Influence of Varying Pre-Culture Conditions on the Level of Population Heterogeneity in Batch Cultures with an Escherichia coli Triple Reporter Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Triple Reporter Strain
2.2. Pre-Cultures
2.3. Bioreactor Cultures
2.4. Sample Analysis
2.5. Data Analysis
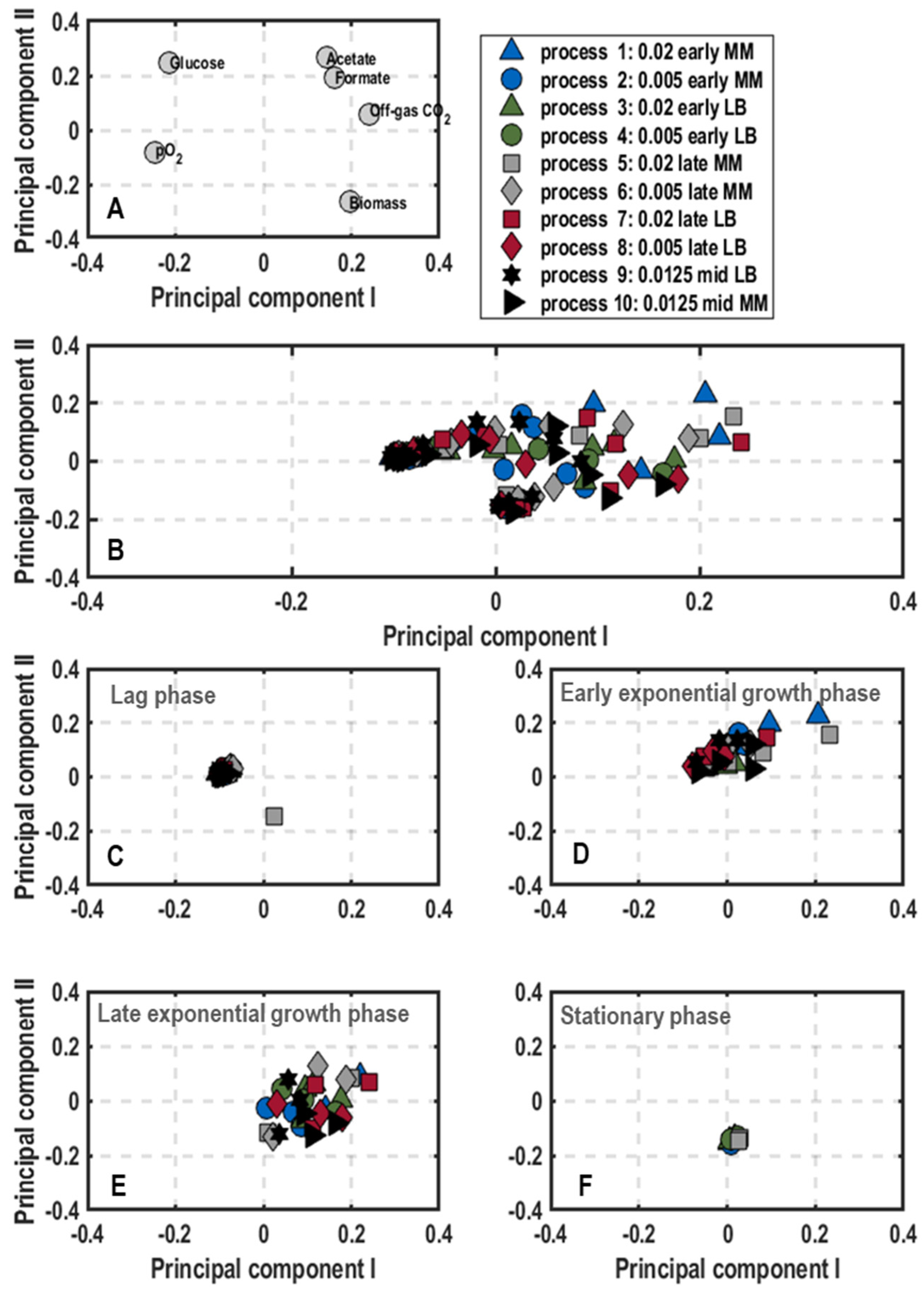
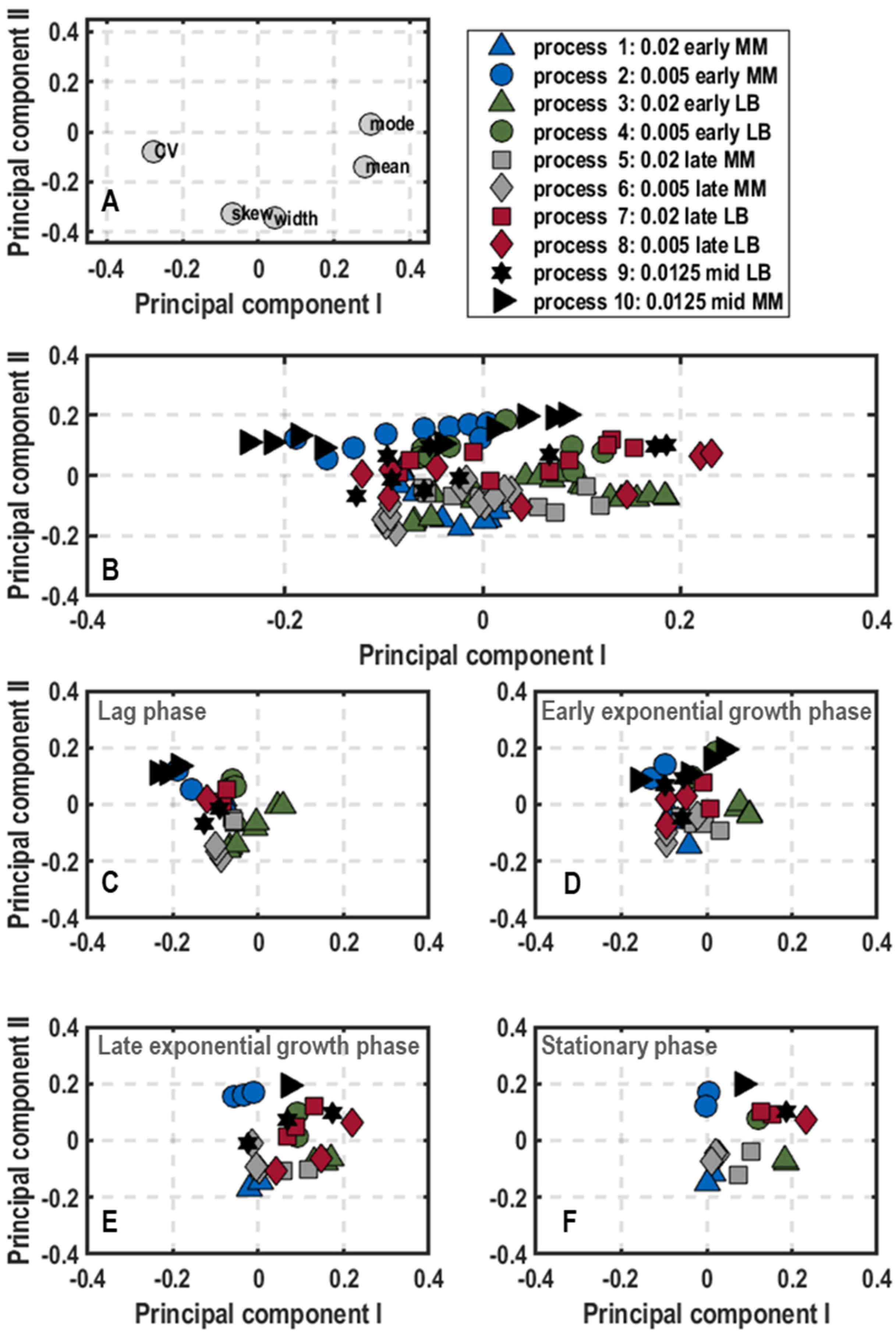
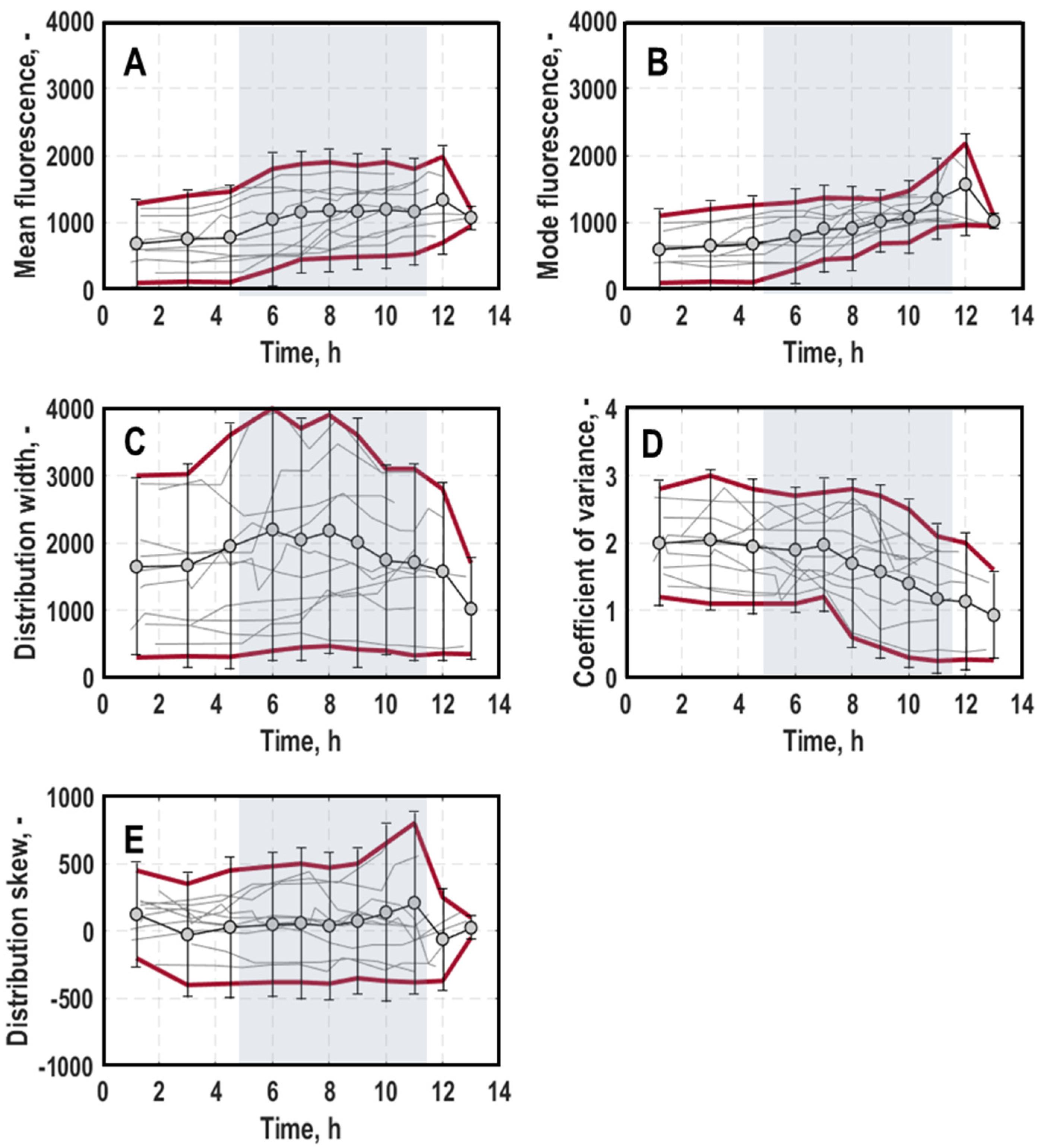
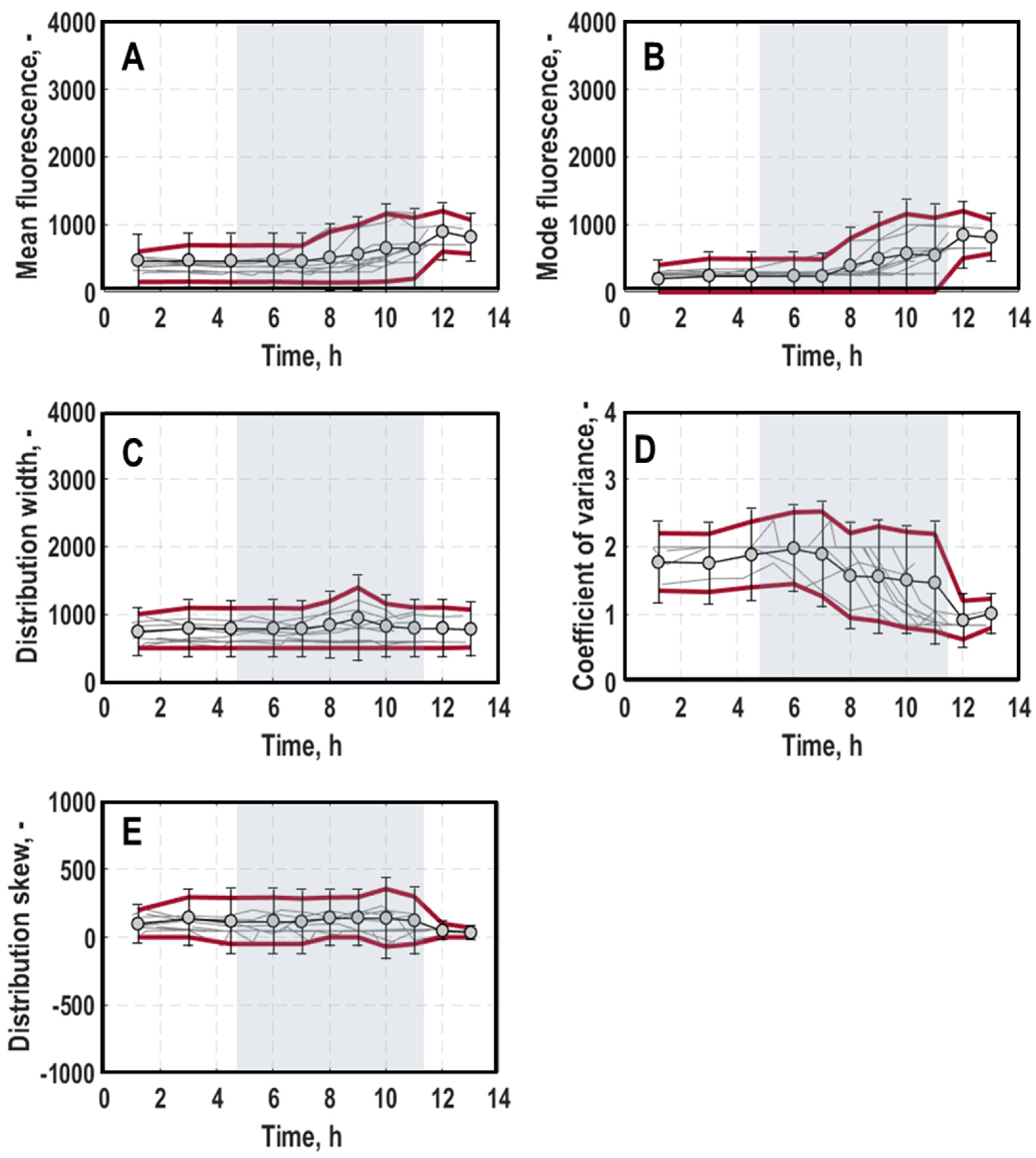
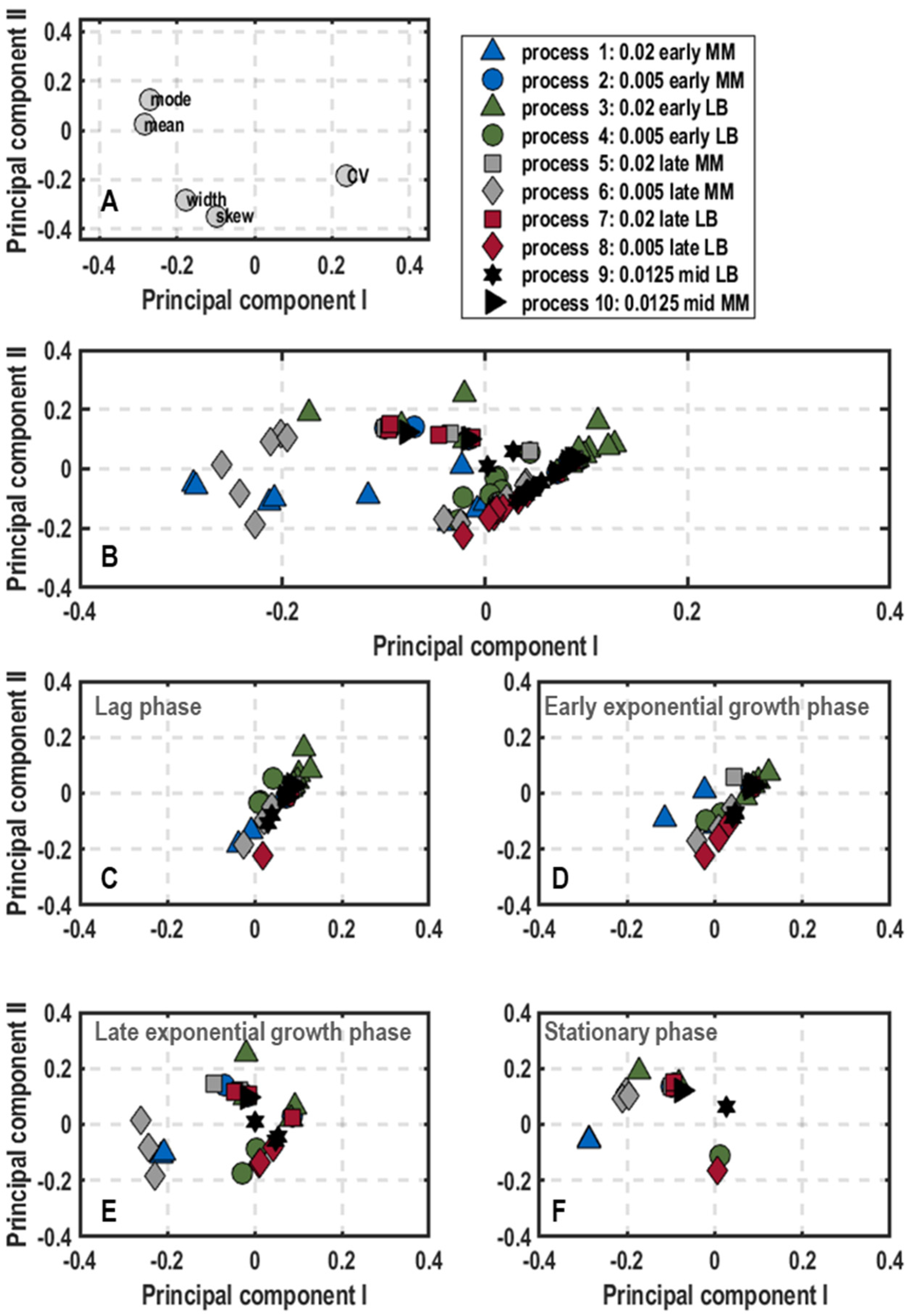
3. Results
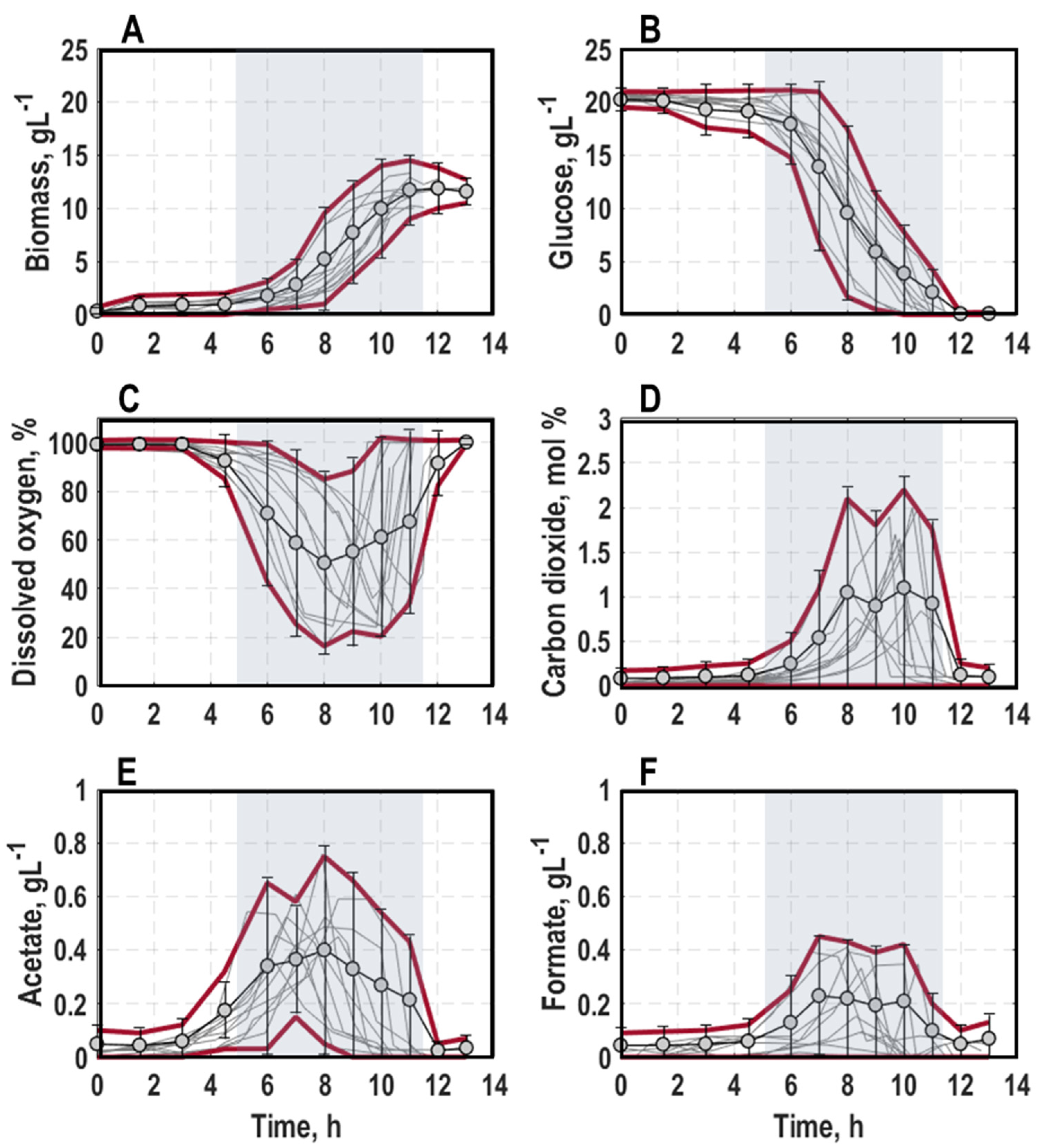
3.1. Population-Level Physiology
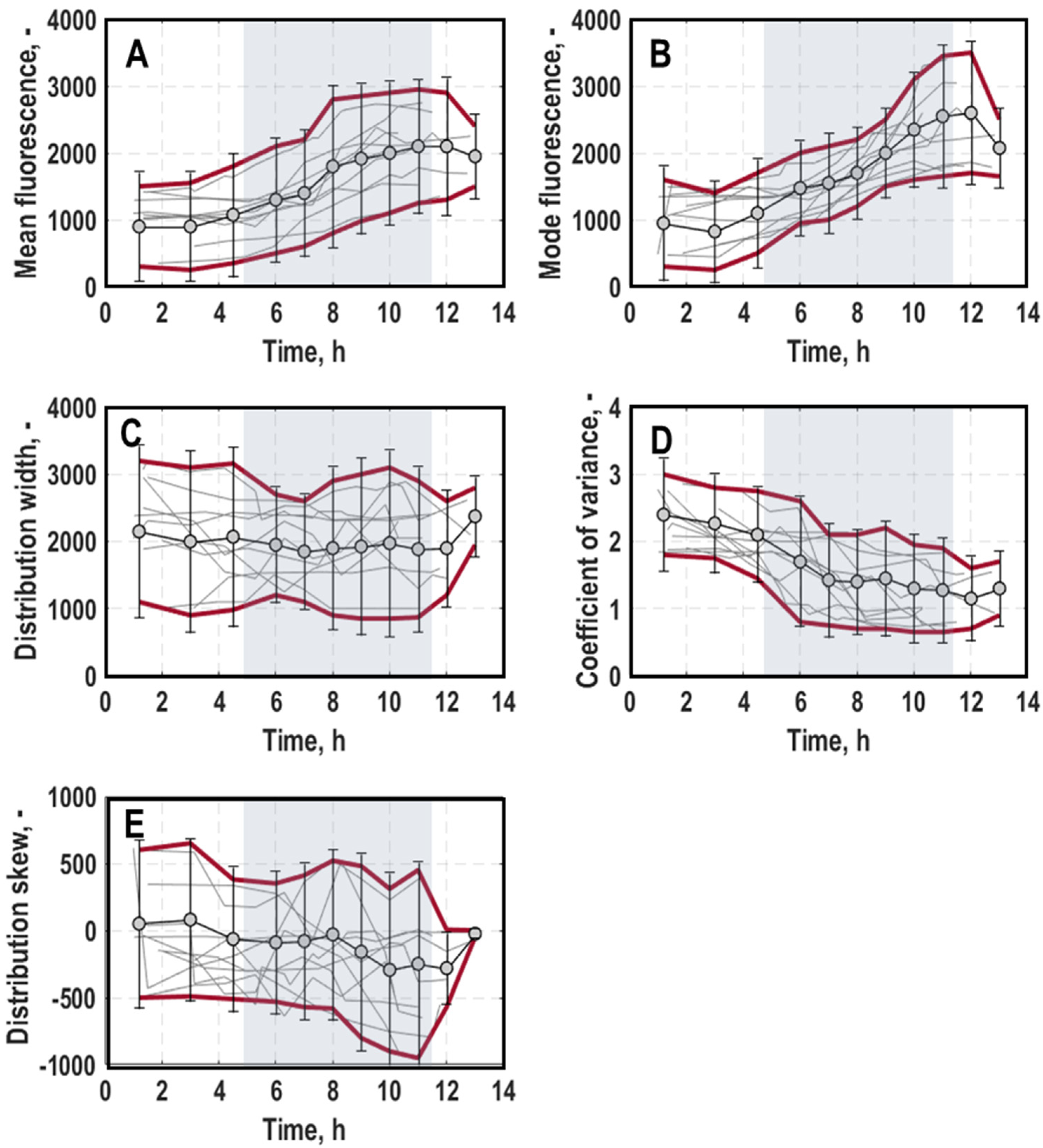
3.2. Single-Cell Physiology
3.2.1. Single-Cell Growth
3.2.2. General Stress Response of Single Cells
3.2.3. Oxygen Limitation of Single Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lemoine, A.; Delvigne, F.; Bockisch, A.; Neubauer, P.; Junne, S. Tools for the determination of population heterogeneity caused by imhomogeneous cultivation conditions. J. Biotechnol. 2017, 251, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Heins, A.-L.; Weuster-Botz, D. Population heterogeneity in microbial bioprocesses: Origin, analysis, mechanisms, and future perspectives. Bioprocess Biosyst. Eng. 2018, 41, 889–916. [Google Scholar] [CrossRef]
- Binder, D.; Drepper, T.; Jaeger, K.-E.; Delvigne, F.; Wiechert, W.; Kohlheyer, D.; Grünberger, A. Homogenizing bacterial cell factories: Analysis and engineering of phenotypic heterogeneity. Metab. Eng. 2017, 42, 145–156. [Google Scholar] [CrossRef]
- Enfors, S.-O.; Jahic, M.; Rozkov, A.; Xu, B.; Hecker, M.; Jürgen, B.; Krüger, E.; Schweder, T.; Hamer, G.; O’Beirne, D.; et al. Physiological responses to mixing in large scale bioreactors. J. Biotechnol. 2001, 85, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Lara, A.R.; Galindo, E.; Ramírez, O.T.; Palomares, L.A. Living With Heterogeneities in Bioreactors. Mol. Biotechnol. 2006, 34, 355–381. [Google Scholar] [CrossRef] [PubMed]
- Delvigne, F.; Goffin, P. Microbial heterogeneity affects bioprocess robustness: Dynamic single-cell analysis contributes to understanding of microbial populations. Biotechnol. J. 2014, 9, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Evans, T.; Zhang, F. Applications and advances of metabolite biosensors for metabolic engineering. Metab. Eng. 2015, 31, 35–43. [Google Scholar] [CrossRef]
- Mahr, R.; Frunzke, J. Transcription factor-based biosensors in biotechnology: Current state and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 79–90. [Google Scholar] [CrossRef]
- Carlquist, M.; Fernandes, R.L.; Helmark, S.; Heins, A.-L.; Lundin, L.; Sørensen, S.J.; Gernaey, K.V.; Lantz, A.E. Physiological heterogeneities in microbial populations and implications for physical stress tolerance. Microb. Cell Factories 2012, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Delvigne, F.; Brognaux, A.; Charbon, G.E.; Sørensen, S.J. Design of growth-dependent biosensors based on destabilized GFP for the detection of physiological behavior of Escherichia coli in heterogeneous bioreactors. Biotechnol. Prog. 2013, 29, 553–563. [Google Scholar] [CrossRef]
- Heins, A.-L.; Reyelt, J.; Schmidt, M.; Kranz, H.; Weuster-Botz, D. Development and characterization of Escherichia coli triple reporter strains for investigation of population heterogeneity in bioprocesses. Microb. Cell Factories 2020, 19, 14. [Google Scholar] [CrossRef] [PubMed]
- Arnoldini, M.; Heck, T.; Blanco-Fernández, A.; Hammes, F. Monitoring of Dynamic Microbiological Processes Using Real-Time Flow Cytometry. PLoS ONE 2013, 8, e80117. [Google Scholar] [CrossRef] [PubMed]
- Delvigne, F.; Boxus, M.; Ingels, S.; Thonart, P. Bioreactor mixing efficiency modulates the activity of a prpoS:GFP reporter gene in E. coli. Microb. Cell Factories 2009, 8, 15. [Google Scholar] [CrossRef]
- Polizzi, K.M.; Kontoravdi, C. Genetically-encoded biosensors for monitoring cellular stress in bioprocessing. Curr. Opin. Biotechnol. 2015, 31, 50–56. [Google Scholar] [CrossRef]
- Mahr, R.; Gätgens, C.; Gätgens, J.; Polen, T.; Kalinowski, J.; Frunzke, J. Biosensor-driven adaptive laboratory evolution of L-valine production in Corynebacterium glutamicum. Metab. Eng. 2015, 32, 184–194. [Google Scholar] [CrossRef]
- Mahr, R.; von Boeselager, R.F.; Wiechert, J.; Frunzke, J. Screening of an Escherichia coli promoter library for a phenylalanine biosensor. Appl. Microbiol. Biotechnol. 2016, 100, 6739–6753. [Google Scholar] [CrossRef]
- Hoang, M.D.; Doan, D.T.; Schmidt, M.; Kranz, H.; Kremling, A.; Heins, A.-L. Application of an Escherichia coli triple reporter strain for at-line monitoring of single-cell physiology during L-phenylalanine production. Eng. Life Sci. 2023, 23, e2100162. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.L.; Nierychlo, M.; Lundin, L.; Pedersen, A.E.; Puentes Tellez, P.E.; Dutta, A.; Carlquist, M.; Bolic, A.; Schäpper, D.; Brunetti, A.C.; et al. Experimental methods and modeling techniques for description of cell population heterogeneity. Biotechnol. Adv. 2011, 29, 575–599. [Google Scholar] [CrossRef]
- Heins, A.-L.; Johanson, T.; Han, S.; Lundin, L.; Carlquist, M.; Gernaey, K.V.; Sørensen, S.J.; Eliasson Lantz, A. Quantitative Flow Cytometry to Understand Population Heterogeneity in Response to Changes in Substrate Availability in Escherichia coli and Saccharomyces cerevisiae Chemostats. Front. Bioeng. Biotechnol. 2019, 7, 187. [Google Scholar] [CrossRef]
- Baert, J.; Delepierre, A.; Telek, S.; Fickers, P.; Toye, D.; Delamotte, A.; Lara, A.R.; Jaén, K.E.; Gosset, G.; Jensen, P.R.; et al. Microbial population heterogeneity versus bioreactor heterogeneity: Evaluation of Redox Sensor Green as an exogenous metabolic biosensor. Eng. Life Sci. 2016, 16, 643–651. [Google Scholar] [CrossRef]
- Liu, J.; François, J.-M.; Capp, J.-P. Use of noise in gene expression as an experimental parameter to test phenotypic effects. Yeast 2016, 33, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.D.; Marques, D.d.A.V.; Malpiedi, L.P.; Maranhão, A.Q.; Parra, D.A.S.; Converti, A.; Junior, A.P. Cultivation of Pichia pastoris carrying the scFv anti LDL (-) antibody fragment. Effect of preculture carbon source. Braz. J. Microbiol. 2017, 48, 419–426. [Google Scholar] [CrossRef] [PubMed]
- da Silva Delabona, P.; Lima, D.J.; Robl, D.; Rabelo, S.C.; Farinas, C.S.; da Cruz Pradella, J.G. Enhanced cellulase production by Trichoderma harzianum by cultivation on glycerol followed by induction on cellulosic substrates. J. Ind. Microbiol. Biotechnol. 2016, 43, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Domingues, F.C.; Queiroz, J.A.; Cabral, J.M.S.; Fonseca, L.P. The influence of culture conditions on mycelial structure and cellulase production by Trichoderma reesei Rut C-30. Enzyme Microb. Technol. 2000, 26, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Masson, F.; Hinrichsen, L.; Talon, R.; Montel, M.C. Factors influencing leucine catabolism by a strain of Staphylococcus carnosus. Int. J. Food Microbiol. 1999, 49, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P. Pre-induced Lac Operon Effect on Non Specific Sugars: Pre-culture Effect is Dependent on Strength of Induction, Exponential Phase and Substrate Concentration. Open Microbiol. J. 2015, 9, 8–13. [Google Scholar] [CrossRef]
- Yue, S.; Liu, Y.; Wang, X.; Xu, D.; Qiu, J.; Liu, Q.; Dong, Q. Modeling the Effects of the Preculture Temperature on the Lag Phase of Listeria monocytogenes at 25 °C. J. Food Prot. 2019, 82, 2100–2107. [Google Scholar] [CrossRef]
- Garcia, J.R.; Cha, H.J.; Rao, G.; Marten, M.R.; Bentley, W.E. Microbial nar-GFP cell sensors reveal oxygen limitations in highly agitated and aerated laboratory-scale fermentors. Microb. Cell Factories 2009, 8, 6. [Google Scholar] [CrossRef]
- Jin, Y.; Qin, S.J.; Huang, Q.; Saucedo, V.; Li, Z.; Meier, A.; Kundu, S.; Lehr, B.; Charaniya, S. Classification and Diagnosis of Bioprocess Cell Growth Productions Using Early-Stage Data. Ind. Eng. Chem. Res. 2019, 58, 13469–13480. [Google Scholar] [CrossRef]
- Patras, A.; Brunton, N.P.; Downey, G.; Rawson, A.; Warriner, K.; Gernigon, G. Application of principal component and hierarchical cluster analysis to classify fruits and vegetables commonly consumed in Ireland base on in vitro antioxidant activity. J. Food Compost. Anal. 2011, 24, 250–256. [Google Scholar] [CrossRef]
- Vigni, M.L.; Durante, C.; Cocchi, M. Exploratory Data Analysis. Data Handl. Sci. Technol. 2013, 28, 55–126. [Google Scholar] [CrossRef]
- Shlens, J. A Tutorial on Principal Component Analysis. arXiv 2014, arXiv:1404.1100. [Google Scholar]
- Skov, T.; Honoré, A.H.; Jensen, H.M.; Næs, T.; Engelsen, S.B. Chemometrics in foodomics: Handling data structures from multiple analytical platforms. Trends Analyt. Chem. 2014, 60, 71–79. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Riesenberg, D.; Schulz, V.; Knorre, W.A.; Pohl, H.-D.; Korz, D.; Sanders, E.A.; Roß, A.; Deckwer, W.-D. High cell density cultivation of Escherichia coli at controlled specific growth rate. J. Biotechnol. 1991, 20, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef]
- Satowa, D.; Fujiwara, R.; Uchio, S.; Nakano, M.; Otomo, C.; Hirata, Y.; Matsumoto, T.; Noda, S.; Tanaka, T.; Kondo, A. Metabolic engineering of E. coli for improving mevalonate production to promote NADPH regeneration and enhance acetyl-CoA supply. Biotechnol. Bioeng. 2020, 117, 2153–2164. [Google Scholar] [CrossRef]
- Shilling, P.J.; Mirzadeh, K.; Cumming, A.J.; Widesheim, M.; Köck, Z.; Daley, D.O. Improved designs for pET expression plasmids increase protein production yield in Escherichia coli. Commun. Biol. 2020, 3, 214. [Google Scholar] [CrossRef]
- Olsson, L.; Rugbjerg, P.; Torello Pianale, L.; Trivellin, C. Robustness: Linking strain design to viable bioprocesses. Trends Biotechnol. 2022, 40, 918–931. [Google Scholar] [CrossRef]
- Becker, L.; Sturm, J.; Eiden, F.; Holtmann, D. Analyzing and understanding the robustness of bioprocesses. Trends Biotechnol. 2023. [Google Scholar] [CrossRef]
- Heim, R.; Prasher, D.C.; Tsien, R.Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 1994, 91, 12501–12504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Patel, H.N.; Lappe, J.W.; Wachter, R.M. Reaction progress of Chromophore Biogenesis in Green Fluorescent Protein. J. Am. Chem. Soc. 2006, 128, 4766–4772. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.E.; Tamplin, M.L.; Phillips, J.G.; Marmer, B.S. Influence of agitation, inoculum density, pH, and strain on the growth parameters of Escherichia coli O157:H7—Relevance to risk assessment. Int. J. Food Microbiol. 2003, 83, 147–160. [Google Scholar] [CrossRef]
- Recorbet, G.; Steinberg, C.; Faurie, G. Survival in soil of genetically engineered Escherichia coli as related to inoculum density, predation and competition. FEMS Microbiol. Ecol. 1992, 101, 251–260. [Google Scholar] [CrossRef]
- Volpato, V.; Webber, C. Addressing variability in iPSC-derived models of human disease: Guidelines to promote reproducibility. Dis. Model. Mech. 2020, 13, dmm042317. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.S.L.; Bezerra, C.C.R.; Neto, R.M.; Morais, C.L.M.; Lima, K.M.G. Identification of resistance in Escherichia coli and Klebsiella pneumoniae using excitation-emission matrix fluorescence spectroscopy and multivariate analysis. Sci. Rep. 2020, 10, 12994. [Google Scholar] [CrossRef] [PubMed]
- Sastry, A.V.; Gao, Y.; Szubin, R.; Hefner, Y.; Xu, S.; Kim, D.; Choudhary, K.S.; Yang, L.; King, Z.A.; Palsson, B.O. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat. Commun. 2019, 10, 5536. [Google Scholar] [CrossRef]
- Ge, Y.; Sealfon, S.C. flowPeaks: A fast unsupervised clustering for flow cytometry data via K-means and density peak finding. Bioinformatics 2012, 28, 2052–2058. [Google Scholar] [CrossRef]
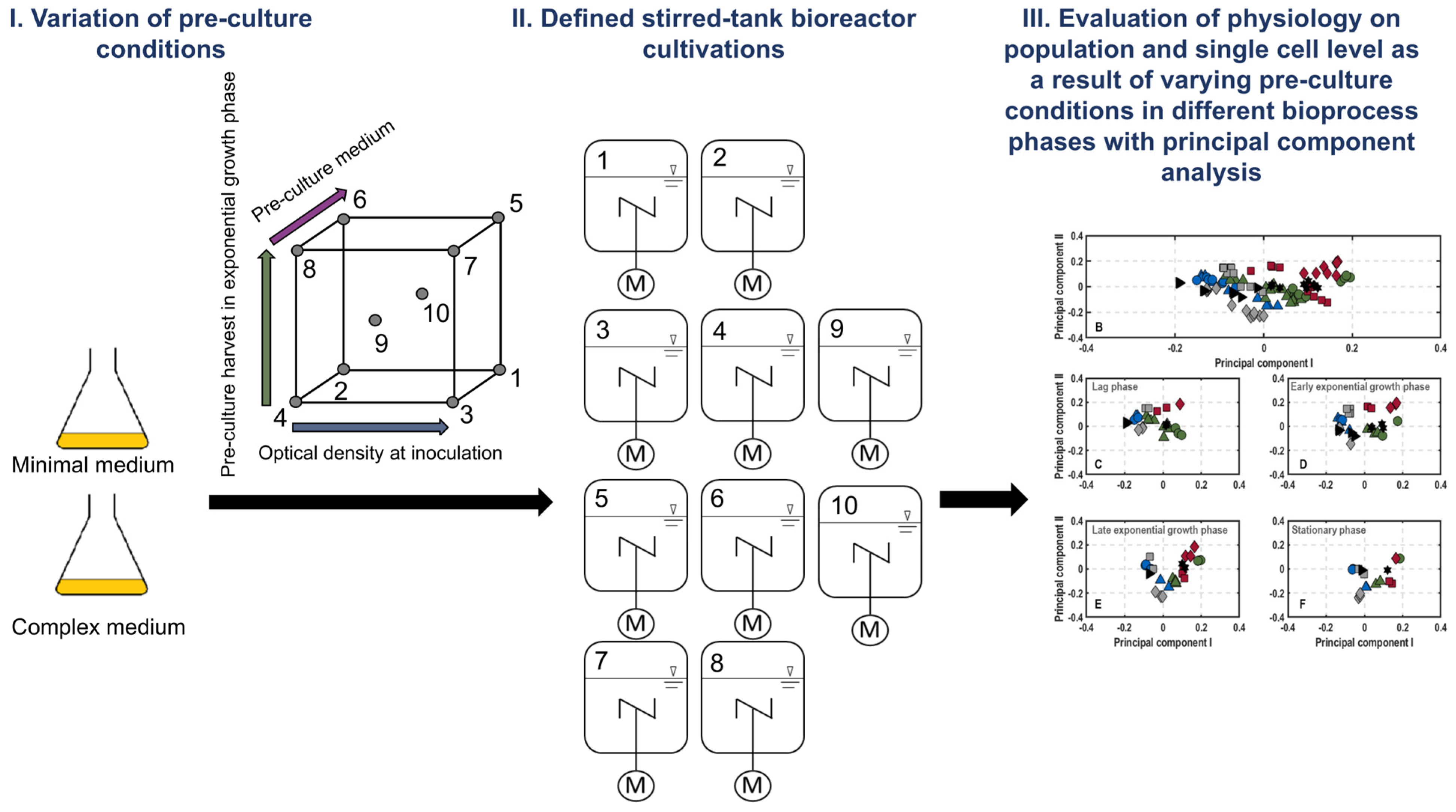

| Process | Inoculation OD600 | Pre-Culture Harvest in Exponential Growth Phase | Pre-Culture Medium |
|---|---|---|---|
| 1 | 0.02 | Early (6 h) | MM |
| 2 | 0.005 | Early (6 h) | MM |
| 3 | 0.02 | Early (3.5 h) | LB |
| 4 | 0.005 | Early (3.5 h) | LB |
| 5 | 0.02 | Late (9 h) | MM |
| 6 | 0.005 | Late (9 h) | MM |
| 7 | 0.02 | Late (7 h) | LB |
| 8 | 0.005 | Late (7 h) | LB |
| 9 | 0.0125 | Mid (5 h) | LB |
| 10 | 0.0125 | Mid (7 h) | MM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoang, M.D.; Riessner, S.; Oropeza Vargas, J.E.; von den Eichen, N.; Heins, A.-L. Influence of Varying Pre-Culture Conditions on the Level of Population Heterogeneity in Batch Cultures with an Escherichia coli Triple Reporter Strain. Microorganisms 2023, 11, 1763. https://doi.org/10.3390/microorganisms11071763
Hoang MD, Riessner S, Oropeza Vargas JE, von den Eichen N, Heins A-L. Influence of Varying Pre-Culture Conditions on the Level of Population Heterogeneity in Batch Cultures with an Escherichia coli Triple Reporter Strain. Microorganisms. 2023; 11(7):1763. https://doi.org/10.3390/microorganisms11071763
Chicago/Turabian StyleHoang, Manh Dat, Sophi Riessner, Jose Enrique Oropeza Vargas, Nikolas von den Eichen, and Anna-Lena Heins. 2023. "Influence of Varying Pre-Culture Conditions on the Level of Population Heterogeneity in Batch Cultures with an Escherichia coli Triple Reporter Strain" Microorganisms 11, no. 7: 1763. https://doi.org/10.3390/microorganisms11071763
APA StyleHoang, M. D., Riessner, S., Oropeza Vargas, J. E., von den Eichen, N., & Heins, A.-L. (2023). Influence of Varying Pre-Culture Conditions on the Level of Population Heterogeneity in Batch Cultures with an Escherichia coli Triple Reporter Strain. Microorganisms, 11(7), 1763. https://doi.org/10.3390/microorganisms11071763







