Clinical and Genomic Characterization of Carbapenem-Resistant Klebsiella pneumoniae with Concurrent Production of NDM and OXA-48-like Carbapenemases in Southern California, 2016–2022
Abstract
1. Introduction
2. Material and Methods
2.1. Identification of CRKP Isolates with NDM and OXA-48-like Carbapenemase Co-Production
2.2. Whole-Genome Sequencing and Genomic Analysis
2.3. Chart Review and Research Ethics
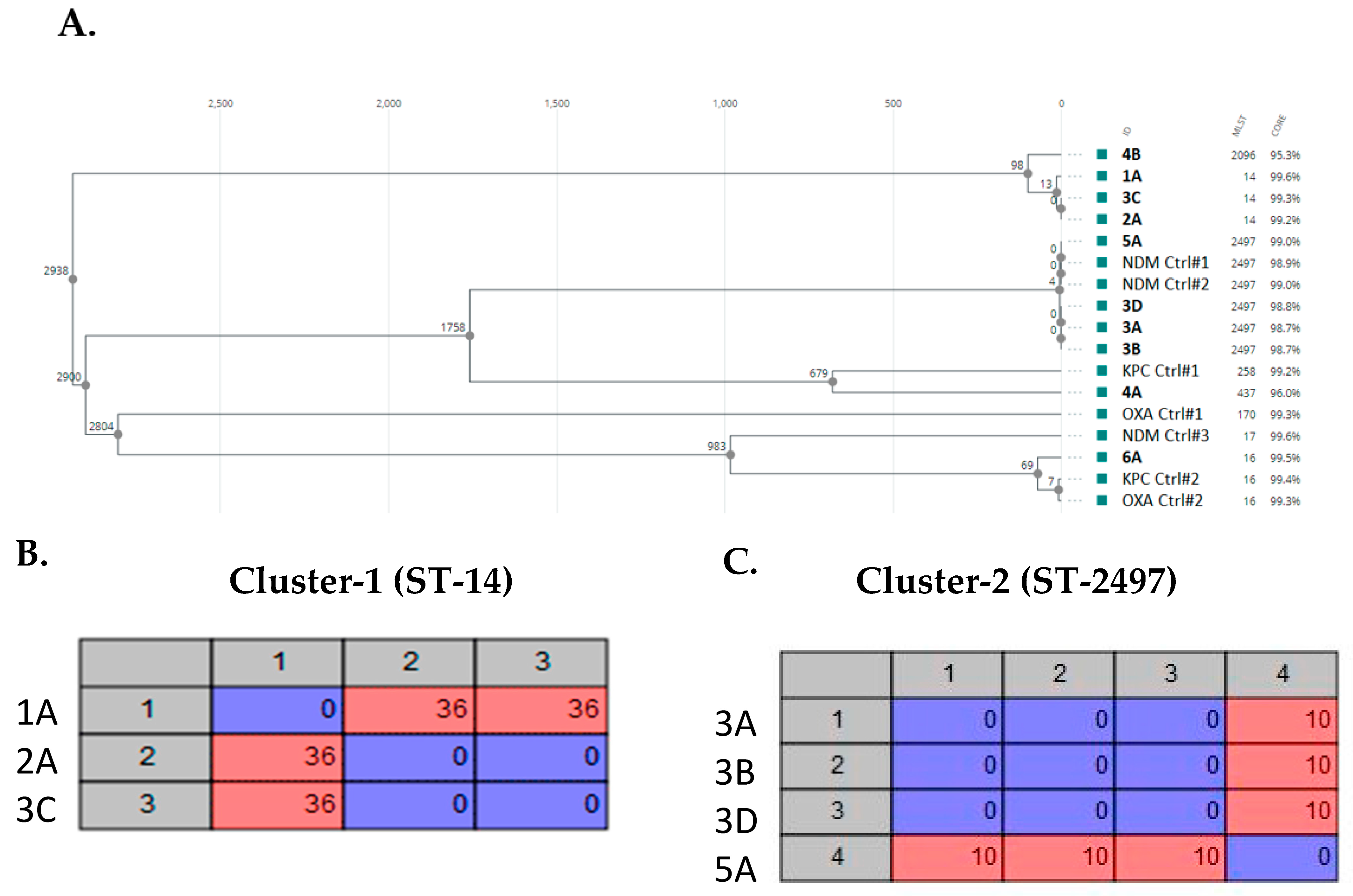
3. Results
3.1. Clinical Characteristics
3.2. Case 1 (2016)
3.3. Case 2 (2017)
3.4. Case 3 (2018)
3.5. Case 4 (2019)
3.6. Case 5 (2020)
3.7. Case 6 (2022)
3.8. Genomic Characteristics
3.9. AMR Profiles
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sheu, C.C.; Chang, Y.T.; Lin, S.Y.; Chen, Y.H.; Hsueh, P.R. Infections Caused by Carbapenem-Resistant Enterobacteriaceae: An Update on Therapeutic Options. Front. Microbiol. 2019, 10, 80. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The Changing Face of the Family Enterobacteriaceae (Order: “Enterobacterales”): New Members, Taxonomic Issues, Geographic Expansion, and New Diseases and Disease Syndromes. Clin. Microbiol. Rev. 2021, 34, e00174-20. [Google Scholar] [CrossRef]
- Novosad, S.A.; Fike, L.; Dudeck, M.A.; Allen-Bridson, K.; Edwards, J.R.; Edens, C.; Sinkowitz-Cochran, R.; Powell, K.; Kuhar, D. Pathogens causing central-line-associated bloodstream infections in acute-care hospitals-United States, 2011–2017. Infect. Control Hosp. Epidemiol. 2020, 41, 313–319. [Google Scholar] [CrossRef]
- Kang, J.; Sickbert-Bennett, E.E.; Brown, V.M.; Weber, D.J.; Rutala, W.A. Relative frequency of health care-associated pathogens by infection site at a university hospital from 1980 to 2008. Am. J. Infect. Control 2012, 40, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Limbago, B.M.; Patel, J.B.; Kallen, A.J. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 2011, 53, 60–67. [Google Scholar] [CrossRef]
- Bowers, D.R.; Huang, V. Emerging Issues and Treatment Strategies in Carbapenem-Resistant Enterobacteriaceae (CRE). Curr. Infect. Dis. Rep. 2016, 18, 48. [Google Scholar] [CrossRef] [PubMed]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, R.G.; Johnson, J.K.; Bork, J.T.; Heil, E.L. Treatment options for extended-spectrum beta-lactamase (ESBL) and AmpC-producing bacteria. Expert Opin. Pharmacother. 2016, 17, 953–967. [Google Scholar] [CrossRef]
- Perez, F.; Van Duin, D. Carbapenem-resistant Enterobacteriaceae: A menace to our most vulnerable patients. Cleve. Clin. J. Med. 2013, 80, 225–233. [Google Scholar] [CrossRef]
- Suay-Garcia, B.; Perez-Gracia, M.T. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics 2019, 8, 122. [Google Scholar] [CrossRef]
- Potter, R.F.; D’Souza, A.W.; Dantas, G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updat. 2016, 29, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular diversity of extended-spectrum beta-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 2020, 8, 13. [Google Scholar] [CrossRef]
- Porreca, A.M.; Sullivan, K.V.; Gallagher, J.C. The Epidemiology, Evolution, and Treatment of KPC-Producing Organisms. Curr. Infect. Dis. Rep. 2018, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.; Nordmann, P.; Poirel, L. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Peirano, G.; Kock, M.M.; Strydom, K.A.; Matsumura, Y. The Global Ascendency of OXA-48-Type Carbapenemases. Clin. Microbiol. Rev. 2019, 33, e00102-19. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe—Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-NET) 2017; ECDC: Stockholm, Sweden, 2018.
- Durante-Mangoni, E.; Andini, R.; Zampino, R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. 2019, 25, 943–950. [Google Scholar] [CrossRef]
- Shields, R.K.; Potoski, B.A.; Haidar, G.; Hao, B.; Doi, Y.; Chen, L.; Press, E.G.; Kreiswirth, B.N.; Clancy, C.J.; Nguyen, M.H. Clinical Outcomes, Drug Toxicity, and Emergence of Ceftazidime-Avibactam Resistance Among Patients Treated for Carbapenem-Resistant Enterobacteriaceae Infections. Clin. Infect. Dis. 2016, 63, 1615–1618. [Google Scholar] [CrossRef]
- King, M.; Heil, E.; Kuriakose, S.; Bias, T.; Huang, V.; El-Beyrouty, C.; McCoy, D.; Hiles, J.; Richards, L.; Gardner, J.; et al. Multicenter Study of Outcomes with Ceftazidime-Avibactam in Patients with Carbapenem-Resistant Enterobacteriaceae Infections. Antimicrob. Agents Chemother. 2017, 61, e00449-17. [Google Scholar] [CrossRef]
- Tumbarello, M.; Trecarichi, E.M.; Corona, A.; De Rosa, F.G.; Bassetti, M.; Mussini, C.; Menichetti, F.; Viscoli, C.; Campoli, C.; Venditti, M.; et al. Efficacy of Ceftazidime-Avibactam Salvage Therapy in Patients with Infections Caused by Klebsiella pneumoniae Carbapenemase-producing K. pneumoniae. Clin. Infect. Dis. 2019, 68, 355–364. [Google Scholar] [CrossRef]
- Rasheed, J.K.; Kitchel, B.; Zhu, W.; Anderson, K.F.; Clark, N.C.; Ferraro, M.J.; Savard, P.; Humphries, R.M.; Kallen, A.J.; Limbago, B.M. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerg. Infect. Dis. 2013, 19, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Hazen, K.C.; Carroll, J.; Yeh, A.J.; Cox, H.L.; Bonomo, R.A.; Sifri, C.D. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: The “menace” arrives in the new world. J. Clin. Microbiol. 2013, 51, 680–683. [Google Scholar] [CrossRef]
- Moubareck, C.A.; Mouftah, S.F.; Pal, T.; Ghazawi, A.; Halat, D.H.; Nabi, A.; AlSharhan, M.A.; AlDeesi, Z.O.; Peters, C.C.; Celiloglu, H.; et al. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int. J. Antimicrob. Agents 2018, 52, 90–95. [Google Scholar] [CrossRef]
- Alhazmi, W.; Al-Jabri, A.; Al-Zahrani, I. The Molecular Characterization of Nosocomial Carbapenem-Resistant Klebsiella pneumoniae Co-Harboring blaNDM and blaOXA-48 in Jeddah. Microbiol. Res. 2022, 13, 753–764. [Google Scholar] [CrossRef]
- Smith, H.Z.; Kendall, B. Carbapenem Resistant Enterobacteriaceae. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Isler, B.; Aslan, A.T.; Akova, M.; Harris, P.; Paterson, D.L. Treatment strategies for OXA-48-like and NDM producing Klebsiella pneumoniae infections. Expert Rev. Anti-Infect. Ther. 2022, 20, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Dyrkell, F.; Giske, C.G.; Fang, H. Epidemiological typing of ST80 vancomycin-resistant Enterococcus faecium: Core genome multilocus sequence typing versus single nucleotide polymorphism-based typing. J. Glob. Antimicrob. Resist. 2021, 25, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, S.A.; Jeraldo, P.R.; Schuetz, A.N.; Heitman, A.A.; Patel, R. Staphylococcus aureus whole genome sequence-based susceptibility and resistance prediction using a clinically amenable workflow. Diagn. Microbiol. Infect. Dis. 2020, 97, 115060. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Hammoudi Halat, D.; Ayoub Moubareck, C. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Boyd, S.E.; Holmes, A.; Peck, R.; Livermore, D.M.; Hope, W. OXA-48-Like beta-Lactamases: Global Epidemiology, Treatment Options, and Development Pipeline. Antimicrob. Agents Chemother. 2022, 66, e0021622. [Google Scholar] [CrossRef] [PubMed]
- Lorenzin, G.; Gona, F.; Battaglia, S.; Spitaleri, A.; Saluzzo, F.; Trovato, A.; di Marco, F.; Cichero, P.; Biancardi, A.; Nizzero, P.; et al. Detection of NDM-1/5 and OXA-48 co-producing extensively drug-resistant hypervirulent Klebsiella pneumoniae in Northern Italy. J. Glob. Antimicrob. Resist. 2022, 28, 146–150. [Google Scholar] [CrossRef]
- Duman, Y.; Ersoy, Y.; Gursoy, N.C.; Altunisik Toplu, S.; Otlu, B. A silent outbreak due to Klebsiella pneumoniae that co-produced NDM-1 and OXA-48 carbapenemases, and infection control measures. Iran. J. Basic. Med. Sci. 2020, 23, 46–50. [Google Scholar] [CrossRef]
- Abe, R.; Akeda, Y.; Takeuchi, D.; Sakamoto, N.; Sugawara, Y.; Yamamoto, N.; Kerdsin, A.; Matsumoto, Y.; Motooka, D.; Leolerd, W.; et al. Clonal dissemination of carbapenem-resistant Klebsiella pneumoniae ST16 co-producing NDM-1 and OXA-232 in Thailand. JAC Antimicrob. Resist. 2022, 4, dlac084. [Google Scholar] [CrossRef] [PubMed]
- Gondal, A.J.; Saleem, S.; Jahan, S.; Choudhry, N.; Yasmin, N. Novel Carbapenem-Resistant Klebsiella pneumoniae ST147 Coharboring bla (NDM-1), bla (OXA-48) and Extended-Spectrum beta-Lactamases from Pakistan. Infect. Drug Resist. 2020, 13, 2105–2115. [Google Scholar] [CrossRef] [PubMed]
- Pirs, M.; Cerar Kisek, T.; Krizan Hergouth, V.; Seme, K.; Mueller Premru, M.; Jeverica, S.; Logar, M.; Mrvic, T.; Znidarsic, B.; Jordan Markocic, O.; et al. Successful control of the first OXA-48 and/or NDM carbapenemase-producing Klebsiella pneumoniae outbreak in Slovenia 2014–2016. J. Hosp. Infect. 2019, 101, 142–149. [Google Scholar] [CrossRef]
- Contreras, D.A.; Fitzwater, S.P.; Nanayakkara, D.D.; Schaenman, J.; Aldrovandi, G.M.; Garner, O.B.; Yang, S. Coinfections of Two Strains of NDM-1- and OXA-232-Coproducing Klebsiella pneumoniae in a Kidney Transplant Patient. Antimicrob. Agents Chemother. 2020, 64, e00948-19. [Google Scholar] [CrossRef]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America 2022 Guidance on the Treatment of Extended-Spectrum beta-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2022, 75, 187–212. [Google Scholar] [CrossRef]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America antimicrobial-resistant treatment guidance: Gram-negative bacterial infections. Infect. Dis. Soc. Am. 2022. [Google Scholar] [CrossRef]
- Al-Marzooq, F.; Ngeow, Y.F.; Tay, S.T. Emergence of Klebsiella pneumoniae producing dual carbapenemases (NDM-1 and OXA-232) and 16S rRNA methylase (armA) isolated from a Malaysian patient returning from India. Int. J. Antimicrob. Agents 2015, 45, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; O’Hara, J.A.; Lando, J.F.; Querry, A.M.; Townsend, B.M.; Pasculle, A.W.; Muto, C.A. Co-production of NDM-1 and OXA-232 by Klebsiella pneumoniae. Emerg. Infect. Dis. 2014, 20, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Jung, Y.H.; Lee, S.; Yun, M.R.; Kim, W.; Kim, D.W. Comparative genomic analysis of Klebsiella pneumoniae subsp. pneumoniae KP617 and PittNDM01, NUHL24835, and ATCC BAA-2146 reveals unique evolutionary history of this strain. Gut. Pathog. 2016, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Baek, J.Y.; Kim, S.Y.; Jo, H.; Kang, K.; Ko, J.H.; Cho, S.Y.; Chung, D.R.; Peck, K.R.; Song, J.H.; et al. Comparison of virulence between matt and mucoid colonies of Klebsiella pneumoniae coproducing NDM-1 and OXA-232 isolated from a single patient. J. Microbiol. 2018, 56, 665–672. [Google Scholar] [CrossRef]
- Al-Baloushi, A.E.; Pal, T.; Ghazawi, A.; Sonnevend, A. Genetic support of carbapenemases in double carbapenemase producer Klebsiella pneumoniae isolated in the Arabian Peninsula. Acta Microbiol. Immunol. Hung. 2018, 65, 135–150. [Google Scholar] [CrossRef]
- Mouftah, S.F.; Pal, T.; Higgins, P.G.; Ghazawi, A.; Idaghdour, Y.; Alqahtani, M.; Omrani, A.S.; Rizvi, T.A.; Sonnevend, A. Diversity of carbapenem-resistant Klebsiella pneumoniae ST14 and emergence of a subgroup with KL64 capsular locus in the Arabian Peninsula. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 1–9. [Google Scholar] [CrossRef]
- Zhang, F.; Li, Z.; Liu, X.; Luo, G.; Wu, Y.; Li, C.; Zhao, J.; Zhang, Y.; Hu, Y.; Lu, B. Molecular Characteristics of an NDM-4 and OXA-181 Co-Producing K51-ST16 Carbapenem-Resistant Klebsiella pneumoniae: Study of Its Potential Dissemination Mediated by Conjugative Plasmids and Insertion Sequences. Antimicrob. Agents Chemother. 2023, 67, e01354-22. [Google Scholar] [CrossRef]
- Emeraud, C.; Birer, A.; Girlich, D.; Jousset, A.B.; Creton, E.; Naas, T.; Bonnin, R.A.; Dortet, L. Polyclonal Dissemination of OXA-232 Carbapenemase-Producing Klebsiella pneumoniae, France, 2013–2021. Emerg. Infect. Dis. 2022, 28, 2304–2307. [Google Scholar] [CrossRef]
- Simner, P.J.; Antar, A.A.R.; Hao, S.; Gurtowski, J.; Tamma, P.D.; Rock, C.; Opene, B.N.A.; Tekle, T.; Carroll, K.C.; Schatz, M.C.; et al. Antibiotic pressure on the acquisition and loss of antibiotic resistance genes in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1796–1803. [Google Scholar] [CrossRef]
- Hishinuma, A.; Yoshida, A.; Suzuki, H.; Okuzumi, K.; Ishida, T. Complete sequencing of an IncFII NDM-1 plasmid in Klebsiella pneumoniae shows structural features shared with other multidrug resistance plasmids. J. Antimicrob. Chemother. 2013, 68, 2415–2417. [Google Scholar] [CrossRef]
- Rojas, L.J.; Hujer, A.M.; Rudin, S.D.; Wright, M.S.; Domitrovic, T.N.; Marshall, S.H.; Hujer, K.M.; Richter, S.S.; Cober, E.; Perez, F.; et al. NDM-5 and OXA-181 Beta-Lactamases, a Significant Threat Continues To Spread in the Americas. Antimicrob. Agents Chemother. 2017, 61, e00454-17. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum beta-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Wachino, J.I.; Arakawa, Y. Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. N. Am. 2016, 30, 523–537. [Google Scholar] [CrossRef]
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | |
|---|---|---|---|---|---|---|
| Year | 2016 | 2017 | 2018 | 2019 | 2020 | 2022 |
| Age (years) | 74 | 74 | 68 | 89 | 64 | 73 |
| Clinical impression | Urinary tract infection | Parotid mass | Intra-abdominal abscess, sepsis | Cardiac arrest with traumatic spinal cord injury, sepsis | Hemoptysis, pneumonia | Weakness, dizziness |
| Antibiotics before CRE diagnosis | Unknown | Piperacillin-Tazobactam | None | Minocycline, Cefepime-Sulbactam, Tigecycline, Colistin, Metronidazole | Vancomycin, Piperacillin-Tazobactam | Ertapenem, Meropenem, Ceftriaxone, Trimethoprim-Sulfamethoxazole |
| Significant medical history | Unknown | Congestive heart failure, ischemic cardiomyopathy | Kidney transplant | Hypertension, Hypersensitivity lung disease | Lung transplant | Vertebrate osteomyelitis, atrial fibrillation |
| Specimen ID | 1A | 2A | 3A, 3B, 3C, 3D | 4A, 4B | 5A | 6A |
| Source | Urine | Parotid tissue | Peritoneal cavity fluid, hematoma, blood, abdominal tissue | Penile, blood | Expectorated sputum | Expectorated sputum |
| Travel | Unknown | Travel and hospitalized in India | No recent travel history | Travel to and hospitalized in India | No recent travel history | Lived in Cambodia for 4 years prior to infection |
| Treatment | Unknown | Meropenem, Polymyxin B | Aztreonam Cefiderocol, Ceftazidime-Avibactam, Plazomicin, Colistin, Polymyxin B, Tigecycline | Ceftazidime-Avibactam, Tigecycline, Piperacillin-Tazobactam, Meropenem, Polymyxin B, Aztreonam | Aztreonam, Polymyxin B | Cefiderocol |
| Outcome | Deceased | Deceased | Deceased | Deceased | Improved | Deceased |
| 1A | 2A | 3A | 3B | 3C | 3D | 4A | 4B | 5A | 6A | |
|---|---|---|---|---|---|---|---|---|---|---|
| MLST | 14 | 14 | 2497 | 2497 | 14 | 2497 | 437 | 2096 | 2497 | 16 |
| Plasmids Types | ColKP3 IncFIA(Hl1) IncFIB(K) IncFII IncR | ColKP3 IncFIB(K) IncFII(K) IncR | Col(pHAD28) ColKP3 IncFIB(pQil) | Col(pHAD28) ColKP3 IncFIB(pQil) | ColKP3 IncFIB(K) IncFII(K) IncR | Col(pHAD28) ColKP3 IncFIB(pQil) | Col(BS512) ColKP3 IncFIB(pQil) IncFII(k) | ColKP3 ColRNAI IncFIB(K) | ColKP3 IncFIB(pQil) | Col4401 Col440ll ColKP3 IncFIB(pKPHS1) IncFII(K) IncX3 |
| Aminoglycosides Resistance Genes | aac(3)-lld aac(6′)-lb-cr ant(3″)-la aph(3″)-lb aph(3′)-Vl aph(6)-ld armA rmtB1 sat2 | aac(6′)-lb ant(3“)-la aph(3″)-lb aph(3′)-Vl aph(6)-ld armA sat2 | aac(6′)-lb-cr ant(3″)-la aph(3′)-Vl armA | aac(6′)-lb-cr ant(3″)-la aph(3′)-VI armA | aac(6′)-lb ant(3“)-la aph(3″)-lb aph(3′)-VI aph(6)-ld armA sat2 | aac(6′)-lb-cr ant(3″)-la aph(3′)-VI armA | aac(6′)-lb-cr ant(3″)-la aph(3′)-VI armA rmtF1 rmtF2 | aac(6′)-lb-cr ant(3″)-la armA sat2 | aac(6′)-lb-cr ant(3″)-la aph(3′)-Vl armA | aac(6′)-lb-cr ant(3″)-la rmtB1 |
| Sulphamethoxazole/Trimethoprim Resistance Genes | sul1 sul2 dfrA1 dfrA12 dfrA14 | sul1 sul2 dfrA1 dfrA12 dfrA14 | sul1 dfrA14 | sul1 dfrA14 | sul1 sul2 dfrA1 dfrA12 dfrA14 | sul1 dfrA14 | sul1 dfrA12 dfrA14 | sul1 dfrA1 dfrA12 dfrA14 | sul1 dfrA14 | sul1 dfrA27 |
| Carbapenemase Genes | NDM-5 OXA-232 | NDM-1 OXA-232 | NDM-1 OXA-232 | NDM-1 OXA-232 | NDM-1 OXA-232 | NDM-1 OXA-232 | NDM-1 OXA-232 | OXA-232 | NDM-1 OXA-232 | NDM-4 OXA-181 |
| Chloramphenicol Resistance Genes | cmlA | catA1 | catA1 cmlA | catA1 cmlA | catA1 | catA1 cmlA | catB floR2 | N.D. | catA1 cmlA | N.D. |
| Chromosomal Beta-lactamase Genes | SHV-11 | SHV-28 | SHV-11 | SHV-11 | SHV-28 | SHV-11 | SHV-11 | SHV-28 | SHV-11 | SHV-1 |
| Plasmid-borne Narrow Spectrum Beta-lactamase Genes | TEM-1 OXA-1 | TEM-1 OXA-1 | TEM-1 OXA-1 | TEM-1 OXA-1 | TEM-1 OXA-1 | TEM-1 OXA-1 | TEM-1 OXA-1 | TEM-1 OXA-1 | TEM-1 | TEM-98 |
| ESBL Genes | CTX-M-15 | CTX-M-15 | CTX-M-15 | CTX-M-15 | CTX-M-15 | CTX-M-15 | CTX-M-15 | CTX-M-15 | CTX-M-15 | CTX-M-15 |
| Fosfomycin Resistance Genes | fosA6 | fosA6 | fosA | fosA | fosA6 | fosA | fosA | fosA6 | fosA | fosA6 |
| Fluroquinolone Resistance Mutations | gyrA(D87G + gyrA(S83Y) + parC(S80I) | gyrA(D87G) + gyrA(S83Y) + parC(S80I) | gyrA(S83I) + parC(S80I) | gyrA(S83l) + parC(S80l) | gyrA(D87G) + gyrA(S83Y) + parC(S80I) | gyrA(S83I) + parC(S80I) | gyrA(S83I) + parC(S80I) | gyrA(D87G) + gyrA(S83Y) + parC(S80I) | gyrA(S83I) + parC(S80I) | gyrA(D87N) + gyrA(S83F) + parC(E84K) |
| Fluroquinolone Resistance Genes | aac(6′)-lb-cr oqxA oqxB20 qnrB1 | oqxA oqxB20 qnrB1 | aac(6′)-lb-cr oqxA oqxB25 qnrB1 | aac(6′)-lb-cr oqxA oqxB25 qnrB1 | oqxA oqxB20 qnrB1 | oqxA oqxB25 qnrB1 | aac(6′)-lb-cr oqxA oqxB25 qnrS1 | aac(6′)-lb-cr oqxA oqxB20 | oqxA oqxB25 qnrB1 | aac(6′)-lb-cr oqxA oqxB32 qnrB6 qnrS1 |
| Macrolide Resistance Genes | ere(A) erm(B) mph(A) mph(E) msr(E) | mph(E) msr(E) | ere(A) mph(E) msr(E) | ere(A) mph(E) msr(E) | mph(E) msr(E) | ere(A) mph(E) msr(E) | mph(E) msr(E) | mph(E) msr(E) | ere(A) mph(E) msr(E) | mph(A) |
| Multidrug Efflux Pump Genes | emrD kdeA | emrD kdeA | emrD kdeA | emrD kdeA | emrD kdeA | emrD kdeA | emrD kdeA | emrD kdeA | emrD kdeA | emrD kdeA |
| Tetracycline Resistance Genes | tet(D) | tet(D) | tet(D) | tet(D) | tet(D) | tet(D) | tet(D) tet(G) | tet(D) | tet(D) | tet(A) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerón, S.; Salem-Bango, Z.; Contreras, D.A.; Ranson, E.L.; Yang, S. Clinical and Genomic Characterization of Carbapenem-Resistant Klebsiella pneumoniae with Concurrent Production of NDM and OXA-48-like Carbapenemases in Southern California, 2016–2022. Microorganisms 2023, 11, 1717. https://doi.org/10.3390/microorganisms11071717
Cerón S, Salem-Bango Z, Contreras DA, Ranson EL, Yang S. Clinical and Genomic Characterization of Carbapenem-Resistant Klebsiella pneumoniae with Concurrent Production of NDM and OXA-48-like Carbapenemases in Southern California, 2016–2022. Microorganisms. 2023; 11(7):1717. https://doi.org/10.3390/microorganisms11071717
Chicago/Turabian StyleCerón, Stacey, Zackary Salem-Bango, Deisy A. Contreras, Elizabeth L. Ranson, and Shangxin Yang. 2023. "Clinical and Genomic Characterization of Carbapenem-Resistant Klebsiella pneumoniae with Concurrent Production of NDM and OXA-48-like Carbapenemases in Southern California, 2016–2022" Microorganisms 11, no. 7: 1717. https://doi.org/10.3390/microorganisms11071717
APA StyleCerón, S., Salem-Bango, Z., Contreras, D. A., Ranson, E. L., & Yang, S. (2023). Clinical and Genomic Characterization of Carbapenem-Resistant Klebsiella pneumoniae with Concurrent Production of NDM and OXA-48-like Carbapenemases in Southern California, 2016–2022. Microorganisms, 11(7), 1717. https://doi.org/10.3390/microorganisms11071717





