ARGs Detection in Listeria Monocytogenes Strains Isolated from the Atlantic Salmon (Salmo salar) Food Industry: A Retrospective Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples: Listeria Monocytogenes Isolates
2.2. Antimicrobial Susceptibility Tests (ASTs)
2.3. DNA Extraction: PCR Assays
2.4. Serotyping and Virulence Factors
2.5. ARGs Detection
2.6. Statistical Analysis
3. Results
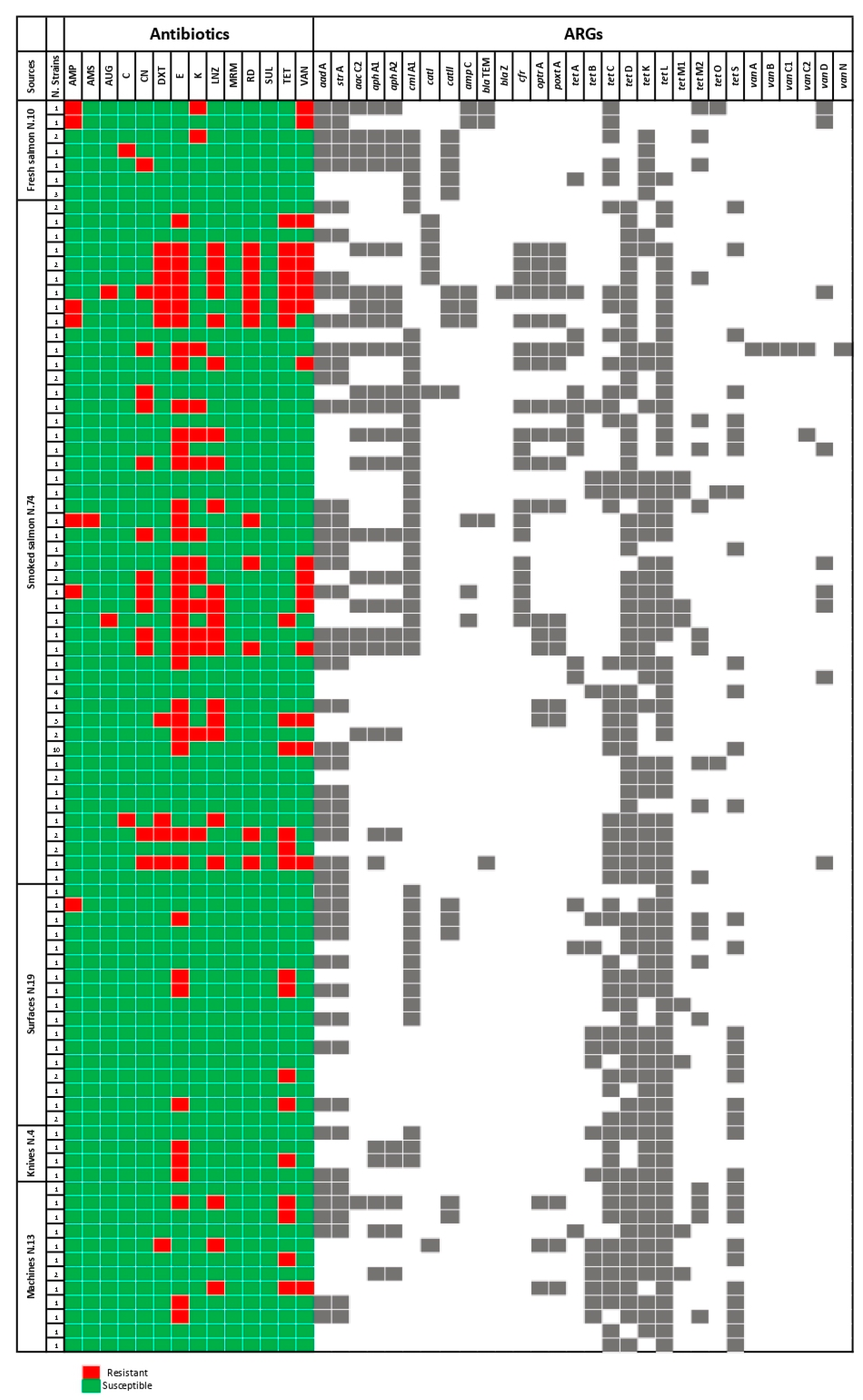
3.1. ASTs
3.2. ARGs Detection
3.3. Serovars and Virulence Factors
3.4. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopez-Valladares, G.; Danielsson-Tham, M.L.; Tham, W. Implicated Food Products for Listeriosis and Changes in Serovars of Listeria monocytogenes Affecting Humans in Recent Decades. Foodborne Pathog. Dis. 2018, 15, 387–397. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA) Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. The public health risk posed by Listeria monocytogenes in frozen fruit and vegetables including herbs, blanched during processing. EFSA J. 2020, 18, e06092. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.K.; Natarajan, V.; Stewart, D.; Fay, M.; Gonsalves, L.J.; Mhetras, T.; Sule, C.; Tortorello, M.L. Listeria monocytogenes growth kinetics in refrigerated ready-to-eat dips and dip components. PLoS ONE 2020, 15, e0235472. [Google Scholar] [CrossRef] [PubMed]
- European Regulation N. 2073/2005. Microbiological Criteria for Foodstuffs. 2005. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32005R2073 (accessed on 1 June 2023).
- Abdeen, E.E.; Mousa, W.S.; Harb, O.H.; Fath-Elbab, G.A.; Nooruzzaman, M.; Gaber, A.; Alsanie, W.F.; Abdeen, A. Prevalence, Antibiogram and Genetic Characterization of Listeria monocytogenes from Food Products in Egypt. Foods 2021, 10, 1381. [Google Scholar] [CrossRef] [PubMed]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes Biofilms in the Wonderland of Food Industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef]
- Mazaheri, T.; Cervantes-Huamán, B.; Bermúdez-Capdevila, M.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Listeria monocytogenes Biofilms in the Food Industry: Is the Current Hygiene Program Sufficient to Combat the Persistence of the Pathogen? Microorganisms 2021, 9, 181. [Google Scholar] [CrossRef]
- Ramires, T.; Kleinubing, N.R.; Iglesias, M.A.; Vitola, H.; Núncio, A.; Kroning, I.S.; Moreira, G.; Fiorentini, Â.M.; da Silva, W.P. Genetic diversity, biofilm and virulence characteristics of Listeria monocytogenes in salmon sushi. Food Res. Int. 2021, 140, 109871. [Google Scholar] [CrossRef]
- Waples, R.S.; Naish, K.A.; Primmer, C.R. Conservation and Management of Salmon in the Age of Genomics. Annu. Rev. Anim. Biosci. 2020, 8, 117–143. [Google Scholar] [CrossRef]
- Garseth, Å.H.; Fritsvold, C.; Svendsen, J.C.; Bang Jensen, B.; Mikalsen, A.B. Cardiomyopathy syndrome in Atlantic salmon Salmo salar L.: A review of the current state of knowledge. J. Fish Dis. 2018, 41, 11–26. [Google Scholar] [CrossRef]
- Chowdhury, S.; Ghosh, S.; Aleem, M.A.; Parveen, S.; Islam, M.A.; Rashid, M.M.; Akhtar, Z.; Chowdhury, F. Antibiotic Usage and Resistance in Food Animal Production: What Have We Learned from Bangladesh? Antibiotics 2021, 10, 1032. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA) Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.M.; El Zowalaty, M.E.; Lundkvist, Å.; Järhult, J.D.; Khan Nayem, M.R.; Tanzin, A.Z.; Badsha, M.R.; Khan, S.A.; Ashour, H.M. Residual antimicrobial agents in food originating from animals. Trends Food Sci. Technol. 2021, 111, 141–150. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency (EMA). Advice on the Designation of Antimicrobials or Groups of Antimicrobials Reserved for Treatment of Certain Infections in Humans—In Relation to Implementing Measures under Article 37(5) of Regulation (EU) 2019/6 on Veterinary Medicinal Products. 2022. Available online: https://www.ema.europa.eu/en/documents/regulatory-procedural-guideline/advice-designation-antimicrobials-groups-antimicrobials-reserved-treatment-certain-infections-humans/6-veterinary-medicinal-products_en.pdf. (accessed on 4 August 2022).
- Guglielmetti, E.; Korhonen, J.M.; Heikkinen, J.; Morelli, L.; von Wright, A. Transfer of plasmid-mediated resistance to tetracycline in pathogenic bacteria from fish and aquaculture environments. FEMS Microbiol. Lett. 2009, 293, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Vitas, A.I.; Sánchez, R.M.; Aguado, V.; García-Jalón, I. Antimicrobial susceptibility of Listeria monocytogenes isolated from food and clinical cases in Navarra, Spain. J. Food Prot. 2007, 70, 2402–2406. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, S.; Chen, H.; Chen, J.; Zhang, J.; Zhang, Z.; Yang, Y.; Xu, Z.; Zhan, L.; Mei, L. Prevalence, Genotypic Characteristics and Antibiotic Resistance of Listeria monocytogenes from Retail Foods in Bulk in Zhejiang Province, China. Front. Microbiol. 2019, 10, 1710. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Assessment of multidrug-resistant Listeria monocytogenes in milk and milk product and One Health perspective. PLoS ONE 2022, 17, e0270993. [Google Scholar] [CrossRef]
- Pensinger, D.A.; Aliota, M.T.; Schaenzer, A.J.; Boldon, K.M.; Ansari, I.U.; Vincent, W.J.; Knight, B.; Reniere, M.L.; Striker, R.; Sauer, J.D. Selective pharmacologic inhibition of a PASTA kinase increases Listeria monocytogenes susceptibility to β-lactam antibiotics. Antimicrob. Agents Chemother. 2014, 58, 4486–4494. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, G.; Yang, J.; Zhao, L.; Jiang, Y.; Guo, D.; Wang, X.; Zhi, S.; Xu, X.; Dong, Q.; et al. Prevalence, antibiotic resistance, and molecular epidemiology of Listeria monocytogenes isolated from imported foods in China during 2018 to 2020. Int. J. Food Microbiol. 2022, 382, 109916. [Google Scholar] [CrossRef]
- Badawy, B.; Gwida, M.; Sadat, A.; El-Toukhy, M.; Sayed-Ahmed, M.; Alam, N.; Ahmad, S.; Ali, M.D.S.; Elafify, M. Prevalence and Antimicrobial Resistance of Virulent Listeria monocytogenes and Cronobacter sakazakii in Dairy Cattle, the Environment, and Dried Milk with the In Vitro Application of Natural Alternative Control. Antibiotics 2022, 11, 1087. [Google Scholar] [CrossRef]
- Kayser, F.H.; Morenzoni, G.; Strässle, A.; Hadorn, K. Activity of meropenem, against gram-positive bacteria. J. Antimicrob. Chemother. 1989, 24, 101–112. [Google Scholar] [CrossRef]
- Conter, M.; Paludi, D.; Zanardi, E.; Ghidini, S.; Vergara, A.; Ianieri, A. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 2009, 128, 497–500. [Google Scholar] [CrossRef] [PubMed]
- ISO 11290-1,2:2017; Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria spp.—Part 1: Detection Method; Part 2: Enumeration Method. ISO: London, UK, 2017. Available online: https://www.iso.org/standard/60313.html (accessed on 2 August 2022).
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2021. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 23 August 2022).
- Chen, M.; Cheng, J.; Wu, Q.; Zhang, J.; Chen, Y.; Xue, L.; Lei, T.; Zeng, H.; Wu, S.; Ye, Q.; et al. Occurrence, Antibiotic Resistance, and Population Diversity of Listeria monocytogenes Isolated from Fresh Aquatic Products in China. Front. Microbiol. 2018, 9, 2215. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, X.; Song, Y.; Yao, S.; Li, K.; Shi, B.; Sun, J.; Liu, Z.; Zhao, W.; Zhao, C.; et al. Genetic diversity, antibiotic resistance, and virulence profiles of Listeria monocytogenes from retail meat and meat processing. Food Res. Int. 2022, 162, 112040. [Google Scholar] [CrossRef]
- Yehia, H.M.; Elkhadragy, M.F.; Aljahani, A.H.; Alarjani, K.M. Prevalence and antibiotic resistance of Listeria monocytogenes in camel meat. Biosci. Rep. 2020, 40, BSR20201062. [Google Scholar] [CrossRef] [PubMed]
- Di Ciccio, P.; Meloni, D.; Festino, A.R.; Conter, M.; Zanardi, E.; Ghidini, S.; Vergara, A.; Mazzette, R.; Ianieri, A. Longitudinal study on the sources of Listeria monocytogenes contamination in cold-smoked salmon and its processing environment in Italy. Int. J. Food Microbiol. 2012, 158, 79–84. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed]
- Border, P.M.; Howard, J.J.; Plastow, G.S.; Siggens, K.W. Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction. Lett. Appl. Microbiol. 1990, 11, 158–162. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Schutze, G.E.; Bhunia, A.K. Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int. J. Food Microbiol. 2002, 76, 1–10. [Google Scholar] [CrossRef]
- Srinivasan, V.; Gillespie, B.E.; Lewis, M.J.; Nguyen, L.T.; Headrick, S.I.; Schukken, Y.H.; Oliver, S.P. Phenotypic and genotypic antimicrobial resistance patterns of Escherichia coli isolated from dairy cows with mastitis. Vet. Microbiol. 2007, 124, 319–328. [Google Scholar] [CrossRef]
- Maynard, C.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Larivière, S.; Harel, J. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J. Clin. Microbiol. 2004, 42, 5444–5452. [Google Scholar] [CrossRef]
- Kishk, R.M.; Anani, M.M.; Nemr, N.A.; Soliman, N.M.; Fouad, M.M. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus in Suez Canal University Hospital, Ismailia, Egypt. J. Infect. Dev. Ctries. 2020, 14, 1281–1287. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.; Grebe, T.; Tait-Kamradt, A.; Wondrack, L. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 1996, 40, 2562–2566. [Google Scholar] [CrossRef] [PubMed]
- Post, V.; Hall, R.M. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 2667–2671. [Google Scholar] [CrossRef]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Commensal Escherichia coli of healthy humans: A reservoir for antibiotic-resistance determinants. J. Med. Microbiol. 2010, 59, 1331–1339. [Google Scholar] [CrossRef]
- Baddour, M.M.; AbuElKheir, M.M.; Fatani, A.J. Comparison of mecA polymerase chain reaction with phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Curr. Microbiol. 2007, 55, 473–479. [Google Scholar] [CrossRef]
- Bender, J.K.; Fleige, C.; Klare, I.; Werner, G. Development of a multiplex-PCR to simultaneously detect acquired linezolid resistance genes cfr, optrA and poxtA in enterococci of clinical origin. J. Microbiol. Methods 2019, 160, 101–103. [Google Scholar] [CrossRef]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Prob. 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Celli, J.; Trieu-Cuot, P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: Characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 1998, 28, 103–117. [Google Scholar] [CrossRef]
- Olsvik, B.; Olsen, I.; Tenover, F.C. Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol. Immunol. 1995, 10, 87–92. [Google Scholar] [CrossRef]
- Trzcinski, K.; Cooper, B.S.; Hryniewicz, W.; Dowson, C.G. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2000, 45, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Hashimoto, Y.; Kurushima, J.; Hirakawa, H.; Tanimoto, K.; Zheng, B.; Ruan, G.; Xue, F.; Liu, J.; Hisatsune, J.; et al. New colony multiplex PCR assays for the detection and discrimination of vancomycin-resistant enterococcal species. J. Microbiol. Methods 2018, 145, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Şanlıbaba, P.; Tezel, B.U.; Çakmak, G.A. Prevalence and Antibiotic Resistance of Listeria monocytogenes Isolated from Ready-to-Eat Foods in Turkey. J. Food Qual. 2018, 9, 7693782. [Google Scholar] [CrossRef]
- European Regulation N. 765/2008. Setting Out the Requirements for Accreditation and Market Surveillance Relating to the Marketing of Products and Repealing Regulation (EEC) No 339/93. 2008. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2008:218:0030:0047:en:PDF (accessed on 1 June 2023).
- Chatzopoulou, S.; Eriksson, N.L.; Eriksson, D. Improving Risk Assessment in the European Food Safety Authority: Lessons from the European Medicines Agency. Front. Plant Sci. 2020, 11, 349. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Matthias, T.; Aminov, R. Potential Effects of Horizontal Gene Exchange in the Human Gut. Front. Immunol. 2017, 8, 1630. [Google Scholar] [CrossRef]
- Basha, K.A.; Kumar, N.R.; Das, V.; Reshmi, K.; Rao, B.M.; Lalitha, K.V.; Joseph, T.C. Prevalence, molecular characterization, genetic heterogeneity and antimicrobial resistance of Listeria monocytogenes associated with fish and fishery environment in Kerala, India. Lett. Appl. Microbiol. 2019, 69, 286–293. [Google Scholar] [CrossRef]
- Bouymajane, A.; Rhazi Filali, F.; Oulghazi, S.; Lafkih, N.; Ed-Dra, A.; Aboulkacem, A.; El Allaoui, A.; Ouhmidou, B.; Moumni, M. Occurrence, antimicrobial resistance, serotyping and virulence genes of Listeria monocytogenes isolated from foods. Heliyon 2021, 7, e06169. [Google Scholar] [CrossRef]
- Skowron, K.; Kwiecińska-Piróg, J.; Grudlewska, K.; Świeca, A.; Paluszak, Z.; Bauza-Kaszewska, J.; Wałecka-Zacharska, E.; Gospodarek-Komkowska, E. The occurrence, transmission, virulence and antibiotic resistance of Listeria monocytogenes in fish processing plant. Int. J. Food Microbiol. 2018, 282, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, K.; Osek, J. Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from fresh and smoked fish in Poland. Food Microbiol. 2017, 64, 164–171. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of Antibiotic Resistance in Listeria monocytogenes Isolated from Food Products: A Comprehensive Review. Comp. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Andriyanov, P.A.; Zhurilov, P.A.; Liskova, E.A.; Karpova, T.I.; Sokolova, E.V.; Yushina, Y.K.; Zaiko, E.V.; Bataeva, D.S.; Voronina, O.L.; Psareva, E.K.; et al. Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from Humans, Animals, and Food Products in Russia in 1950–1980, 2000–2005, and 2018–2021. Antibiotics 2021, 10, 1206. [Google Scholar] [CrossRef]
- Baquero, F.; Lanza, V.; Duval, M.; Coque, T.M. Ecogenetics of antibiotic resistance in Listeria monocytogenes. Mol. Microbiol. 2020, 113, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.Y.; Pyla, R.; Kim, T.J.; Silva, J.L.; Jung, Y.S. Antibiotic resistance in Listeria species isolated from catfish fillets and processing environment. Lett. Appl. Microbiol. 2010, 50, 626–632. [Google Scholar] [CrossRef]
- Jamali, H.; Paydar, M.; Ismail, S.; Looi, C.Y.; Wong, W.F.; Radmehr, B.; Abedini, A. Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol. 2015, 15, 144. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, P.; Zakrzewski, A.J.; Zadernowska, A.; Chajęcka-Wierzchowska, W. Antimicrobial Resistance and Virulence Characterization of Listeria monocytogenes Strains Isolated from Food and Food Processing Environments. Pathogens 2022, 11, 1099. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Holý, O.; Bustamante, F.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Castillo, C.; Ovalle, C.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; et al. Virulence and Antibiotic Resistance Genes in Listeria monocytogenes Strains Isolated from Ready-to-Eat Foods in Chile. Front. Microbiol. 2022, 12, 796040. [Google Scholar] [CrossRef]
- Park, M.; Horn, L.; Lappi, V.; Boxrud, D.; Hedberg, C.; Jeon, B. Antimicrobial Synergy between Aminoglycosides and Licorice Extract in Listeria monocytogenes. Pathogens 2022, 11, 440. [Google Scholar] [CrossRef]
- Hanes, R.M.; Huang, Z. Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021. Int. J. Environ. Res. Public Health 2022, 19, 5506. [Google Scholar] [CrossRef]
- Wu, J.W.F.W.; Redondo-Solano, M.; Uribe, L.; WingChing-Jones, R.; Usaga, J.; Barboza, N. First characterization of the probiotic potential of lactic acid bacteria isolated from Costa Rican pineapple silages. PeerJ 2021, 9, e12437. [Google Scholar] [CrossRef]
- Odjadjare, E.E.O.; Okoh, A.I. Prevalence and distribution of Listeria pathogens in the final effluents of a rural wastewater treatment facility in the Eastern Cape Province of South Africa. World J. Microbiol. Biotechnol. 2010, 26, 297–307. [Google Scholar] [CrossRef]
- Su, X.; Zhang, S.; Shi, W.; Yang, X.; Li, Y.; Pan, H.; Kuang, D.; Xu, X.; Shi, X.; Meng, J. Molecular characterization and antimicrobial susceptibility of Listeria monocytogenes isolated from foods and humans. Food Control 2016, 70, 96–102. [Google Scholar] [CrossRef]
- Swetha, C.S.; Porteen, K.; Elango, A.; Ronald, B.S.M.; Senthil Kumar, T.M.A.; Milton, A.P.; Sureshkannan, S. Genetic diversity, virulence and distribution of antimicrobial resistance among Listeria monocytogenes isolated from milk, beef, and bovine farm environment. Iranian J. Vet. Res. 2021, 22, 1–8. [Google Scholar] [CrossRef]
- Painset, A.; Björkman, J.T.; Kiil, K.; Guillier, L.; Mariet, J.F.; Félix, B.; Amar, C.; Rotariu, O.; Roussel, S.; Perez-Reche, F.; et al. LiSEQ—Whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 2019, 5, e000257. [Google Scholar] [CrossRef] [PubMed]
- Abril, A.G.; Carrera, M.; Böhme, K.; Barros-Velázquez, J.; Calo-Mata, P.; Sánchez-Pérez, A.; Villa, T.G. Proteomic Characterization of Antibiotic Resistance in Listeria and Production of Antimicrobial and Virulence Factors. Int. Mol. Sci. 2021, 22, 8141. [Google Scholar] [CrossRef] [PubMed]
- Osman, K.M.; Kappell, A.D.; Fox, E.M.; Orabi, A.; Samir, A. Prevalence, Pathogenicity, Virulence, Antibiotic Resistance, and Phylogenetic Analysis of Biofilm-Producing Listeria monocytogenes Isolated from Different Ecological Niches in Egypt: Food, Humans, Animals, and Environment. Pathogens 2019, 9, 5. [Google Scholar] [CrossRef] [PubMed]

| Antibiotic Class | Genes | Nucleotide Sequences | Amplicon Sizes | M/U | References |
|---|---|---|---|---|---|
| Aminoglycosides (CN; KAN) | aadA | F: GTGGATGGCGGCCTGAAGCC R: AATGCCCAGTCGGCAGCG | 525 bp | M | [33] |
| strA | F: CTTGGTGATAACGGCAATTC R: CCAATCGCAGATAGAAGGC | 348 bp | |||
| aacC2 | F: CGGAAGGCAATAACGGAG R: TCGAACAGGTAGCACTGAG | 428 bp | M | [34] | |
| aphA1 | F: ATGGGCTCGCGATAATGTC R: CTCACCGAGGCAGTTCCAT | 600 bp | |||
| aphA2 | F: GAACAAGATGGATTGCACGC R: GCTCTTCAGCAATATCACGG | 510 bp | |||
| Lincomycin (CLI) | ermA | F: GTTCAAGAACAATCAATACAGGAG R: GGATCAGGAAAAGGACATTTTAC | 421 bp | M | [35] |
| ermC | F: GCTAATATTGTTTAAATCGTCAATTCC R: GGATCAGGAAAAGGACATTTTAC | 572 bp | |||
| ermB | F: GAAAAGGTACTCAACCAAATA R: AGTAACGGTACTTAAATTGTTTAC | 639 bp | U | [36] | |
| Macrolides (C; E) | cmlA1 | F: CACCAATCATGACCAAG R: GGCATCACTCGGCATGGACATG | 115 bp | U | [37] |
| catI | F: AGTTGCTCAATGTACCTATAACC R: TTGTAATTCATTAAGCATTCTGCC | 320 bp | M | [33] | |
| catII | F: ACACTTTGCCCTTTATCGTC R: TGAAAGCCATCACATACTGC | 543 bp | |||
| Beta-lactams (AMP; AMS; AUG) | ampC | F: TTCTATCAAMACTGGCARCC R: CCYTTTTATGTACCCAYGA | 550 bp | U | [32] |
| blaTEM | F: TTTCGTGTCGCCCTTATTCC R: CCGGCTCCAGATTTATCAGC | 690 bp | U | [37] | |
| blaZ | F: ACTTCAACACCTGCTGCTTTC R: TGACCACTTTTATCAGCAACC | 490 bp | U | [38] | |
| Oxazolidinone (LNZ) | cfr | F: TGAAGTATAAAGCAGGTTGGGAGTC R: AACCATATAATTGACCACAAGCAGC | 746 bp | M | [39] |
| optrA | F: TACTTGATGAACCTACTAACCA R: CCTTGAACTACTGATTCTCGG | 422 bp | |||
| poxtA | F: AAAGCTACCCATAAAATATC R: TCATCAAGCTGTTCGAGTTC | 533 bp | |||
| Tetracycline (DXT; TET) | tetA | F: GCTACATCCTGCTTGCCTTC R: CATAGATCGCCGTGAAGAGG | 210 bp | M | [40] |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | 659 bp | |||
| tetC | F: CTTGAGAGCCTTCAACCCAG R: ATGGTCGTCATCTACCTGCC | 418 bp | |||
| tetD | F: AAA CCA TTA CGG CAT TCT GC R: GACCGGATACACCATCCATC | 787 bp | |||
| tetS | F: CATAGACAAGCCGTTGACC R: ATGTTTTTGGAACGCCAGAG | 667 bp | |||
| tetM(1) | F: GGTACTTGAAAAGAACGGGAG R: TTCACCTTAGTATTTTTCCACTG | 630 bp | M | [41] | |
| tetM(2) | F: GGTACTTGAAAAGAACGGGAG R: ATACGAGTTTGTGCTTGTACGCC | 740 bp | |||
| tetK | F: TATTTTGGCTTTGTATTCTTTCAT R: GCTATACCTGTTCCCTCTGATAA | 1159 bp | M | [42] | |
| tetL | F: ATAAATTGTTTCGGGTCGGTAAT R: AACCAGCCAACTAATGACAATGAT | 1077 bp | |||
| tetO | F: AACTTAGGCATTCTGGCTCAC R: TCCCACTGTTCCATATCGTCA | 519 bp | U | [43,44] | |
| Glycopeptide (VAN) | vanA | F: GCAAGTCAGGTGAAGATGGA R: GCTAATACGATCAAGCGGTC | 171 bp | M | [45] |
| vanB | F: GATGTGTCGGTAAAATCCGC R: CCACTTCGCCGACAATCAAA | 271 bp | |||
| vanC1 | F: GTATCAAGGAAACCTCGCGA R: CGTAGGATAACCCGACTTCC A | 836 bp | |||
| vanC2 | F: GCAAACGTTGGTACCTGATG R: GGTGATTTTGGCGCTGATCA | 523 bp | |||
| vanD | F: TGGAATCACAAAATCCGGCG R: TWCCCGCATTTTTCACAACS | 311 bp | |||
| vanM | F: GGCAGAGATTGCCAACAACA R: AGGTAAACGAATCTGCCGCT | 425 bp | |||
| vanN | F: CCTCAAATCAGCAGCTAGTG R: GCTCCTGATAAGTGATACCC | 941 bp |
| Sources | N. Strains | Resistant Strains | Antibiotic Molecules | |
|---|---|---|---|---|
| Food matrices 84/120 | Fresh salmon | 10/84 | 3/10 | K |
| 2/10 | AMP, VAN | |||
| 1/10 | C | |||
| 1/10 | CN | |||
| Smoked salmon | 74/84 | 30/74 | E | |
| 19/74 | LNZ | |||
| 15/74 | VAN | |||
| 13/74 | CN, TET | |||
| 12/74 | K | |||
| 11/74 | RD | |||
| 10/74 | DXT | |||
| 0/74 | CLI | |||
| 4/74 | AMP | |||
| 2/74 | AUG | |||
| 1/74 | AMS, C | |||
| Environment 36/120 | Surfaces | 19/36 | 4/19 | E, TET |
| 1/19 | AMP | |||
| Knives | 4/36 | 3/4 | E | |
| 1/4 | TET | |||
| Machines | 13/36 | 4/13 | TET | |
| 3/13 | E, LNZ | |||
| 1/13 | DXT, VAN | |||
| Isolates | N. L. monocytogenes | Sources | Serovars |
|---|---|---|---|
| n. 120 L. monocytogenes strains | n. 10 | Fresh salmon | n. 5 serovars 4d |
| n. 5 serovars 1/2c | |||
| n. 74 | Smoked salmon | n. 39 serovars 1/2a | |
| n. 19 serovars 1/2b | |||
| n. 6 serovars 4e | |||
| n. 5 serovars 4b | |||
| n. 5 serovars 4d | |||
| n. 19 | Surfaces | n. 18 serovars 1/2a | |
| n. 1 serovar 1/2c | |||
| n. 4 | Knives | n. 4 serovars 1/2a | |
| n. 13 | Machines | n. 9 serovars 1/2a | |
| n. 3 serovars 1/2c | |||
| n. 1 serovar 4b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferri, G.; Lauteri, C.; Festino, A.R.; Vergara, A. ARGs Detection in Listeria Monocytogenes Strains Isolated from the Atlantic Salmon (Salmo salar) Food Industry: A Retrospective Study. Microorganisms 2023, 11, 1509. https://doi.org/10.3390/microorganisms11061509
Ferri G, Lauteri C, Festino AR, Vergara A. ARGs Detection in Listeria Monocytogenes Strains Isolated from the Atlantic Salmon (Salmo salar) Food Industry: A Retrospective Study. Microorganisms. 2023; 11(6):1509. https://doi.org/10.3390/microorganisms11061509
Chicago/Turabian StyleFerri, Gianluigi, Carlotta Lauteri, Anna Rita Festino, and Alberto Vergara. 2023. "ARGs Detection in Listeria Monocytogenes Strains Isolated from the Atlantic Salmon (Salmo salar) Food Industry: A Retrospective Study" Microorganisms 11, no. 6: 1509. https://doi.org/10.3390/microorganisms11061509
APA StyleFerri, G., Lauteri, C., Festino, A. R., & Vergara, A. (2023). ARGs Detection in Listeria Monocytogenes Strains Isolated from the Atlantic Salmon (Salmo salar) Food Industry: A Retrospective Study. Microorganisms, 11(6), 1509. https://doi.org/10.3390/microorganisms11061509






