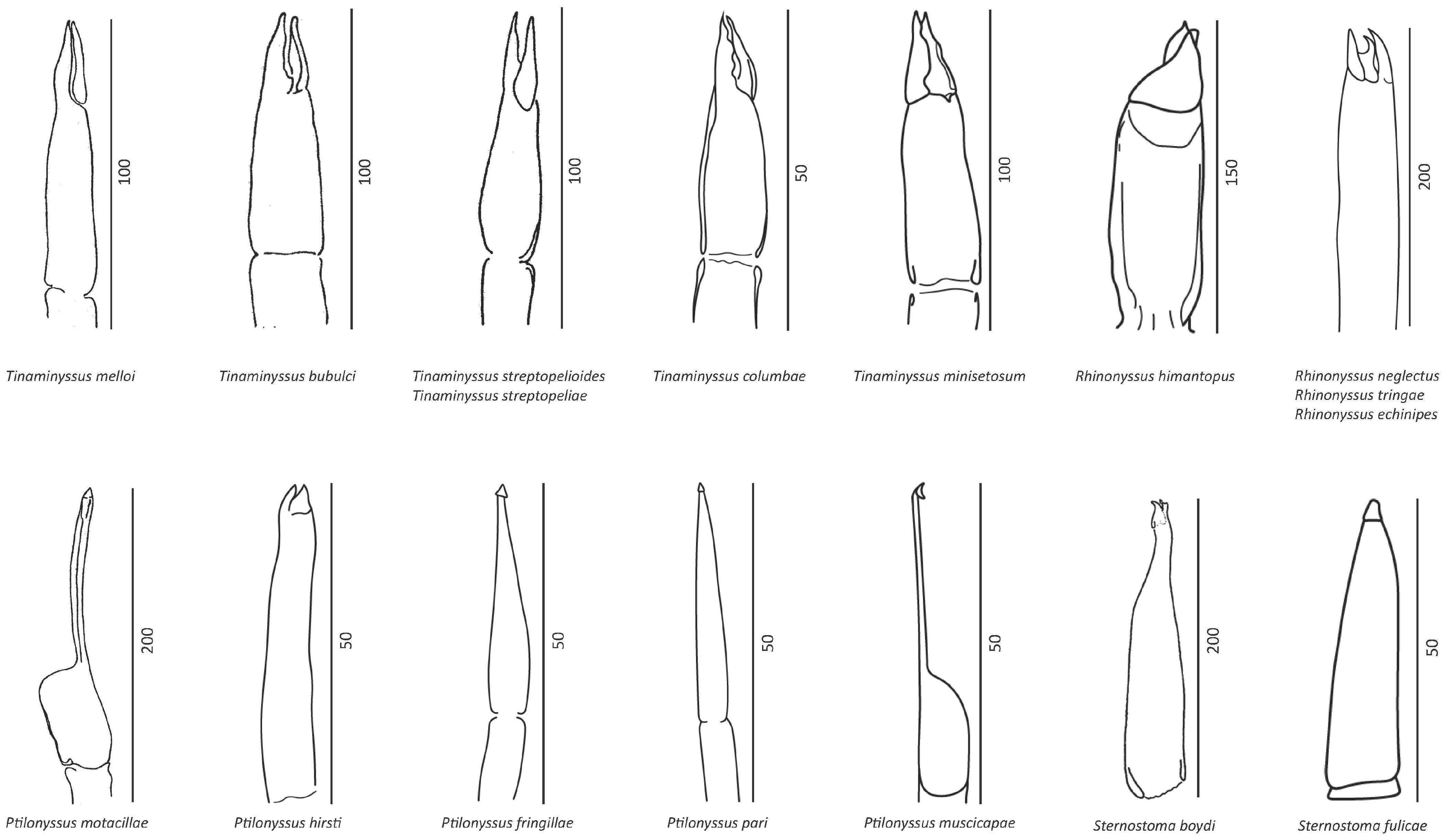
3. Results
3.1. Morphometrical Identification
Molecular Results
3.2. Phylogenetic Study of the Species Included in the Alignment
4. Discussion
4.1. Morphological Analysis
4.2. Molecular Analysis
4.3. Analysis of Phylogenetic and Genetic Distances
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knee, W.; Galloway, T.D. New Host and Locality Records for Endoparasitic Nasal Mites (Acari: Rhinonyssidae, Turbinoptidae, and Ereynetidae) Infesting Birds in Manitoba, Canada. Can. Entomol. 2017, 149, 89–103. [Google Scholar] [CrossRef]
- Dimov, I. Rhinonyssidosis avium. VetPharma 2011, 3–4, 88–90. [Google Scholar]
- Knee, W. Five New Species of Rhinonyssidae (Mesostigmata) and One New Species of Dermanyssus (Mesostigmata: Dermanyssidae) from Birds of Alberta and Manitoba, Canada. J. Parasitol. 2008, 94, 348–374. [Google Scholar] [CrossRef] [PubMed]
- Strandtmann, R.W. The Mesostigmatic Nasal Mites of Birds. I. Two New Genera from Shore and Marsh Birds. J. Parasitol. 1948, 34, 505. [Google Scholar] [CrossRef]
- Bregetova, N.G. Generic grouping of the rhinonyssid mites (Mesostigmata, Rhinonyssidae), parasites of birds. I. The new genus Sternostomoides. Ent. Obozr. 1965, 44, 709–713. [Google Scholar]
- Fain, A. Comparative Morphology of Rhinonyssidae. Bull. Ann. Soc. Roy. Ent. Belg. 1960, 96, 303–313. [Google Scholar]
- Vitzthum, H.G. Milben aus der Nasenhöhle von Vögeln. J. Fur Ornithol. 1935, 83, 563–587. [Google Scholar] [CrossRef]
- Berlese, A.; Trouessart, E. Diagnoses of New or Little-Known Mites. Bull. Bibl. Sci. Quest 1889, 2, 121–143. [Google Scholar]
- Strandtmann, R.W.; Wharton, G.W. A Manual of Mesostigmatid Mites Parasitic on Vertebrates; Institute of Allergy: College Park, MD, USA, 1958; Volume 4, 330p. [Google Scholar]
- Cooreman, J. Rhinoecius oti n. Gen., n. Sp. Acarien: Rhinonyssidae. Bull. Mus. Hist. Nat. Begl. 1946, 22, 1–4. [Google Scholar]
- Trouessart, E.L. Note sur les Acariens Parasites des Fosses Nasales des Oiseaux. C R Soc. Biol. IDe Ser. 1894, 1, 723–724. [Google Scholar]
- Strandtmann, R.W. Host Specificity of Bird Nasal Mites (Rhinonyssidae) Is a Function of the Gregariousness of the Host. In Proceedings of the 10th International Congress of Entomology, Montreal, QC, Canada, 17–25 August 1956; Volume 1, pp. 909–911. [Google Scholar]
- Castro, M.P. Reestruturacao Generica of the Family Rhinonyssidae Vitzhum, 1935 (Acari: Mesostigmata: Gamasides) and Descricao of Some Species Novas. Arch. Int. Biol. 1948, 18, 253–284. [Google Scholar]
- Giebel, C. Ueber einige Milben. Z. Ges. Naturw. 1871, 38, 29–32. [Google Scholar]
- Strandtmann, R.W. The Mesostigmatic Nasal Mites of Birds. II. New and Poorly Known Species of Rhinonyssidae. J. Parasitol. 1951, 37, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Fain, A. Essai de clasiffication des Rhinonyssidae (Acari: Mesostigmata) avec description de deux genres nouveaux. Ann. Parasitol. Hum. Comp. 1957, 31, 145–157. [Google Scholar]
- Pence, D.B.; Casto, S.D. Studies on the Variation and Morphology of the Ptilonyssus “Sairae” Complex (Agarina: Rhinonyssinae) from North American Passeriform Birds. J. Med. Entomol. 1976, 13, 71–95. [Google Scholar] [CrossRef]
- Fain, A. Les Rhinonyssides Parasites Des Pigeons. Rev. Zool. Bot. Afr. 1962, 65, 305–324. [Google Scholar]
- Fain, A. Le Complexe “Rhinonyssus Coniventris”(Rhinonyssidae: Mesostigmata). Bull. Ann. Soc. R Entomolog. Belg. 1963, 99, 86–100. [Google Scholar]
- Huyse, T.; Poulin, R.; Théron, A. Speciation in Parasites: A Population Genetics Approach. Trends Parasitol. 2005, 21, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Sunantaraporn, S.; Sanprasert, V.; Pengsakul, T. Molecular Survey of the Head Louse Pediculus Humanus Capitis in Thailand and Its potential Role for Transmitting Acinetobacter spp. Parasites Vectors 2015, 8, 127. [Google Scholar] [CrossRef]
- Kelomey, A.E.; Paraiso, A.; Sina, H.; Legout, H.; Garnery, L.; Baba-Moussa, L. Genetic Characterization of the Honeybee Ectoparasitic Mite Varroa destructor from Benin (West Africa) Using Mitochondrial and Microsatellite Markers. Exp. Appl. Acarol. 2017, 72, 61–67. [Google Scholar] [CrossRef]
- Baulieu, F.; Knee, W.; Nowell, V.; Schwarzfeld, M.; Lindo, Z.; Behan-Pelletier, V.M.; Lumley, L.; Young, M.R.; Smith, I.; Proctor, H.C.; et al. Acari of Canada. Zookeys 2019, 819, 77–168. [Google Scholar] [CrossRef]
- Morelli, M.; Spicer, G.S. Cospeciation between the Nasal Mite Ptilonyssus sairae (Acari: Rhinonyssidae) and Its Bird Hosts. Syst. Appl. Acarol. 2007, 12, 3. [Google Scholar] [CrossRef]
- De Rojas, M.; Mora, M.D.; Ubeda, J.M.; Cutillas, C.; Navajas, M.; Guevara, D.C. Phylogenetic Relationships in Rhinonyssid Mites (Acari: Rhinonyssidae) Based on Mitochondrial 16S RDNA Sequences. Exp. Appl. Acarol. 2001, 25, 957–967. [Google Scholar] [CrossRef]
- Rojas, D.; Doña, M.; Jovani, J.; Dimov, R.; Zurita, I.; Callejon, A. Evidence of Cryptic Species in the Genus Tinaminyssus (Acari: Rhinonyssidae) Based on Morphometrical and Molecular Data. Exp. Appl. Acarol. 2018, 75, 355–391. [Google Scholar] [CrossRef]
- Domrow, R. The Nasal Mites of Queensland Birds (Acari: Dermanyssidae, Ereynetidae, and Epidermoptidae). Proc. Linn. Soc. 1969, 93, 297–426. [Google Scholar]
- Pence, D.B.; Casto, S. Two New Species and New Records of Nasal Mites of the Genus Sternostoma (Acarina: Rhinonyssinae) from Birds in Texas. J. Parasitol. 1975, 61, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Braza, R.; Úbeda Ontiveros, J.M.; Guevara Benítez, D.C. Ácaros del Género Tinaminyssus Strandtmann y Wharton, 1958, Parásitos Nasícolas de Aves Ciconiformes y Columbiformes Españolas. Rev. Ibérica Parasitol. 1990, 50, 73–80. [Google Scholar]
- Dimov, I. A New Nasal Mite of the Genus Ptilonyssus (Rhinonyssidae) from Parus caeruleus (Passeriformes) from Russia. J. Hell. Vet. Med. Soc. 2017, 63, 25. [Google Scholar] [CrossRef]
- Dimov, I. Three New Species of Nasal Mites Ptilonyssus (Mesostigmata, Rhinonyssidae) from Russia. Arhimed-J. Sci. Pract. 2020, 9, 41–52. [Google Scholar]
- Butenko, O.M. Rhinonyssid Mites of Non-Passerine Birds of the USSR; Academy of Sciences of USSR: Moscow, Russia, 1984; p. 188. [Google Scholar]
- Kaneko, K.; Matsudaira, Y.; Masahito, P. Endoparasitic mites of anatid birds collected in Chiba and Saitama prefectures, Japan (Acarina: Rhinonyssidae and Ereynetidae). Jpn. Soc. Med. Entomol. Zool. 1978, 29, 147–154. [Google Scholar] [CrossRef]
- Strandtmann, R.V. The Mesostigmatic Nasal Mites of Birds. IV. The Species and Hosts of the Genus Rhinonyssus (Acarina, Rhinonyssidae). Proc. Entom. Soc. Wash. 1956, 58, 129–142. [Google Scholar]
- Dimov, I. Taxonomic Diversity and Morphology of Mites of the Family Rhinonyssidae of the Northwest of Russia; ZIN RAS: St. Petersburg, Russia, 2000; 214p, ISBN 978-5-6044687-1-5. [Google Scholar]
- Navajas, M.; Lagnel, J.; Fauvel, G.; De Moraes, G. Sequence Variation of Ribosomal Internal Transcribed Spacers (ITS) in Commercially Important Phytoseiidae Mites. Exp. Appl. Acarol. 1999, 23, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. DAMBE5: A Comprehensive Software Package for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate max-imum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Rannala, B. Phylogenetic Methods Come of Age: Testing Hypotheses in an Evolutionary Context. Science 1997, 276, 227–232. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches over Likelihood Ratio Tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Domrow, R. Acari Spinturnicidae from Australia and New Guinea. Acarologia 1972, 13, 552–584. [Google Scholar]
- Pence, D.B. The Nasal Mites of Birds from Louisiana. I. Dermanyssids (Rhinonyssinae) from Shore and Marsh Birds. J. Parasitol. 1972, 58, 153–168. [Google Scholar] [CrossRef]
- Pence, D.B. The Nasal Mites of Birds from Louisiana. IX. Synopsis. J. Parasitol. 1973, 59, 881. [Google Scholar] [CrossRef]
- Zumpt, F.; Till, W.M. Nasal Mites of Birds Hitherto Known from the Ethiopian Region, with Keys and Descriptions of Nine New Species (Acarina: Laelaptidae). J. Entomol. Soc. South. Afr. 1955, 18, 50–59. [Google Scholar]
- Domrow, R. Fourteen Species of Ptilonyssus from Australian Birds (Acarina, Laelaptidae). Acarologia 1964, 6, 595–623. [Google Scholar]
- Fain, A.; Sixl, W. Two New Rhinonyssids from Austrian Birds. Bull. Ann. Soc. Roy. Ent. Belg. 1971, 107, 89–93. [Google Scholar]
- Fain, A.; Sixl, W.; Moritsch, C. The Nasal Mites of the Family Rhinonyssidae with Description of a New Species (Acarina). Mitteilungen der Abteilung Zool. Bot. Landesmus. Joanneum Graz 1974, 3, 1–9. [Google Scholar]
- Fain, A.; Hyland, K. On Three Species of Rhinonyssids Described by Hirst. Ann. Mag. Nat. Hist. 1962, 5, 341–348. [Google Scholar] [CrossRef]
- Fain, A. La spermatheque et ses canaux adducteurs chez les acariens mesostigmatiques parasites des voies respiratoires. Acarologia 1963, 5, 463–479. [Google Scholar]
- Gretillat, S. Description de Deux Nouvelles Especes de Rhinonyssidae (Acarina, Mesostigmata) Rallinyssus strandtmanni et Larinyssus petiti. Vie et Milleu 1961, 12, 151–160. [Google Scholar]
- Bregetova, N.G. Mites from the nasal cavity of Fly carchers of the genus Muscicapa. Parazitologiya 1970, 4, 59–62. [Google Scholar]
- Hirst, S. On Some New Parasitic Mites. Proc. Zool. Soc. 1921, 1922, 769–802. [Google Scholar] [CrossRef]
- Guevara Benítez, D.; Úbeda Ontiveros, J.M.; Morillas Márquez, F. Acaros Del Genero Ptilonyssus Berlese y Trouessart, 1889 (Mesostigmata: Rhinonyssidae) Parasito de Fosas Nasales de Aves Paseriformes Españolas, 1: Ptilonyssus hirsti (Castro y Pereira, 1947) Pereira y Castro. Rev. Ibérica Parasitol. 1949, 46, 75–80. [Google Scholar]
- Bregetova, N. Ticks Parasitizing in the Nasal Cavity of Birds. Parasit Sb. Zool. Inst. Acad. Sci. USSR 1951, 13, 111–119. [Google Scholar]
- TerBush, L.E. Incidence of Nasal Mites in Different Age Classes of Herring Gulls (Larus argentatus). J. Parasitol. 1963, 49, 525. [Google Scholar] [CrossRef]
- Spicer, G.S. Two New Nasal Mites of the Genus Ptilonyssus (Mesostigmata: Rhinonyssidae) from Texas. Acarologia 1977, 18, 595–601. [Google Scholar]
- Fain, A.; Bafort, J. Deux Nouveaux Acariens Parasites Nasicoles Du Rossignol Du Japon Leioth Rix Lutea Swainson. Bull. La Soc. R. Zool. D’anvers 1963, 31, 7–11. [Google Scholar]
- Guevara, D.C.; Ontiveros, J.M. Descripción de La Larva y Redescripción de La Hembra de Sternostoma fulicae Fain y Bafort, 1963 (Acarina: Rhinonyssidae). Rev. Iber. Parasitol. 1975, 35, 81–93. [Google Scholar]
- de Rojas, M.; Mora, M.D.; Ubeda, J.M.; Cutillas, C.; Navajas, M.; Guevara, D.C. Phylogenetic Relationships in Rhinonyssid Mites (Acari: Rhinonyssidae) Based on Ribosomal DNA Sequences: Insights for the Discrimination of Closely Related Species. Parasitol. Res. 2002, 88, 675–681. [Google Scholar] [CrossRef]
- Dobler, S.; Farrell, B.D. Host Use Evolution in Chrysochus Milkweed Beetles: Evidence from Behaviour, Population Genetics and Phylogeny. Mol. Ecol. 1999, 8, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Úbeda Ontiveros, J.M.; Guevara Benítez, D.C.; Cutillas Barrios, C.; Ariza Astolfi, C.; de Rojas Alvarez, M. Ácaros de las vías respiratorias de vertebrados. In “In Memoriam” al Profesor Doctor D. Ignacio Navarrete López-Cózar; Tovar Andrada, J.J., Reina Esojo, D., Eds.; Facultad de Veterinaria, Universidad de Extremadura: Cáceres, Spain, 2006; ISBN 84-690-2894-4. [Google Scholar]
- Dimov, I. Kleshchi-Rinonissidy Ptic Severo-Zapada Rossii; LLC Zhigulin.: Saint Petersburg, Russia, 2018. [Google Scholar]
- Pence, D.B. Congruent Inter-Relationships of the Rhinonyssinae (Dermanyssidae) with Their Avian Hosts. In Recent Advances in Acarology; Elsevier: Amsterdam, The Netherlands, 1979; pp. 371–377. ISBN 9780125922029. [Google Scholar]
- Strandtmann, R.W. New Records for Rhinonyssus himantopus and Notes on Other Species of the Genus. J. Kans. Entomol. Soc. 1959, 32, 1. [Google Scholar]
| Species | ITS1 | 5.8S | ITS2 |
|---|---|---|---|
| Tinaminyssus melloi (Zaragoza) Castro, 1948 | 218 | 184 | - |
| Tinaminyssus melloi (Utrera) Castro, 1948 | 218 | 182 | 90 |
| Tinaminyssus columbae (Utrera) Castro, 1948 | 200 | 182 | 90 |
| Tinaminyssus columbae (Zaragoza) Castro, 1948 | 200 | 182 | 90 |
| Tinaminyssus minisetosum Butenko, 1984 | 201 | 181 | 90 |
| Tinaminyssus streptopelioides Butenko, 1984 | 218 | 182 | 90 |
| Tinaminyssus streptopeliae Fain, 1962 | 218 | 182 | 94 |
| Tinaminyssus bubulci Zumpt & Till, 1955 | 198 | 181 | - |
| Ptilonyssus hirsti (Montellano) Castro, 1948 | 214 | 189 | - |
| Ptilonyssus hirsti (Utrera) Castro, 1948 | 214 | 189 | 87 |
| Ptilonyssus motacillae Fain, 1966 | 214 | 191 | 82 |
| Ptilonyssus muscicapae Bregetova, 1970 | 213 | 189 | 82 |
| Ptilonyssus motacillae phoenicuri Fain, 1966 | 213 | 189 | 82 |
| Ptilonyssus fringillae (Montellano) Fain & Sixl, 1971 | 226 | 188 | 87 |
| Ptilonyssus pari (Russia) Fain & Hyland, 1963 | 226 | 188 | 89 |
| Rhinonyssus echinipes Hirst, 1921 | 206 | 177 | 89 |
| Rhinonyssus himantopus Strandtmann, 1951 | 197 | 181 | 90 |
| Rhinonyssus neglectus Hirst, 1921 | 210 | 177 | 89 |
| Rhinonyssus tringae Fain, 1963 | 193 | 177 | 89 |
| Sternostoma boydi Strandtmann, 1951 | 236 | 184 | 90 |
| Sternostoma fulicae Guevara & Úbeda, 1975 | 232 | 184 | 90 |
| Level | Genetic Distance (K2P) | % |
|---|---|---|
| Genera | ||
| Tinaminyssus–Sternostoma | 0.26–0.34 | 77.6–81.1 |
| Tinaminyssus–Rhinonyssus | 0.33–0.54 | 70.5–79.5 |
| Tinaminyssus–Ptilonyssus | 0.22–0.39 | 73.8–82.8 |
| Sternostoma–Rhinonyssus | 0.32–0.42 | 72.7–77.8 |
| Sternostoma–Ptilonyssus | 0.23–0.32 | 76.4–82.5 |
| Ptilonyssus–Rhinonyssus | 0.31–0.48 | 71.7–79.3 |
| Species | ||
| Tinaminyssus | 0.05–0.27 | 82.5–96 |
| Sternostoma | 0.1 | 91.5 |
| Ptilonyssus | 0.08–0.30 (0.01 *) | 72.6–94.1 |
| Rhinonyssus | 0.08–0.48 | 78.3–93.2 |
| Populations | ||
| T. melloi (Zaragoza–Utrera) | 0 | 100 |
| T. columbae (Utrera–Zaragoza) | 0 | 100 |
| P. hirsti (Montellano–Utrera) | 0.04 | 96.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Carrión, S.A.; Dimov, I.; Márquez Jiménez, F.J.; de Rojas Álvarez, M. Morphometrical Identification and Phylogenetic Analysis of Rhinonyssidae (Acari: Mesostigmata) Parasitizing Avian Hosts: New Molecular Data. Microorganisms 2023, 11, 1783. https://doi.org/10.3390/microorganisms11071783
Sánchez-Carrión SA, Dimov I, Márquez Jiménez FJ, de Rojas Álvarez M. Morphometrical Identification and Phylogenetic Analysis of Rhinonyssidae (Acari: Mesostigmata) Parasitizing Avian Hosts: New Molecular Data. Microorganisms. 2023; 11(7):1783. https://doi.org/10.3390/microorganisms11071783
Chicago/Turabian StyleSánchez-Carrión, Susana A., Ivan Dimov, Francisco J. Márquez Jiménez, and Manuel de Rojas Álvarez. 2023. "Morphometrical Identification and Phylogenetic Analysis of Rhinonyssidae (Acari: Mesostigmata) Parasitizing Avian Hosts: New Molecular Data" Microorganisms 11, no. 7: 1783. https://doi.org/10.3390/microorganisms11071783
APA StyleSánchez-Carrión, S. A., Dimov, I., Márquez Jiménez, F. J., & de Rojas Álvarez, M. (2023). Morphometrical Identification and Phylogenetic Analysis of Rhinonyssidae (Acari: Mesostigmata) Parasitizing Avian Hosts: New Molecular Data. Microorganisms, 11(7), 1783. https://doi.org/10.3390/microorganisms11071783





