Role of the Solute-Binding Protein CuaD in the Signaling and Regulating Pathway of Cellobiose and Cellulose Utilization in Ruminiclostridium cellulolyticum
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media Vectors
2.2. Transcriptional Analysis
2.3. Cloning of the Gene Encoding Truncated CuaSEC in E. coli
2.4. Production and Purification of the Recombinant Protein
2.5. Complementation of MTLcuaD Strain
2.6. Preparation and Western Blot Analysis of Membrane Proteins from R. cellulolyticum
2.7. Isothermal Titration Calorimetry
2.8. Bacterial Two-Hybrid Assays
2.9. Model Building and Docking Experiments
3. Results
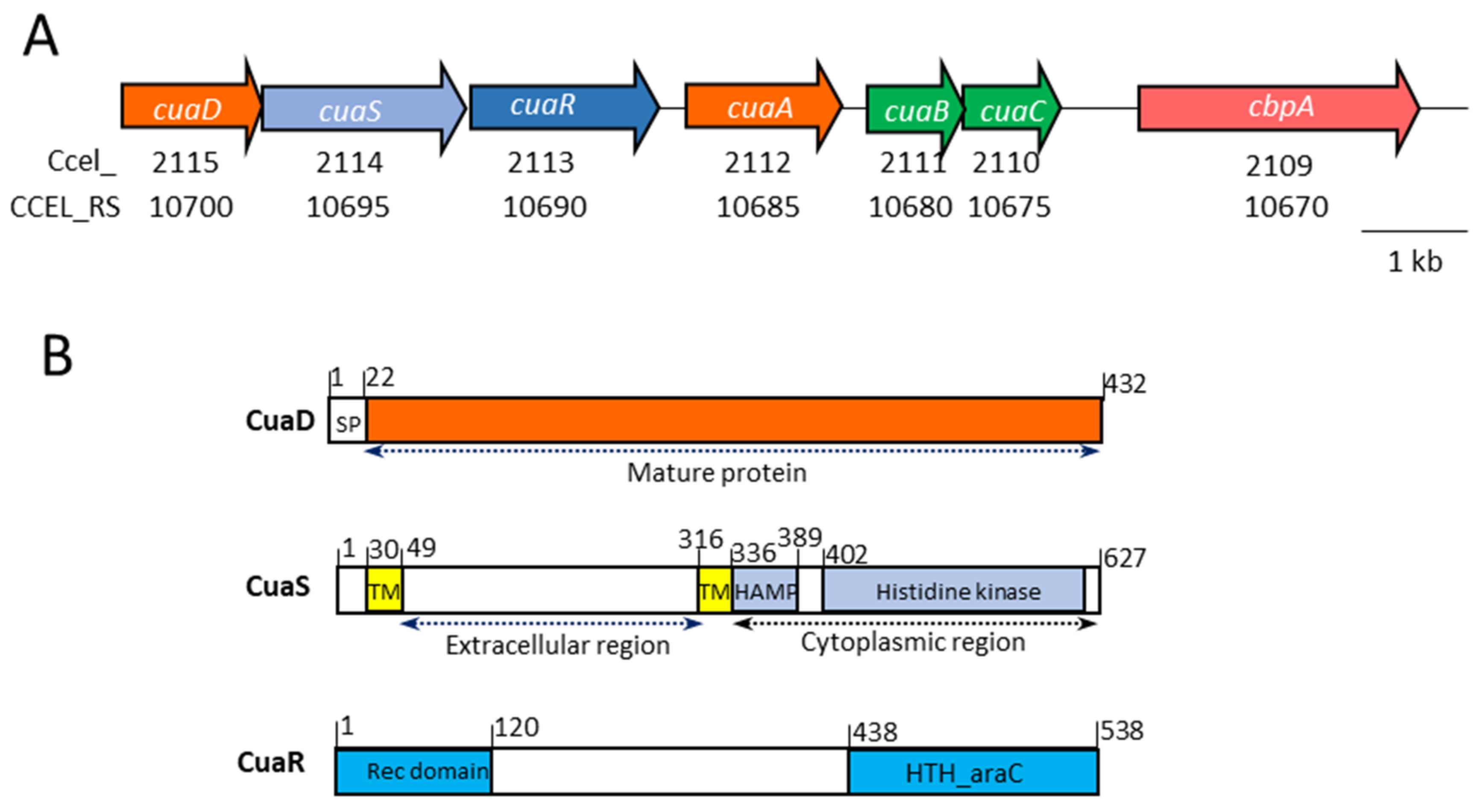
3.1. Proteins of the Putative Three-Component System
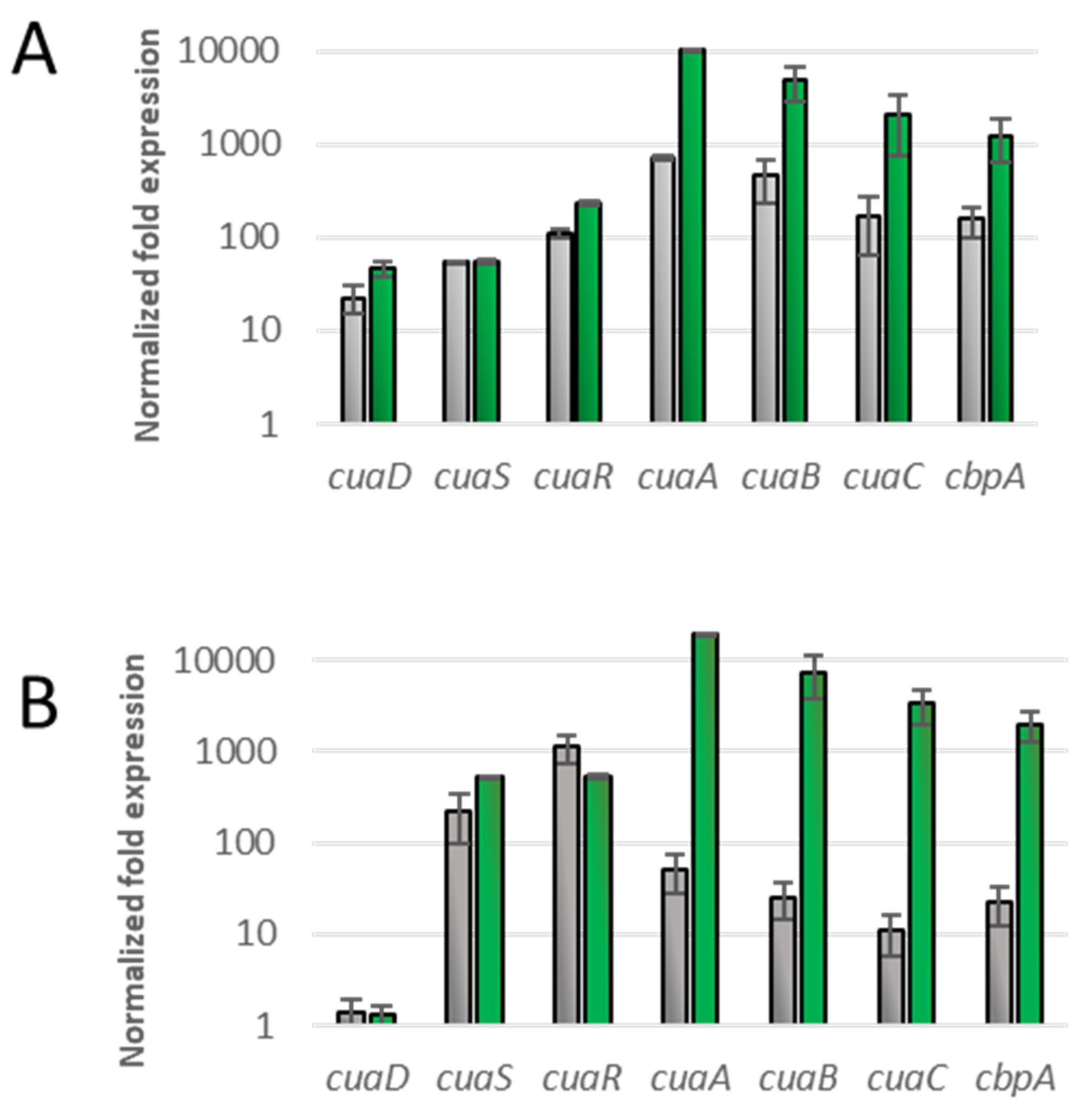
3.2. Overproduction of CuaR in cuaD Mutant Strain
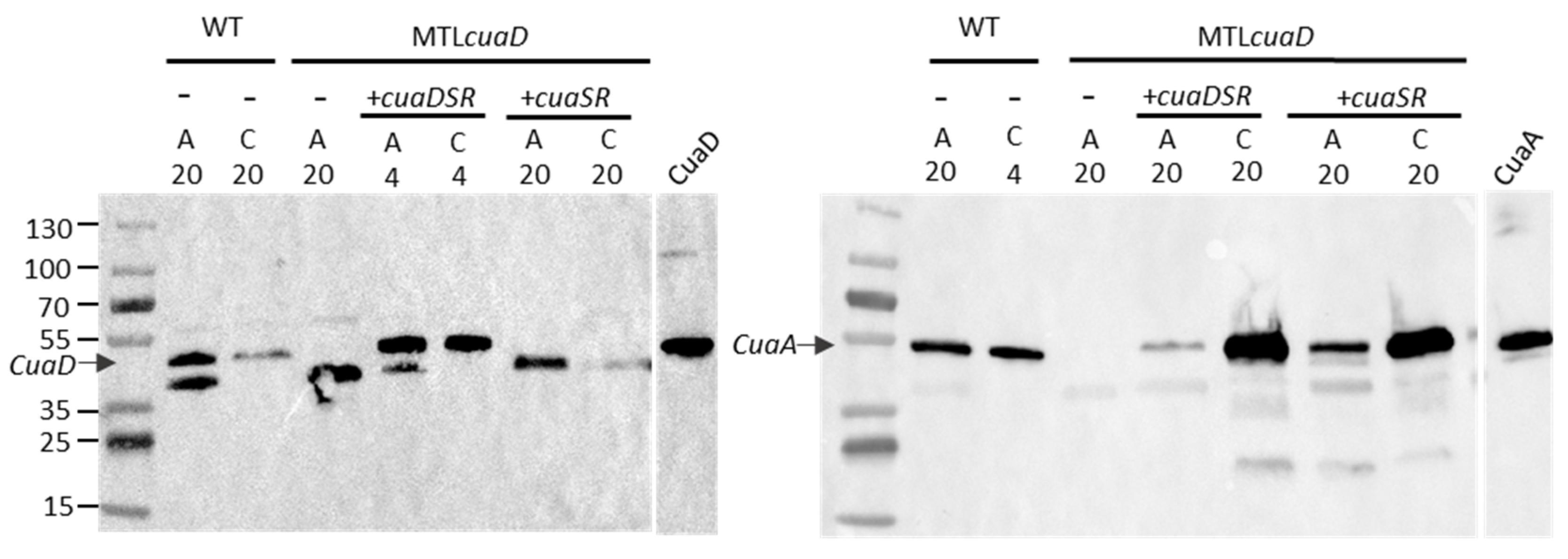
3.3. Role of CuaD in the Signaling System
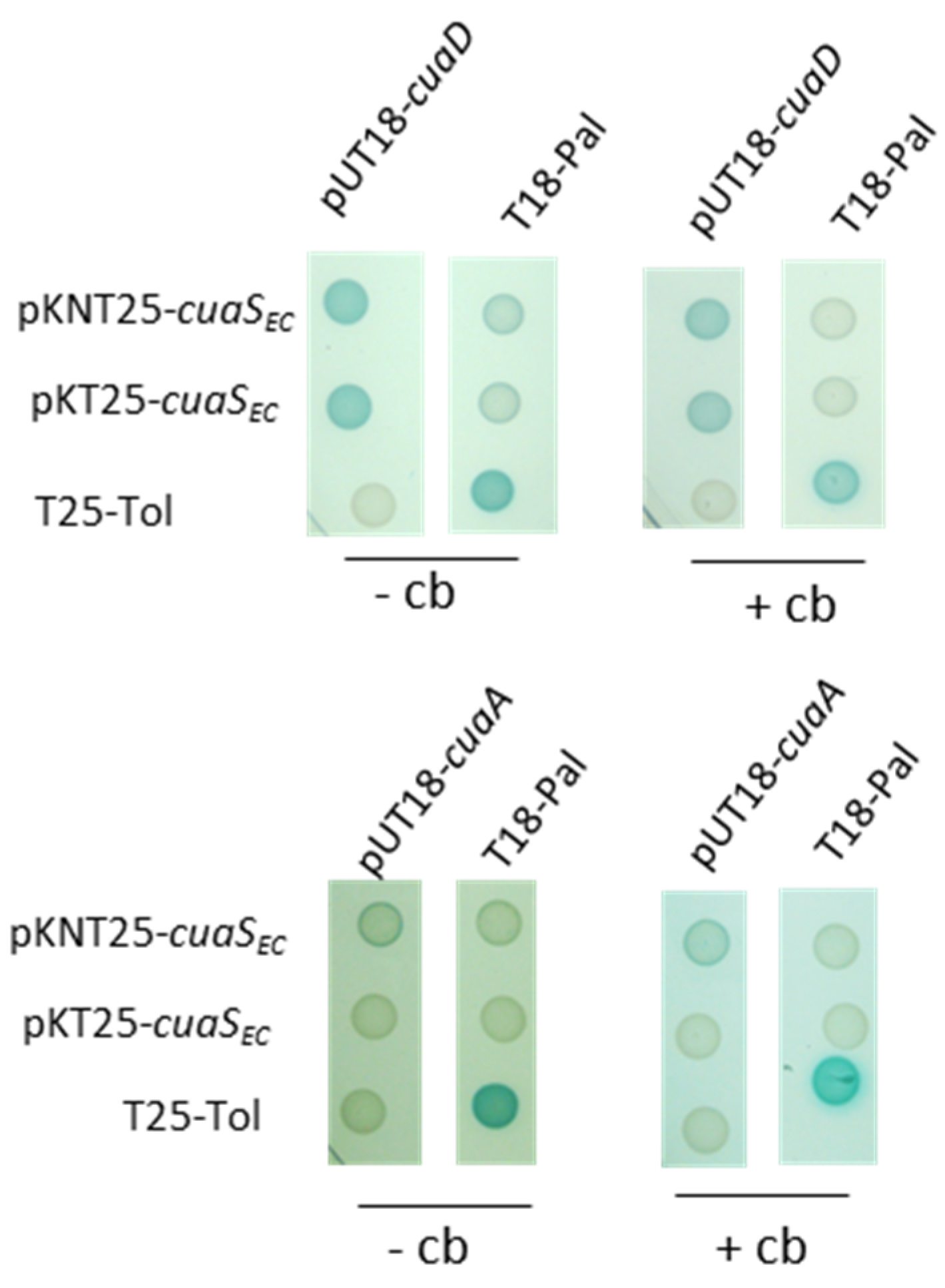
3.4. Binding of SBPs with CuaS
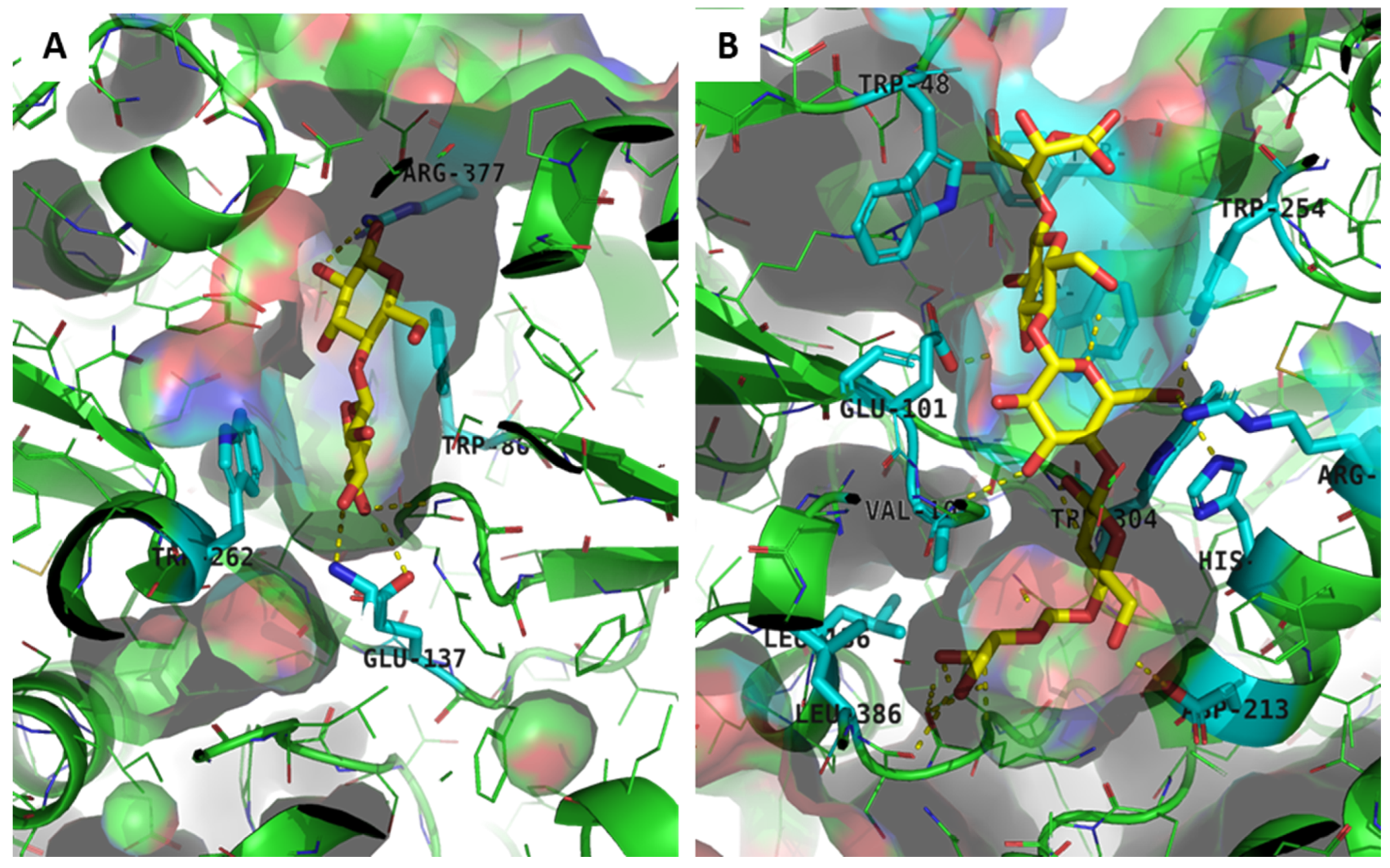
3.5. Model of the Quaternary Structure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Artzi, L.; Bayer, E.A.; Moraïs, S. Cellulosomes: Bacterial Nanomachines for Dismantling Plant Polysaccharides. Nat. Rev. Microbiol. 2017, 15, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Dong, S.; Liu, Y.-J.; Yao, X.; Chen, C.; Xiao, Y.; Bayer, E.A.; Shoham, Y.; You, C.; Cui, Q.; et al. Deciphering Cellodextrin and Glucose Uptake in Clostridium thermocellum. mBio 2022, 13, e01476-22. [Google Scholar] [CrossRef] [PubMed]
- Nataf, Y.; Yaron, S.; Stahl, F.; Lamed, R.; Bayer, E.A.; Scheper, T.-H.; Sonenshein, A.L.; Shoham, Y. Cellodextrin and Laminaribiose ABC Transporters in Clostridium thermocellum. J. Bacteriol. 2009, 191, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Fosses, A.; Maté, M.; Franche, N.; Liu, N.; Denis, Y.; Borne, R.; de Philip, P.; Fierobe, H.-P.; Perret, S. A Seven-Gene Cluster in Ruminiclostridium cellulolyticum Is Essential for Signalization, Uptake and Catabolism of the Degradation Products of Cellulose Hydrolysis. Biotechnol. Biofuels 2017, 10, 250. [Google Scholar] [CrossRef]
- Blouzard, J.-C.; Coutinho, P.M.; Fierobe, H.-P.; Henrissat, B.; Lignon, S.; Tardif, C.; Pagès, S.; de Philip, P. Modulation of Cellulosome Composition in Clostridium cellulolyticum: Adaptation to the Polysaccharide Environment Revealed by Proteomic and Carbohydrate-Active Enzyme Analyses. Proteomics 2010, 10, 541–554. [Google Scholar] [CrossRef]
- Liu, N.; Fosses, A.; Kampik, C.; Parsiegla, G.; Denis, Y.; Vita, N.; Fierobe, H.-P.; Perret, S. In Vitro and in Vivo Exploration of the Cellobiose and Cellodextrin Phosphorylases Panel in Ruminiclostridium cellulolyticum: Implication for Cellulose Catabolism. Biotechnol. Biofuels 2019, 12, 208. [Google Scholar] [CrossRef]
- Liu, N.; Gagnot, S.; Denis, Y.; Byrne, D.; Faulds, C.; Fierobe, H.-P.; Perret, S. Selfish Uptake versus Extracellular Arabinoxylan Degradation in the Primary Degrader Ruminiclostridium cellulolyticum, a New String to Its Bow. Biotechnol. Biofuels 2022, 15, 127. [Google Scholar] [CrossRef]
- Ravachol, J.; de Philip, P.; Borne, R.; Mansuelle, P.; Maté, M.J.; Perret, S.; Fierobe, H.-P. Mechanisms Involved in Xyloglucan Catabolism by the Cellulosome-Producing Bacterium Ruminiclostridium cellulolyticum. Sci. Rep. 2016, 6, 22770. [Google Scholar] [CrossRef]
- Matilla, M.A.; Ortega, Á.; Krell, T. The Role of Solute Binding Proteins in Signal Transduction. Comput. Struct. Biotechnol. J. 2021, 19, 1786–1805. [Google Scholar] [CrossRef]
- Sun, Z.; Chen, Y.; Yang, C.; Yang, S.; Gu, Y.; Jiang, W. A Novel Three-Component System-Based Regulatory Model for d-Xylose Sensing and Transport in Clostridium beijerinckii: A Novel d-Xylose Sensing and Transport Model. Mol. Microbiol. 2015, 95, 576–589. [Google Scholar] [CrossRef]
- Li, X.; Wang, L.-X.; Wang, X.; Roseman, S. The Chitin Catabolic Cascade in the Marine Bacterium Vibrio cholerae: Characterization of a Unique Chitin Oligosaccharide Deacetylase. Glycobiology 2007, 17, 1377–1387. [Google Scholar] [CrossRef]
- Baraquet, C.; Théraulaz, L.; Guiral, M.; Lafitte, D.; Méjean, V.; Jourlin-Castelli, C. TorT, a Member of a New Periplasmic Binding Protein Family, Triggers Induction of the Tor Respiratory System upon Trimethylamine N-Oxide Electron-Acceptor Binding in Escherichia coli. J. Biol. Chem. 2006, 281, 38189–38199. [Google Scholar] [CrossRef] [PubMed]
- Giallo, J.; Gaudin, C.; Belaich, J.P.; Petitdemange, E.; Caillet-Mangin, F. Metabolism of Glucose and Cellobiose by Cellulolytic Mesophilic Clostridium sp. Strain H10. Appl. Environ. Microbiol. 1983, 45, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Ravachol, J.; Borne, R.; Meynial-Salles, I.; Soucaille, P.; Pagès, S.; Tardif, C.; Fierobe, H.-P. Combining Free and Aggregated Cellulolytic Systems in the Cellulosome-Producing Bacterium Ruminiclostridium cellulolyticum. Biotechnol. Biofuels 2015, 8, 114. [Google Scholar] [CrossRef]
- Ferdinand, P.-H.; Borne, R.; Trotter, V.; Pagès, S.; Tardif, C.; Fierobe, H.-P.; Perret, S. Are Cellulosome Scaffolding Protein CipC and CBM3-Containing Protein HycP, Involved in Adherence of Clostridium cellulolyticum to Cellulose? PLoS ONE 2013, 8, e69360. [Google Scholar] [CrossRef] [PubMed]
- Jennert, K.C.B.; Tardif, C.; Young, D.I.; Young, M. Gene Transfer to Clostridium cellulolyticum ATCC 35319. Microbiology 2000, 146, 3071–3080. [Google Scholar] [CrossRef]
- Tardif, C.; Maamar, H.; Balfin, M.; Belaich, J. Electrotransformation Studies in Clostridium cellulolyticum. J. Ind. Microbiol. Biotechnol. 2001, 27, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Battesti, A.; Bouveret, E. The Bacterial Two-Hybrid System Based on Adenylate Cyclase Reconstitution in Escherichia coli. Methods 2012, 58, 325–334. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Mirdita, M.; Steinegger, M.; Söding, J. MMseqs2 Desktop and Local Web Server App for Fast, Interactive Sequence Searches. Bioinformatics 2019, 35, 2856–2858. [Google Scholar] [CrossRef] [PubMed]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons Learned in Empirical Scoring with Smina from the CSAR 2011 Benchmarking Exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 32, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and Sequence Analysis Tools Services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Hertig, C.; Li, R.Y.; Louarn, A.M.; Garnerone, A.M.; David, M.; Batut, J.; Kahn, D.; Boistard, P. Rhizobium meliloti Regulatory Gene FixJ Activates Transcription of R. meliloti NifA and FixK Genes in Escherichia coli. J. Bacteriol. 1989, 171, 1736–1738. [Google Scholar] [CrossRef]
- Li, J.; Wang, C.; Yang, G.; Sun, Z.; Guo, H.; Shao, K.; Gu, Y.; Jiang, W.; Zhang, P. Molecular Mechanism of Environmental d-Xylose Perception by a XylFII-LytS Complex in Bacteria. Proc. Natl. Acad. Sci. USA 2017, 114, 8235–8240. [Google Scholar] [CrossRef]
- Berntsson, R.P.-A.; Smits, S.H.J.; Schmitt, L.; Slotboom, D.-J.; Poolman, B. A Structural Classification of Substrate-Binding Proteins. FEBS Lett. 2010, 584, 2606–2617. [Google Scholar] [CrossRef]
- Scheepers, G.H.; Lycklama A Nijeholt, J.A.; Poolman, B. An Updated Structural Classification of Substrate-Binding Proteins. FEBS Lett. 2016, 590, 4393–4401. [Google Scholar] [CrossRef]
- Han, Y.; Agarwal, V.; Dodd, D.; Kim, J.; Bae, B.; Mackie, R.I.; Nair, S.K.; Cann, I.K.O. Biochemical and Structural Insights into Xylan Utilization by the Thermophilic Bacterium Caldanaerobius polysaccharolyticus. J. Biol. Chem. 2012, 287, 34946–34960. [Google Scholar] [CrossRef]
- Shulami, S.; Raz-Pasteur, A.; Tabachnikov, O.; Gilead-Gropper, S.; Shner, I.; Shoham, Y. The l -Arabinan Utilization System of Geobacillus stearothermophilus. J. Bacteriol. 2011, 193, 2838–2850. [Google Scholar] [CrossRef] [PubMed]
- Antoine, R.; Huvent, I.; Chemlal, K.; Deray, I.; Raze, D.; Locht, C.; Jacob-Dubuisson, F. The Periplasmic Binding Protein of a Tripartite Tricarboxylate Transporter Is Involved in Signal Transduction. J. Mol. Biol. 2005, 351, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Klancher, C.A.; Yamamoto, S.; Dalia, T.N.; Dalia, A.B. ChiS Is a Noncanonical DNA-Binding Hybrid Sensor Kinase that Directly Regulates the Chitin Utilization Program in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2020, 117, 20180–20189. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fosses, A.; Franche, N.; Parsiegla, G.; Denis, Y.; Maté, M.; de Philip, P.; Fierobe, H.-P.; Perret, S. Role of the Solute-Binding Protein CuaD in the Signaling and Regulating Pathway of Cellobiose and Cellulose Utilization in Ruminiclostridium cellulolyticum. Microorganisms 2023, 11, 1732. https://doi.org/10.3390/microorganisms11071732
Fosses A, Franche N, Parsiegla G, Denis Y, Maté M, de Philip P, Fierobe H-P, Perret S. Role of the Solute-Binding Protein CuaD in the Signaling and Regulating Pathway of Cellobiose and Cellulose Utilization in Ruminiclostridium cellulolyticum. Microorganisms. 2023; 11(7):1732. https://doi.org/10.3390/microorganisms11071732
Chicago/Turabian StyleFosses, Aurélie, Nathalie Franche, Goetz Parsiegla, Yann Denis, Maria Maté, Pascale de Philip, Henri-Pierre Fierobe, and Stéphanie Perret. 2023. "Role of the Solute-Binding Protein CuaD in the Signaling and Regulating Pathway of Cellobiose and Cellulose Utilization in Ruminiclostridium cellulolyticum" Microorganisms 11, no. 7: 1732. https://doi.org/10.3390/microorganisms11071732
APA StyleFosses, A., Franche, N., Parsiegla, G., Denis, Y., Maté, M., de Philip, P., Fierobe, H.-P., & Perret, S. (2023). Role of the Solute-Binding Protein CuaD in the Signaling and Regulating Pathway of Cellobiose and Cellulose Utilization in Ruminiclostridium cellulolyticum. Microorganisms, 11(7), 1732. https://doi.org/10.3390/microorganisms11071732





