Evaluation of Antibacterial Activity of Selenium Nanoparticles against Food-Borne Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of SeNPs
2.2. UV–Vis Spectroscopy
2.3. Fourier Transform Infrared (FTIR) Spectrometer Analysis of SeNPs
2.4. Zeta Potential Measurements of SeNPs
2.5. Scanning Transmission Electron Microscopic Characterization
2.6. Antibacterial Activity of SeNPs
2.7. Preparation of Bacteria for SEM Analysis
2.8. Statistical Methods
3. Results
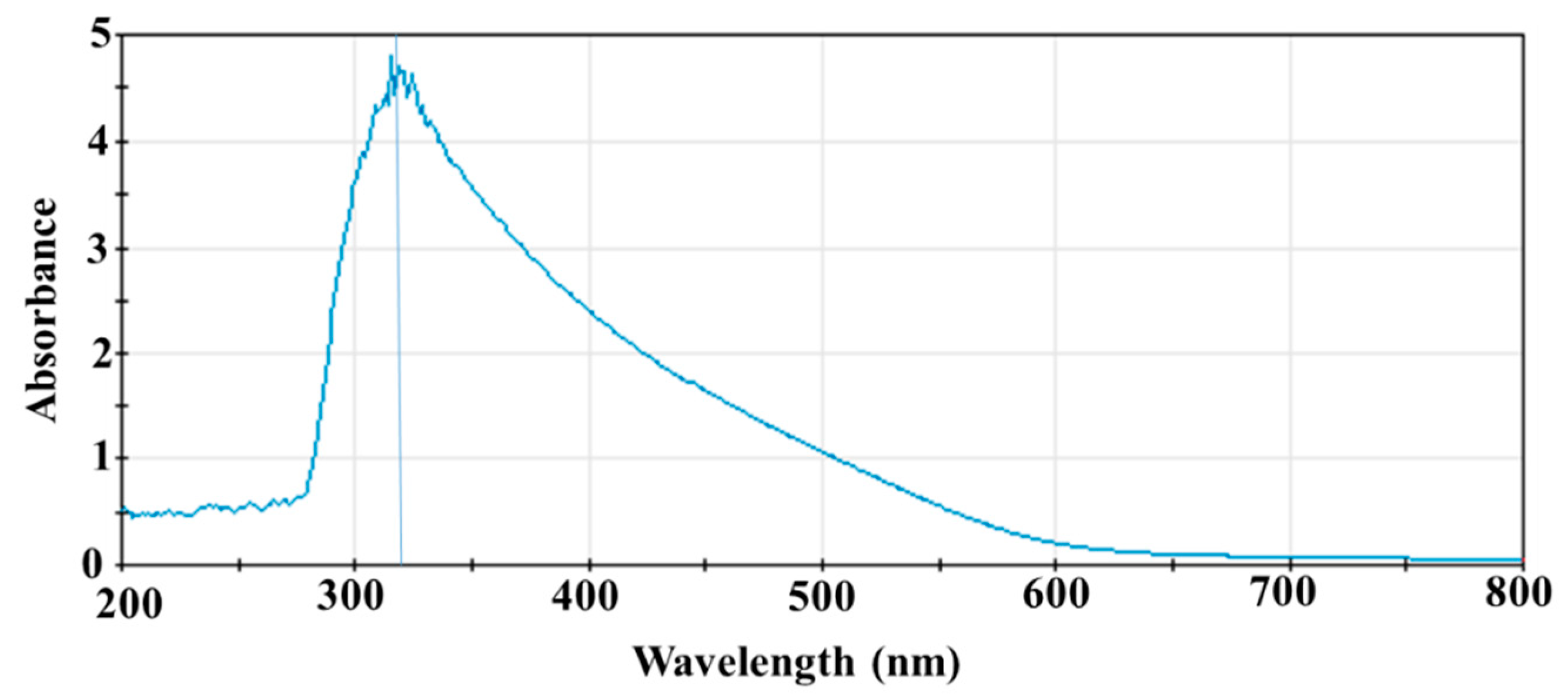
3.1. UV-Vis Spectrum of SeNPs
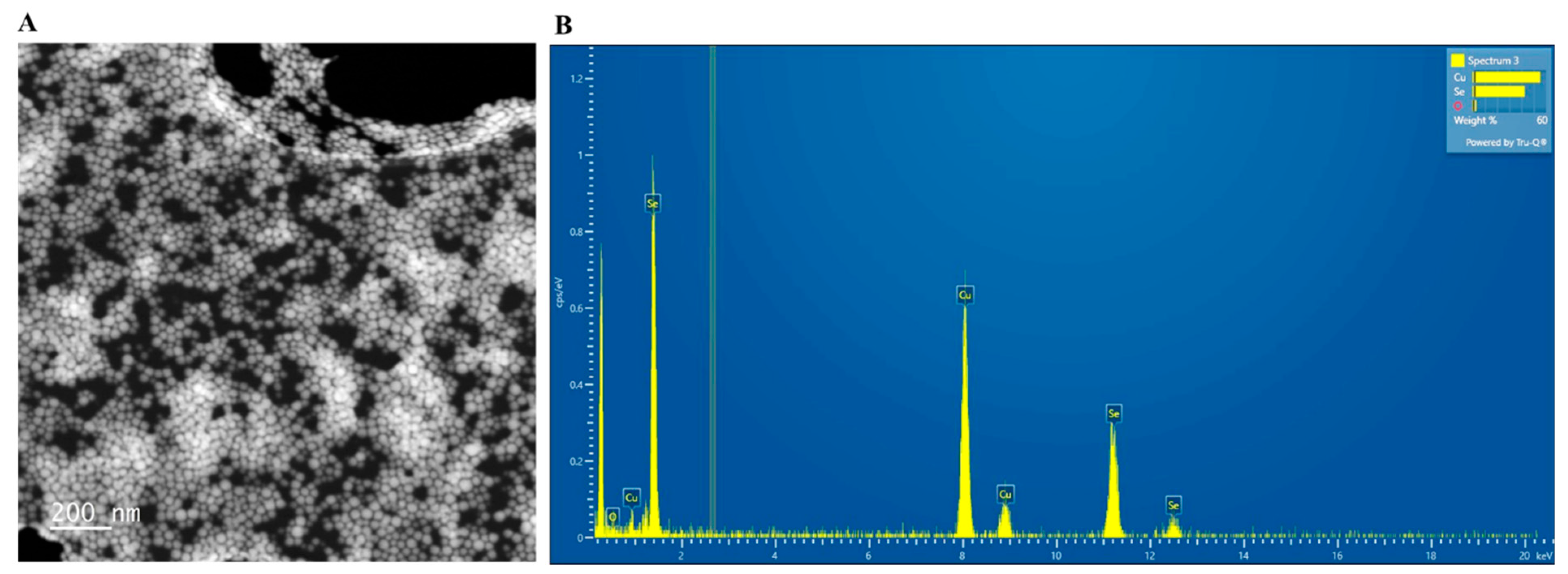
3.2. TEM and EDS Examination of SeNPs
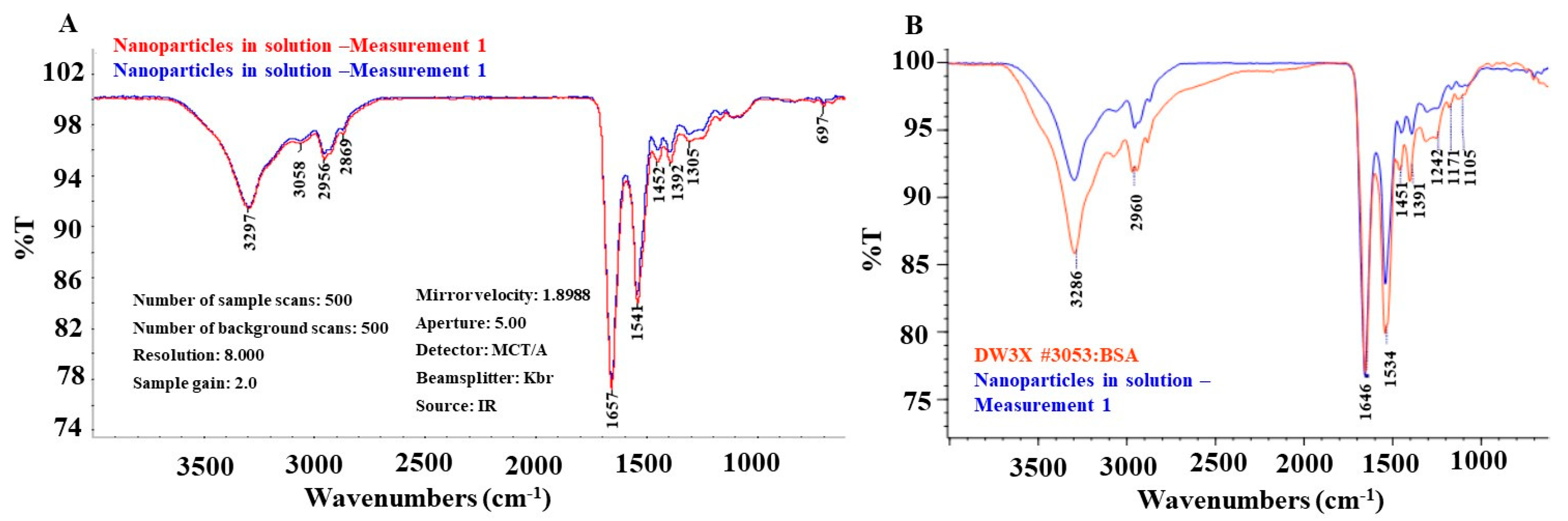
3.3. FTIR of SeNPs
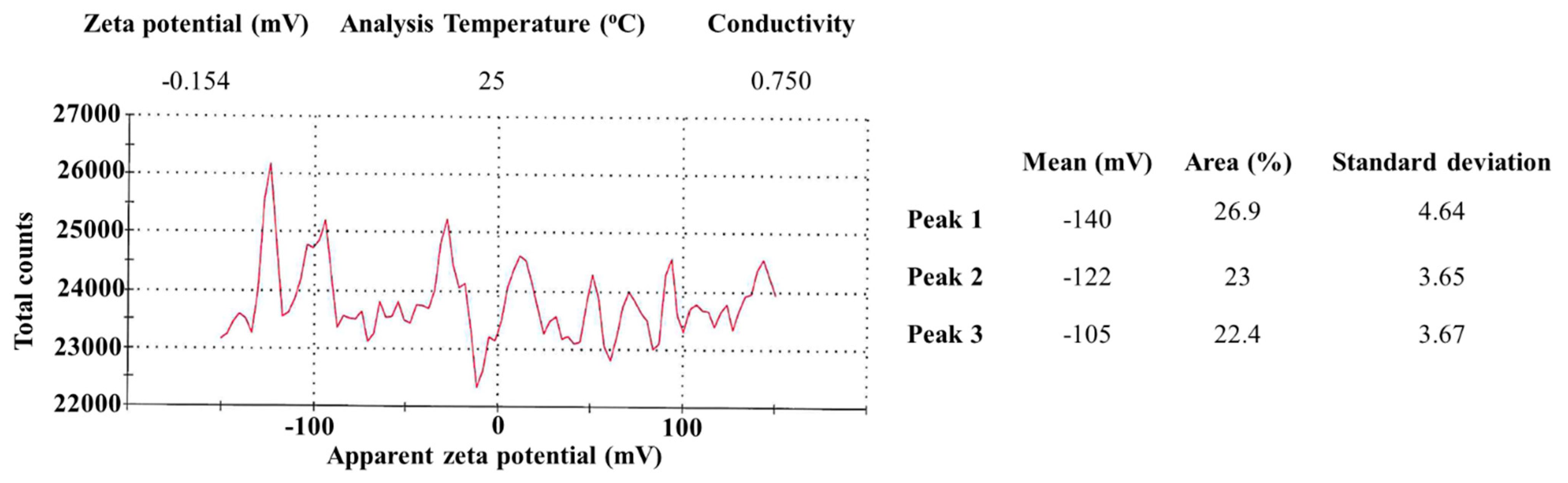
3.4. Zeta Potential Analysis
3.5. Antibacterial Activity of SeNPs
3.6. SEM Analysis of Bacteria Treated with SeNPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium nanoparticles as a nutritional supplement. Nutrition 2017, 33, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Gunti, L.; Dass, R.S.; Kalagatur, N.K. Phytofabrication of Selenium Nanoparticles from Emblica officinalis Fruit Extract and Exploring Its Biopotential Applications: Antioxidant, Antimicrobial, and Biocompatibility. Front. Microbiol. 2019, 10, 931. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, X.; Wang, Y.; Deng, X.; Miao, J.; Zhao, D.; Sun, K.; Li, M.; Wang, X.; Sun, W.; et al. Construction of Selenium Nanoparticle-Loaded Mesoporous Silica Nanoparticles with Potential Antioxidant and Antitumor Activities as a Selenium Supplement. ACS Omega 2022, 7, 44851–44860. [Google Scholar] [CrossRef]
- Filipovic, N.; Ušjak, D.; Milenković, M.T.; Zheng, K.; Liverani, L.; Boccaccini, A.R.; Stevanovic, M.M. Comparative Study of the Antimicrobial Activity of Selenium Nanoparticles with Different Surface Chemistry and Structure. Front. Bioeng. Biotechnol. 2021, 8, 624621. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhang, J.; Xu, J.-F.; Pi, J. The Advancing of Selenium Nanoparticles Against Infectious Diseases. Front. Pharmacol. 2021, 12, 682284. [Google Scholar] [CrossRef] [PubMed]
- Sentkowska, A.; Pyrzyńska, K. The Influence of Synthesis Conditions on the Antioxidant Activity of Selenium Nanoparticles. Molecules 2022, 27, 2486. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Bano, I.; Zare, H. A Comprehensive Review on Selenium and Its Effects on Human Health and Distribution in Middle Eastern Countries. Biol. Trace Elem. Res. 2022, 200, 971–987. [Google Scholar] [CrossRef]
- Yang, J.; Wang, J.; Yang, K.; Liu, M.; Qi, Y.; Zhang, T.; Fan, M.; Wei, X. Antibacterial activity of selenium-enriched lactic acid bacteria against common food-borne pathogens in vitro. J. Dairy Sci. 2018, 101, 1930–1942. [Google Scholar] [CrossRef]
- Mates, I.; Antoniac, I.; Laslo, V.; Vicas, S.; Brocks, M.; Fritea, L.; Milea, C.; Mohan, A.; Cavalu, S. Selenium nanoparticles: Production, characterization and possible applications in biomedicine and food science. UPB Sci. Bull. Ser. B Chem. Mater. Sci. 2019, 81, 205–216. [Google Scholar]
- Salem, S.S. Bio-fabrication of Selenium Nanoparticles Using Baker’s Yeast Extract and Its Antimicrobial Efficacy on Food Borne Pathogens. Appl. Biochem. Biotechnol. 2022, 194, 1898–1910. [Google Scholar] [CrossRef]
- Gonca, S. Selenium: A Micronutrient Essential for Maintaining Human Health. Med. Sci. 2013, 2, 649–664. [Google Scholar] [CrossRef]
- Mehdi, Y.; Hornick, J.-L.; Istasse, L.; Dufrasne, I. Selenium in the Environment, Metabolism and Involvement in Body Functions. Molecules 2013, 18, 3292–3311. [Google Scholar] [CrossRef]
- Kieliszek, M. Selenium–Fascinating Microelement, Properties and Sources in Food. Molecules 2019, 24, 1298. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.F.; Hill, K.E. Selenoprotein P– Expression, Functions, and Roles in Mammals. Biochim. Biophys. Acta 2009, 1790, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular Pathways and Physiological Roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef]
- Weekley, C.M.; Harris, H.H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 2013, 42, 8870–8894. [Google Scholar] [CrossRef]
- Hatfield, D.L.; Yoo, M.H.; Carlson, B.A.; Gladyshev, V.N. Selenoproteins that function in cancer prevention and promotion. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 1541–1545. [Google Scholar] [CrossRef]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef]
- Larsen, P.R.; Zavacki, A.M. Role of the Iodothyronine Deiodinases in the Physiology and Pathophysiology of Thyroid Hormone Action, Translational Thyroidology/Review. Eur. Thyroid J. 2012, 1, 232–242. [Google Scholar]
- Xia, Y.; Tang, G.; Wang, C.; Zhong, J.; Chen, Y.; Hua, L.; Li, Y.; Liu, H.; Zhu, B. Functionalized selenium nanoparticles for targeted siRNA delivery silence Derlin1 and promote antitumor efficacy against cervical cancer. Drug Deliv. 2020, 27, 15–25. [Google Scholar] [CrossRef]
- Varlamova, E.G.; Turovsky, E.A.; Blinova, E.V. Therapeutic Potential and Main Methods of Obtaining Selenium Nanoparticles. Int. J. Mol. Sci. 2021, 22, 10808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, X.; Xu, T. Elemental Selenium at Nano Size (Nano-Se) as a Potential Chemopreventive Agent with Reduced Risk of Selenium Toxicity: Comparison with Se-Methylselenocysteine in Mice. Toxicol. Sci. 2008, 101, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.L.C.; Hinchman, A.; Williams, R.; Tran, P.A.; Fox, K. Nanostructured biomedical selenium at the biological interface (Review). Biointerphases 2018, 13, 06D301. [Google Scholar] [CrossRef] [PubMed]
- Estevez, H.; Garcia-Lidon, J.C.; Luque-Garcia, J.L.; Camara, C. Effects of chitosan-stabilized selenium nanoparticles on cell proliferation, apoptosis and cell cycle pattern in HepG2 cells: Comparison with other selenospecies. Colloids Surf. B Biointerfaces 2014, 122, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Hosnedlova, B.; Kepinska, M.; Skalickova, S.; Fernandez, C.; Ruttkay-Nedecky, B.; Peng, Q.; Baron, M.; Melcova, M.; Opatrilova, R.; Zidkova, J.; et al. Nano-selenium and its nanomedicine applications: A critical review. Int. J. Nanomed. 2018, 13, 2107–2128. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, K.; Mvs, S.; Koganti, S.; Burgula, S. Selenium Nanoparticles Synthesized Using Pseudomonas stutzeri (MH191156) Show Antiproliferative and Anti-angiogenic Activity Against Cervical Cancer Cells. Int. J. Nanomed. 2020, 15, 4523–4540. [Google Scholar] [CrossRef] [PubMed]
- El-Deeba, B.; Al-Talhib, A.; Mostafac, N.; Abou-assyd, R. Biological Synthesis and Structural Characterization of Selenium Nanoparticles and Assessment of Their Antimicrobial Properties. Am. Sci. Res. J. Eng. Technol. Sci. (ASRJETS) 2018, 45, 135–170. [Google Scholar]
- Mosallam, F.M.; El-Sayyad, G.S.; Fathy, R.M.; El-Batal, A.I. Biomolecules-mediated synthesis of selenium nanoparticles using Aspergillus oryzae fermented Lupin extract and gamma radiation for hindering the growth of some multidrug-resistant bacteria and pathogenic fungi. Microb. Pathog. 2018, 122, 108–116. [Google Scholar] [CrossRef]
- Alghuthaymi, M.A.; Diab, A.M.; Elzahy, A.F.; Mazrou, K.E.; Tayel, A.A.; Moussa, S.H. Green Biosynthesized Selenium Nanoparticles by Cinnamon Extract and Their Antimicrobial Activity and Application as Edible Coatings with Nano-Chitosan. J. Food Qual. 2021, 2021, 6670709. [Google Scholar] [CrossRef]
- Chung, S.; Zhou, R.; Webster, T.J. Green Synthesized BSA-Coated Selenium Nanoparticles Inhibit Bacterial Growth While Promoting Mammalian Cell Growth. Int. J. Nanomed. 2020, 15, 115–124. [Google Scholar] [CrossRef]
- Fouda, A.; Hassan, S.E.D.; Eid, A.M.; Abdel-Rahman, M.A.; Hamza, M.F. Light enhanced the antimicrobial, anticancer, and catalytic activities of selenium nanoparticles fabricated by endophytic fungal strain, Penicillium crustosum EP-1. Sci. Rep. 2022, 12, 11834. [Google Scholar] [CrossRef] [PubMed]
- Kamble, S.; Agrawal, S.; Cherumukkil, S.; Sharma, V.; Jasra, R.V.; Munshi, P. Revisiting Zeta Potential, the Key Feature of Interfacial Phenomena, with Applications and Recent Advancements. ChemistrySelect 2022, 7, e202103084. [Google Scholar] [CrossRef]
- Liu, J. Scanning transmission electron microscopy and its application to the study of nanoparticles and nanoparticle systems. Microscopy 2005, 54, 251–278. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Saha, S.; Kostina, O.; Muravnik, L.; Mitra, A. Replacing critical point drying with a low-cost chemical drying provides comparable surface image quality of glandular trichomes from leaves of Millingtonia hortensis L. f. in scanning electron micrograph. Appl. Microsc. 2020, 50, 15. [Google Scholar] [CrossRef]
- Alagesan, V.; Venugopal, S. Green Synthesis of SeNPs Using Leaves Extract of Withania somnifera and Its Biological Applications and Photocatalytic Activities. BioNanoSci 2019, 9, 105–116. [Google Scholar] [CrossRef]
- Sheeana, G.; Dragana, S.; Robert, J.H.; Moore, R.J.; Chapman, J. The synthesis and characterization of highly stable and reproducible selenium nanoparticles. Inorg. Nano-Met. Chem. 2017, 47, 1568–1576. [Google Scholar]
- Mu, X.; Yan, C.; Tian, Q.W.; Lin, J.; Yang, S. BSA-assisted synthesis of ultrasmall gallic acid–Fe (III) coordination polymer nanoparticles for cancer theranostics. Int. J. Nanomed. 2017, 12, 7207–7223. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Toubhans, B.; Gazze, S.A.; Bissardon, C.; Bohic, S.; Gourlan, A.T.; Gonzalez, D.; Charlet, L.; Conlan, R.S.; Francis, L.W. Selenium nanoparticles trigger alterations in ovarian cancer cell biomechanics. Nanomed. Nanotechnol. Biol. Med. 2020, 29, 102258. [Google Scholar] [CrossRef]
- Escobar-Ramírez, M.C.; Castañeda-Ovando, A.; Pérez-Escalante, E.; Rodríguez-Serrano, G.M.; Ramírez-Moreno, E.; Quintero-Lira, A.; Contreras-López, E.; Añorve-Morga, J.; Jaimez-Ordaz, J.; González-Olivares, L.G. Antimicrobial Activity of Se-Nanoparticles from Bacterial Biotransformation. Fermentation 2021, 7, 130. [Google Scholar] [CrossRef]
- Yuan, Q.; Bomma, M.; Hill, H.; Xiao, Z. Expression of Rhizobium tropici phytochelatin synthase in Escherichia coli resulted in increased bacterial selenium nanoparticle synthesis. J. Nanopart. Res. 2022, 22, 369. [Google Scholar] [CrossRef]
- Sentkowska, A.; Pyrzyńska, K. Antioxidant Properties of Selenium Nanoparticles Synthesized Using Tea and Herb Water Extracts. Appl. Sci. 2023, 13, 1071. [Google Scholar] [CrossRef]
- Toprakcioglu, Z.; Wiita, E.G.; Jayaram, A.K.; Gregory, R.C.; Knowles, T.P.J. Selenium Silk Nanostructured Films with Antifungal and Antibacterial Activity. ACS Appl. Mater. Interfaces 2023, 15, 10452–10463. [Google Scholar] [CrossRef] [PubMed]
- Varlamova, E.G.; Gudkov, S.V.; Plotnikov, E.Y.; Turovsky, E.A. Size-Dependent Cytoprotective Effects of Selenium Nanoparticles during Oxygen-Glucose Deprivation in Brain Cortical Cells. Int. J. Mol. Sci. 2022, 23, 7464. [Google Scholar] [CrossRef] [PubMed]
- Foroozandeh, P.; Aziz, A.A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Touliabah, H.E.; El-Sheekh, M.M.; Makhlof, M.E.M. Evaluation of Polycladia myrica mediated selenium nanoparticles (PoSeNPS) cytotoxicity against PC-3 cells and antiviral activity against HAV HM175 (Hepatitis A), HSV-2 (Herpes simplex II), and Adenovirus strain 2. Front. Mar. Sci. 2022, 9, 1092343. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. [Google Scholar] [CrossRef]
- Chung, Y.C.; Su, Y.P.; Chen, C.C.; Jia, G.; Wang, H.L.; Wu, J.C.G.; Lin, J.G. Relationship between antibacterial activity of chitosan and surface characteristics of cell wall. Acta Pharmacol. Sin. 2004, 25, 932–936. [Google Scholar]
- Gottenbos, B.; Grijpma, D.W.; Van Der Mei, H.C.; Feijen, J.; Busscher, H.J. Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2001, 48, 7–13. [Google Scholar] [CrossRef]
- Dziarski, R. Cell-bound albumin is the 70-kDa peptidoglycan-, lipopolysaccharide-, and lipoteichoic acid-binding protein on lymphocytes and macrophages. J. Biol. Chem. 1994, 269, 20431–20436. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.R.; Basheer, S.M.; Gupta, I.R.; Elyas, K.K.; Joseph, A. Investigation on Bovine Serum Albumin (BSA) binding efficiency and antibacterial activity of ZnO nanoparticles. Mater. Chem. Phys. 2020, 240, 122115. [Google Scholar] [CrossRef]
- Espinosa-Cristóbal, L.F.; Martínez-Castañón, G.A.; Loyola-Rodríguez, J.P.; Niño-Martínez, N.; Ruiz, F.; Zavala-Alonso, N.V.; Lara, R.H.; Reyes-López, S.Y. Bovine serum albumin and chitosan coated silver nanoparticles and its antimicrobial activity against oral and nonoral bacteria. J. Nanomater. 2016, 16, 366. [Google Scholar] [CrossRef]
- Rao, C.K.; Mangamuri, U.K.; Sikharam, A.S.; Devaraj, K.; Kalagatur, N.K.; Kadirvelu, K. Biosynthesis of Selenium Nanoparticles from Annona muricata Fruit Aqueous Extract and Investigation of their Antioxidant and Antimicrobial potentials. Curr. Trends Biotechnol. Pharm. 2022, 16, 101–107. [Google Scholar] [CrossRef]
- El-Zayat, M.M.; Eraqi, M.M.; Alrefai, H.; El-Khateeb, A.Y.; Ibrahim, M.A.; Aljohani, H.M.; Aljohani, M.M.; Elshaer, M.M. The Antimicrobial, Antioxidant, and Anticancer Activity of Greenly Synthesized Selenium and Zinc Composite Nanoparticles Using Ephedra aphylla Extract. Biomolecules 2021, 11, 470. [Google Scholar] [CrossRef]
- Bian, S.; Zeng, W.; Li, Q.; Li, Y.; Wong, N.K.; Jiang, M.; Zuo, L.; Hu, Q.; Li, L. Genetic Structure, Function, and Evolution of Capsule Biosynthesis Loci in Vibrio parahaemolyticus. Front. Microbiol. 2021, 11, 546150. [Google Scholar] [CrossRef]
- MacFarquhar, J.K.; Broussard, D.L.; Melstrom, P.; Hutchinson, R.; Wolkin, A.; Martin, C.; Burk, R.F.; Dunn, J.R.; Green, A.L.; Hammond, R.; et al. Acute selenium toxicity associated with a dietary supplement. Arch. Intern. Med. 2010, 170, 256–261. [Google Scholar] [CrossRef]

| Treatment | Kanamycin (µg/mL) | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|---|---|---|---|---|---|---|---|---|---|---|
| SeNPs (µg/mL) | 0 | 0 | 0.5 | 1 | 2.5 | 5 | 10 | 15 | 30 | |
| CFU (×107) after treatment | L. Monocytogens (ATCC15313) | 0 | 65.0 ± 13.1 | 54.5 ± 6.3 ** | 32.5 ± 8 *** | 14.2 ± 4.7 *** | 17.8 ± 16.6 *** | 12.0 ± 8.0 *** | 10.0 ± 3.6 *** | 10.8 ± 6.0 *** |
| S. aureus (ATCC12600) | 0 | 68.0 ± 11.5 | 57.8 ± 13.4 | 74.4 ± 9.8 | 51.8 ± 11.3 | 63.0 ± 17.3 | 39.2 ± 5.7 * | 33.2 ± 10.1 * | 27.6 ± 16.7 *** | |
| S. epidermidis (ATCC 700583) | 0 | 57.7 ± 9.3 | 41.9 ± 13.0 | 36.1 ± 7.7 ** | 26.9 ± 7.9 *** | 24.1 ± 6.2 *** | 26.6 ± 9.3 ** | 31.8 ± 9.2 ** | 31.1 ± 8.4 ** | |
| E. faecalis (ATCC 19433) | 0 | 45.1 ± 10.4 | 51.5 ± 19.9 | 50.9 ± 20.6 | 43.1 ± 16.6 | 38.0 ± 19.7 | 41.7 ± 10.6 | 41.5 ± 10.8 | 42.5 ± 12.8 | |
| V. alginolyticus (ATCC 33787) | 0 | 65.0 ± 13.9 | 57.5 ± 10.9 | 47.0 ± 11.9 | 36.5 ± 15.2 * | 38.9 ± 10.7 ** | 33.8 ± 11.8 ** | 31.9 ± 13.2 ** | 31.2 ± 13.3 ** | |
| S. enterica (ATCC19585) | 0 | 56.3 ± 8.8 | 57.2 ± 5.9 | 64.0 ± 13.5 | 50.8 ± 11.1 | 52.3 ± 6.7 | 48.5 ± 10.4 | 39.0 ± 13.1 * | 39.6 ± 8.1 * | |
| V. Parahaemolyticus (ATCC43996) | 0 | 18.5 ± 7.5 | 21.2 ± 8.7 | 24.4 ± 13.8 | 17.7 ± 6.8 | 18.9 ± 6.6 | 16.1 ± 5.8 | 18.4 ± 5.5 | 24.5 ± 6.8 | |
| S. enterica (ATCC49284) | 0 | 33.1 ± 10.5 | 46.2 ± 7.0 | 39.2 ± 11.7 | 35.6 ± 18.7 | 35.6 ± 11.7 | 32.9 ± 4.4 | 28.1 ± 9.1 | 34.7 ± 12.5 | |
| E. cloacae (ATCC BAA3044) | 126.0 ± 21.2 # | 186.8 ± 50.8 | 142.1 ± 25.1 | 182.3 ± 19.1 | 175.2 ± 58.7 | 182.1 ± 55.0 | 181.9 ± 50.7 | 188.9 ± 29.7 | 202.6 ± 62.8 | |
| E. coli (ATCC BAA2326) | 0 | 196.1 ± 37.2 | 191.6 ± 59.6 | 218.9 ± 9.31 | 201.1 ± 39.7 | 173.5 ± 6.30 | 172.9 ± 54.3 | 210.9 ± 9.63 | 240.6 ± 92.2 |
| Bacterium | L. Monocytogens (ATCC15313) | Enterobacter cloacae (ATCC BAA3044) | ||||||
|---|---|---|---|---|---|---|---|---|
| SeNP Concentration (µg/mL) | 0 | 5 | 10 | 30 | 0 | 5 | 10 | 30 |
| Width of bacteria (nm) | 384.4 ± 45.0 | 391.0 ± 37.7 | 395.9 ± 46.1 | 378.8 ± 42.6 | 640.8 ± 47.6 | 661.8 ± 205.6 | 638.6 ± 74.2 | 594.9 ± 73.3 * |
| Number of bacteria (n) | 64 | 66 | 66 | 43 | 54 | 45 | 44 | 65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Q.; Xiao, R.; Afolabi, M.; Bomma, M.; Xiao, Z. Evaluation of Antibacterial Activity of Selenium Nanoparticles against Food-Borne Pathogens. Microorganisms 2023, 11, 1519. https://doi.org/10.3390/microorganisms11061519
Yuan Q, Xiao R, Afolabi M, Bomma M, Xiao Z. Evaluation of Antibacterial Activity of Selenium Nanoparticles against Food-Borne Pathogens. Microorganisms. 2023; 11(6):1519. https://doi.org/10.3390/microorganisms11061519
Chicago/Turabian StyleYuan, Qunying, Rong Xiao, Mojetoluwa Afolabi, Manjula Bomma, and Zhigang Xiao. 2023. "Evaluation of Antibacterial Activity of Selenium Nanoparticles against Food-Borne Pathogens" Microorganisms 11, no. 6: 1519. https://doi.org/10.3390/microorganisms11061519
APA StyleYuan, Q., Xiao, R., Afolabi, M., Bomma, M., & Xiao, Z. (2023). Evaluation of Antibacterial Activity of Selenium Nanoparticles against Food-Borne Pathogens. Microorganisms, 11(6), 1519. https://doi.org/10.3390/microorganisms11061519









