The Microbiome of Things: Appliances, Machines, and Devices Hosting Artificial Niche-Adapted Microbial Communities
Abstract
1. Microorganisms: From General to Specialized Metabolisms
2. Microbiomes on Artificial Devices
2.1. Microbial Diversity of Sun-Exposed Artificial Outdoor Surfaces
2.2. Microbial Diversity of Indoor Artificial Devices and Appliances
| Artificial Devices | Extreme Condition of the Device | Most Abundant Genera | Ref. |
|---|---|---|---|
| Photovoltaic panels | UV-irradiation Desiccation Low nutrition | Bacteria: Hymenobacter Sphingomonas Deinococcus Methylobacterium Roseomonas Novosphingobium | [20,21,24,25,26] |
| Coffee machines | Thermal stress Water pressure Particular nutrient availability Alkaloids (caffeine) | Bacteria: Enterococcus Pseudomonas Stenotrophomonas Sphingobacterium Acinetobacter Coprococcus Paenibacillus Agrobacterium Sphingobium | [36] |
| Dishwashers | Thermal stress High salt concentrations Presence of detergents pH variations Water pressure | Fungi: Ascomycota Basidiomycota Bacteria: Gordonia Micrococcus Chryseobacterium Exiguobacterium Meiothermus Staphylococcus Streptococcus Lactobacillus Corynebacterium Enterococcus Acinetobacter Escherichia/Shigella Pseudomonas | [39,40] |
| Washing machines | Thermal stress Presence of detergents pH variations Constant water pressure | Bacteria: Pseudomonas Enhydrobacter Leptospira Sphingomonas Legionella Moraxellaceae family Acinetobacter | [55,56,57,58] |
| Water heating systems | High temperature Conductivity and oxygen concentration | Bacteria: Thermus Acidovorax Agrobacterium Roseococcus Flavobacterium Sphingomonas Brochothrix Buchnera Polynucleobacter Ralstonia Thermicanus Parascardovia Micrococcus Rothia Brachybacterium Methylobacterium Sejonia Moraxellaceae family | [59,61,75] |
| Saunas | High temperature Low nutrient | Bacteria: Bacillus Virgibacillus Tepidomonas Pseudoxanthomonas Stenotrophomonas Janthinobacterium Aquaspirillum Chromobacterium Aquabacterium Gulbenkiania Pelomonas Aquitalea Deinococcus Moraxellaceae family | [62,63,64] |
| Refrigerators | Low temperature | Fungi: Saccharomyces Candida Bacteria: Pseudomonas Pantoea Bacillus Acinetobacter Enterococcus Citrobacter Exiguobacterium Staphylococcus Enterobacter | [65,66] |
| Air conditioning systems | Low temperature Low nutrient | Fungi: Malassezia Cladosporium Leotiomycetes Bacteria: Agaricomycetes Pseudomonas Sphingomonas Propionibacterium Methylobacterium Enhydrobacter Moraxellaceae family Perlucidibaca Acinetobacter | [69,70] |
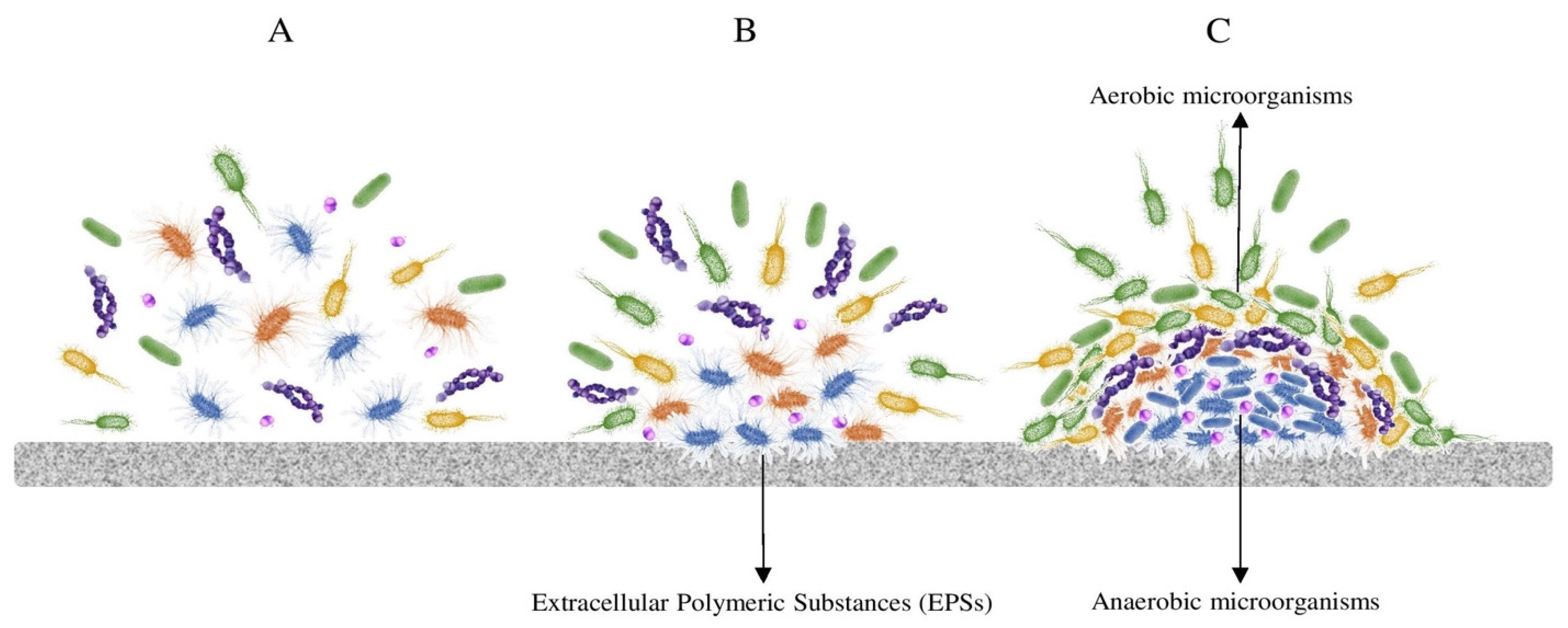
3. How to Live in a Machine: Microbial Adaptations
4. Biotechnological Potential of the Microbiome of Things
5. As Conclusions: The Microbiome of Things
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lennon, J.T.; Locey, K.J. More Support for Earth’s Massive Microbiome. Biol. Direct 2020, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Gupta, R.; Singh, R.L. Microbes and Environment. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Singh, R.L., Ed.; Springer: Singapore, 2017; pp. 43–84. ISBN 978-981-10-1866-4. [Google Scholar]
- Chandra, N.; Kumar, S. Antibiotics Producing Soil Microorganisms. In Antibiotics and Antibiotics Resistance Genes in Soils: Monitoring, Toxicity, Risk Assessment and Management; Hashmi, M.Z., Strezov, V., Varma, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–18. ISBN 978-3-319-66260-2. [Google Scholar]
- Ramakrishna, W.; Yadav, R.; Li, K. Plant Growth Promoting Bacteria in Agriculture: Two Sides of a Coin. Appl. Soil Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]
- Christofi, N.; Philp, J.C. European Microbiology Related to the Subsurface Disposal of Nuclear Waste. In The Microbiology of the Terrestrial Deep Subsurface; CRC Press: Boca Raton, FL, USA, 2018; pp. 267–298. ISBN 1-351-07456-3. [Google Scholar]
- Lee, H.B.; Jeong, D.H.; Cho, B.C.; Park, J.S. The Diversity Patterns of Rare to Abundant Microbial Eukaryotes Across a Broad Range of Salinities in a Solar Saltern. Microb. Ecol. 2021, 84, 1103–1121. [Google Scholar] [CrossRef]
- Sharma, P.; Pandey, A.K.; Kim, S.-H.; Singh, S.P.; Chaturvedi, P.; Varjani, S. Critical Review on Microbial Community during In-Situ Bioremediation of Heavy Metals from Industrial Wastewater. Environ. Technol. Innov. 2021, 24, 101826. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, W.; Zhang, Z.; Grossart, H.-P.; Gadd, G.M. Microplastics Provide New Microbial Niches in Aquatic Environments. Appl. Microbiol. Biotechnol. 2020, 104, 6501–6511. [Google Scholar] [CrossRef] [PubMed]
- Bahl, S.; Dolma, J.; Jyot Singh, J.; Sehgal, S. Biodegradation of Plastics: A State of the Art Review. Mater. Today Proc. 2021, 39, 31–34. [Google Scholar] [CrossRef]
- Weaver, J.L.; DePriest, P.T.; Plymale, A.E.; Pearce, C.I.; Arey, B.; Koestler, R.J. Microbial Interactions with Silicate Glasses. Npj Mater. Degrad. 2021, 5, 11. [Google Scholar] [CrossRef]
- Lekbach, Y.; Liu, T.; Li, Y.; Moradi, M.; Dou, W.; Xu, D.; Smith, J.A.; Lovley, D.R. Microbial Corrosion of Metals-the Corrosion Microbiome. Adv. Microb. Physiol. 2021, 78, 317–390. [Google Scholar]
- Satari, L.; Guillén, A.; Vidal-Verdú, À.; Porcar, M. The Wasted Chewing Gum Bacteriome. Sci. Rep. 2020, 10, 16846. [Google Scholar] [CrossRef]
- Ghatge, S.; Yang, Y.; Ahn, J.-H.; Hur, H.-G. Biodegradation of Polyethylene: A Brief Review. Appl. Biol. Chem. 2020, 63, 27. [Google Scholar] [CrossRef]
- Jeon, J.-M.; Park, S.-J.; Choi, T.-R.; Park, J.-H.; Yang, Y.-H.; Yoon, J.-J. Biodegradation of Polyethylene and Polypropylene by Lysinibacillus Species JJY0216 Isolated from Soil Grove. Polym. Degrad. Stab. 2021, 191, 109662. [Google Scholar] [CrossRef]
- Richardson, P.M.; Gottel, N.; Gilbert, J.A.; Lax, S.; Bailey, M.J. Microbial Similarity between Students in a Common Dormitory Environment Reveals the Forensic Potential of Individual Microbial Signatures. mBio 2019, 10, e01054-19. [Google Scholar] [CrossRef] [PubMed]
- Hernández, A.M.; Vargas-Robles, D.; Alcaraz, L.D.; Peimbert, M. Station and Train Surface Microbiomes of Mexico City’s Metro (Subway/Underground). Sci. Rep. 2020, 10, 8798. [Google Scholar] [CrossRef] [PubMed]
- Klimenko, N.S.; Tyakht, A.V.; Toshchakov, S.V.; Shevchenko, M.A.; Korzhenkov, A.A.; Afshinnekoo, E.; Mason, C.E.; Alexeev, D.G. Co-Occurrence Patterns of Bacteria within Microbiome of Moscow Subway. Comput. Struct. Biotechnol. J. 2020, 18, 314–322. [Google Scholar] [CrossRef]
- Hartmann, B.; Benson, M.; Junger, A.; Quinzio, L.; Röhrig, R.; Fengler, B.; Färber, U.W.; Wille, B.; Hempelmann, G. Computer Keyboard and Mouse as a Reservoir of Pathogens in an Intensive Care Unit. J. Clin. Monit. Comput. 2004, 18, 7–12. [Google Scholar] [CrossRef]
- Nepomuceno, D.B.; Lima, D.V.; Silva, M.O.; Porto, J.C.S.; Mobin, M. Evaluation of Disinfectants in Order to Eliminate Fungal Contamination in Computer Keyboards of an Integrated Health Center in Piauí, Brazil. Environ. Monit. Assess. 2018, 190, 608. [Google Scholar] [CrossRef]
- Tanner, K.; Martí, J.M.; Belliure, J.; Fernández-Méndez, M.; Molina-Menor, E.; Peretó, J.; Porcar, M. Polar Solar Panels: Arctic and Antarctic Microbiomes Display Similar Taxonomic Profiles. Environ. Microbiol. Rep. 2018, 10, 75–79. [Google Scholar] [CrossRef]
- Moura, J.B.; Delforno, T.P.; do Prado, P.F.; Duarte, I.C. Extremophilic Taxa Predominate in a Microbial Community of Photovoltaic Panels in a Tropical Region. FEMS Microbiol. Lett. 2021, 368, fnab105. [Google Scholar] [CrossRef]
- Vidal-Verdú, À.; Gómez-Martínez, D.; Latorre-Pérez, A.; Peretó, J.; Porcar, M. The Car Tank Lid Bacteriome: A Reservoir of Bacteria with Potential in Bioremediation of Fuel. Npj Biofilms Microbiomes 2022, 8, 32. [Google Scholar] [CrossRef]
- Park, C.; Jung, H.S.; Park, S.; Jeon, C.O.; Park, W.; McMahon, K. Dominance of Gas-Eating, Biofilm-Forming Methylobacterium Species in the Evaporator Cores of Automobile Air-Conditioning Systems. mSphere 2019, 5, e00761-19. [Google Scholar] [CrossRef]
- Porcar, M.; Louie, K.B.; Kosina, S.M.; Van Goethem, M.W.; Bowen, B.P.; Tanner, K.; Northen, T.R. Microbial Ecology on Solar Panels in Berkeley, CA, United States. Front. Microbiol. 2018, 9, 3043. [Google Scholar] [CrossRef]
- Dorado-Morales, P.; Vilanova, C.; Peretó, J.; Codoñer, F.M.; Ramón, D.; Porcar, M. A Highly Diverse, Desert-like Microbial Biocenosis on Solar Panels in a Mediterranean City. Sci. Rep. 2016, 6, 29235. [Google Scholar] [CrossRef] [PubMed]
- Tanner, K.; Molina-Menor, E.; Latorre-Pérez, A.; Vidal-Verdú, À.; Vilanova, C.; Peretó, J.; Porcar, M. Extremophilic Microbial Communities on Photovoltaic Panel Surfaces: A Two-year Study. Microb. Biotechnol. 2020, 13, 1819–1830. [Google Scholar] [CrossRef] [PubMed]
- Tanner, K.; Mancuso, C.P.; Peretó, J.; Khalil, A.S.; Vilanova, C.; Pascual, J. Sphingomonas Solaris sp. nov., Isolated from a Solar Panel in Boston, Massachusetts. Int. J. Syst. Evol. Microbiol. 2020, 70, 1814–1821. [Google Scholar] [CrossRef] [PubMed]
- Romani, M.; Carrion, C.; Fernandez, F.; Intertaglia, L.; Pecqueur, D.; Lebaron, P.; Lami, R. High Bacterial Diversity in Pioneer Biofilms Colonizing Ceramic Roof Tiles. Int. Biodeterior. Biodegrad. 2019, 144, 104745. [Google Scholar] [CrossRef]
- Gutman, J.; Kaufman, Y.; Kawahara, K.; Walker, S.L.; Freger, V.; Herzberg, M. Interactions of Glycosphingolipids and Lipopolysaccharides with Silica and Polyamide Surfaces: Adsorption and Viscoelastic Properties. Biomacromolecules 2014, 15, 2128–2137. [Google Scholar] [CrossRef]
- Olsen, M.; Nassar, R.; Senok, A.; Albastaki, A.; Leggett, J.; Lohning, A.; Campos, M.; Jones, P.; McKirdy, S.; Tajouri, L.; et al. A Pilot Metagenomic Study Reveals That Community Derived Mobile Phones Are Reservoirs of Viable Pathogenic Microbes. Sci. Rep. 2021, 11, 14102. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current Understanding of the Human Microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Fujiyoshi, S.; Tanaka, D.; Maruyama, F. Transmission of Airborne Bacteria across Built Environments and Its Measurement Standards: A Review. Front. Microbiol. 2017, 8, 2336. [Google Scholar] [CrossRef]
- Rai, S.; Singh, D.K.; Kumar, A. Microbial, Environmental and Anthropogenic Factors Influencing the Indoor Microbiome of the Built Environment. J. Basic Microbiol. 2021, 61, 267–292. [Google Scholar] [CrossRef] [PubMed]
- Lax, S.; Smith, D.P.; Hampton-Marcell, J.; Owens, S.M.; Handley, K.M.; Scott, N.M.; Gibbons, S.M.; Larsen, P.; Shogan, B.D.; Weiss, S.; et al. Longitudinal Analysis of Microbial Interaction between Humans and the Indoor Environment. Science 2014, 345, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Vilanova, C.; Iglesias, A.; Porcar, M. The Coffee-Machine Bacteriome: Biodiversity and Colonisation of the Wasted Coffee Tray Leach. Sci. Rep. 2015, 5, 17163. [Google Scholar] [CrossRef] [PubMed]
- Zalar, P.; Novak, M.; de Hoog, G.S.; Gunde-Cimerman, N. Dishwashers—A Man-Made Ecological Niche Accommodating Human Opportunistic Fungal Pathogens. Fungal Biol. 2011, 115, 997–1007. [Google Scholar] [CrossRef]
- Zupančič, J.; Raghupathi, P.K.; Houf, K.; Burmølle, M.; Sørensen, S.J.; Gunde-Cimerman, N. Synergistic Interactions in Microbial Biofilms Facilitate the Establishment of Opportunistic Pathogenic Fungi in Household Dishwashers. Front. Microbiol. 2018, 9, 21. [Google Scholar] [CrossRef]
- Raghupathi, P.K.; Zupančič, J.; Brejnrod, A.D.; Jacquiod, S.; Houf, K.; Burmølle, M.; Gunde-Cimerman, N.; Sørensen, S.J.; Dudley, E.G. Microbial Diversity and Putative Opportunistic Pathogens in Dishwasher Biofilm Communities. Appl. Environ. Microbiol. 2018, 84, e02755-17. [Google Scholar] [CrossRef]
- Zupančič, J.; Turk, M.; Črnigoj, M.; Ambrožič, A.J.; Gunde-Cimerman, N. The Dishwasher Rubber Seal Acts as a Reservoir of Bacteria in the Home Environment. BMC Microbiol. 2019, 19, 300. [Google Scholar] [CrossRef]
- Döğen, A.; Kaplan, E.; Öksüz, Z.; Serin, M.S.; Ilkit, M.; de Hoog, G.S. Dishwashers Are a Major Source of Human Opportunistic Yeast-like Fungi in Indoor Environments in Mersin, Turkey. Med. Mycol. 2013, 51, 493–498. [Google Scholar] [CrossRef]
- Gümral, R.; Özhak-Baysan, B.; Tümgör, A.; Saraçlı, M.A.; Yıldıran, Ş.T.; Ilkit, M.; Zupančič, J.; Novak-Babič, M.; Gunde-Cimerman, N.; Zalar, P.; et al. Dishwashers Provide a Selective Extreme Environment for Human-Opportunistic Yeast-like Fungi. Fungal Divers. 2016, 76, 1–9. [Google Scholar] [CrossRef]
- Zupančič, J.; Novak Babič, M.; Zalar, P.; Gunde-Cimerman, N. The Black Yeast Exophiala Dermatitidis and Other Selected Opportunistic Human Fungal Pathogens Spread from Dishwashers to Kitchens. PLoS ONE 2016, 11, e0148166. [Google Scholar] [CrossRef]
- Kalyani, D.; Munk, L.; Mikkelsen, J.; Meyer, A. Molecular and Biochemical Characterization of a New Thermostable Bacterial Laccase from Meiothermus Ruber DSM 1279. Rsc Adv. 2016, 6, 3910–3918. [Google Scholar] [CrossRef]
- Yolmeh, M.; Khomeiri, M.; Ghorbani, M.; Ghaemi, E.; Ramezanpour, S.S. High Efficiency Pigment Production from Micrococcus Roseus (PTCC 1411) under Ultraviolet Irradiation. Biocatal. Agric. Biotechnol. 2017, 9, 156–161. [Google Scholar] [CrossRef]
- Gurbanov, R.; Tunçer, S.; Mingu, S.; Severcan, F.; Gozen, A.G. Methylation, Sugar Puckering and Z-Form Status of DNA from a Heavy Metal-Acclimated Freshwater Gordonia sp. J. Photochem. Photobiol. B Biol. 2019, 198, 111580. [Google Scholar] [CrossRef]
- Pandey, N. Chapter 10—Exiguobacterium. In Beneficial Microbes in Agro-Ecology; Amaresan, N., Senthil Kumar, M., Annapurna, K., Kumar, K., Sankaranarayanan, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 169–183. ISBN 978-0-12-823414-3. [Google Scholar]
- Flemming, H.-C.; Wingender, J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Kılıç, N.K.; Dönmez, G. Environmental Conditions Affecting Exopolysaccharide Production by Pseudomonas Aeruginosa, Micrococcus sp., and Ochrobactrum sp. J. Hazard. Mater. 2008, 154, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Fusconi, R.; Maria Nascimento Assunção, R.; de Moura Guimarães, R.; Rodrigues Filho, G.; Eduardo da Hora Machado, A. Exopolysaccharide Produced by Gordonia Polyisoprenivorans CCT 7137 in GYM Commercial Medium and Sugarcane Molasses Alternative Medium: FT-IR Study and Emulsifying Activity. Carbohydr. Polym. 2010, 79, 403–408. [Google Scholar] [CrossRef]
- Hu, X.; Pan, Y.; Bao, M.; Zhang, X.; Luo, C.; Han, X.; Li, F. The Structure, Properties and Rheological Characterisation of Exopolysaccharides Produced by Chryseobacterium Cucumeris AP-2 from Deteriorated Milk. Int. Dairy J. 2022, 126, 105253. [Google Scholar] [CrossRef]
- Idris, F.N.; Nadzir, M.M. Application of Microbes in Household Products. In Application of Microbes in Environmental and Microbial Biotechnology; Inamuddin, Ahamed, M.I., Prasad, R., Eds.; Springer: Singapore, 2022; pp. 219–233. ISBN 978-981-16-2225-0. [Google Scholar]
- Babič, M.N.; Zalar, P.; Ženko, B.; Schroers, H.-J.; Džeroski, S.; Gunde-Cimerman, N. Candida and Fusarium Species Known as Opportunistic Human Pathogens from Customer-Accessible Parts of Residential Washing Machines. Fungal Biol. 2015, 119, 95–113. [Google Scholar] [CrossRef]
- Nix, I.D.; Frontzek, A.; Bockmühl, D.P. Characterization of Microbial Communities in Household Washing Machines. Tenside Surfactants Deterg. 2015, 52, 432–440. [Google Scholar] [CrossRef]
- Jacksch, S.; Kaiser, D.; Weis, S.; Weide, M.; Ratering, S.; Schnell, S.; Egert, M. Influence of Sampling Site and Other Environmental Factors on the Bacterial Community Composition of Domestic Washing Machines. Microorganisms 2020, 8, 30. [Google Scholar] [CrossRef]
- Callewaert, C.; Van Nevel, S.; Kerckhof, F.-M.; Granitsiotis, M.S.; Boon, N. Bacterial Exchange in Household Washing Machines. Front. Microbiol. 2015, 6, 1381. [Google Scholar] [CrossRef] [PubMed]
- Kubota, H.; Mitani, A.; Niwano, Y.; Takeuchi, K.; Tanaka, A.; Yamaguchi, N.; Kawamura, Y.; Hitomi, J. Moraxella Species Are Primarily Responsible for Generating Malodor in Laundry. Appl. Environ. Microbiol. 2012, 78, 3317–3324. [Google Scholar] [CrossRef] [PubMed]
- Bockmühl, D.P.; Schages, J.; Rehberg, L. Laundry and Textile Hygiene in Healthcare and Beyond. Microb Cell 2019, 6, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Brock, T.D.; Boylen, K.L. Presence of Thermophilic Bacteria in Laundry and Domestic Hot-Water Heaters. Appl. Microbiol. 1973, 25, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Ram, S.; Mitra, M.; Shah, F.; Tirkey, S.R.; Mishra, S. Bacteria as an Alternate Biofactory for Carotenoid Production: A Review of Its Applications, Opportunities and Challenges. J. Funct. Foods 2020, 67, 103867. [Google Scholar] [CrossRef]
- Kjellerup, B.; Thomsen, T.; Nielsen, J.; Olesen, B.; Frølund, B.; Nielsen, P. Microbial Diversity in Biofilms from Corroding Heating Systems. Biofouling 2005, 21, 19–29. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, D.H. Characterization of Bacterial Community Contaminating Floor of A Hot and Dry Sauna. J. Bacteriol. Virol. 2012, 42, 313–320. [Google Scholar] [CrossRef][Green Version]
- Kim, B.S.; Seo, J.R.; Park, D.H. Variation and Characterization of Bacterial Communities Contaminating Two Saunas Operated at 64 °C and 76 °C. J. Bacteriol. Virol. 2013, 43, 195–203. [Google Scholar] [CrossRef][Green Version]
- Tanner, K.; Vilanova, C.; Porcar, M. Bioprospecting Challenges in Unusual Environments. Microb. Biotechnol. 2017, 10, 671–673. [Google Scholar] [CrossRef]
- Jeon, Y.-S.; Chun, J.; Kim, B.-S. Identification of Household Bacterial Community and Analysis of Species Shared with Human Microbiome. Curr. Microbiol. 2013, 67, 557–563. [Google Scholar] [CrossRef]
- Ye, K.; Wang, J.; Han, Y.; Wang, C.; Qi, C.; Ge, X. Investigation on Microbial Contamination in the Cold Storage Room of Domestic Refrigerators. Food Control 2019, 99, 64–67. [Google Scholar] [CrossRef]
- Schmidt, M.G.; Attaway, H.H.; Terzieva, S.; Marshall, A.; Steed, L.L.; Salzberg, D.; Hamoodi, H.A.; Khan, J.A.; Feigley, C.E.; Michels, H.T. Characterization and Control of the Microbial Community Affiliated with Copper or Aluminum Heat Exchangers of HVAC Systems. Curr. Microbiol. 2012, 65, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Bakker, A.; Siegel, J.A.; Mendell, M.J.; Peccia, J. Building and Environmental Factors That Influence Bacterial and Fungal Loading on Air Conditioning Cooling Coils. Indoor Air 2018, 28, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Acerbi, E.; Chénard, C.; Miller, D.; Gaultier, N.E.; Heinle, C.E.; Chang, V.W.-C.; Uchida, A.; Drautz-Moses, D.I.; Schuster, S.C.; Lauro, F.M. Ecological Succession of the Microbial Communities of an Air-Conditioning Cooling Coil in the Tropics. Indoor Air 2017, 27, 345–353. [Google Scholar] [CrossRef]
- Bakker, A.; Siegel, J.A.; Mendell, M.J.; Prussin, A.J.; Marr, L.C.; Peccia, J. Bacterial and Fungal Ecology on Air Conditioning Cooling Coils Is Influenced by Climate and Building Factors. Indoor Air 2020, 30, 326–334. [Google Scholar] [CrossRef]
- Hugenholtz, P. An Ecological and Phylogenetic Investigation of a Bacterial Biofilm Growing on the Cooling Coils in an Air-Handling System. Ph.D. Thesis, The University of Queensland, Brisbane, Australia, 1994. [Google Scholar]
- Simmons, R.B.; Rose, L.J.; Crow, S.A.; Ahearn, D.G. The Occurrence and Persistence of Mixed Biofilms in Automobile Air Conditioning Systems. Curr. Microbiol. 1999, 39, 141–145. [Google Scholar] [CrossRef]
- Hanson, B.; Zhou, Y.; Bautista, E.J.; Urch, B.; Speck, M.; Silverman, F.; Muilenberg, M.; Phipatanakul, W.; Weinstock, G.; Sodergren, E.; et al. Characterization of the Bacterial and Fungal Microbiome in Indoor Dust and Outdoor Air Samples: A Pilot Study. Environ. Sci. Process. Impacts 2016, 18, 713–724. [Google Scholar] [CrossRef]
- Wilson, S.C.; Palmatier, R.N.; Andriychuk, L.A.; Martin, J.M.; Jumper, C.A.; Holder, H.W.; Straus, D.C. Mold Contamination and Air Handling Units. J. Occup. Environ. Hyg. 2007, 4, 483–491. [Google Scholar] [CrossRef]
- Savage, A.M.; Hills, J.; Driscoll, K.; Fergus, D.J.; Grunden, A.M.; Dunn, R.R. Microbial Diversity of Extreme Habitats in Human Homes. PeerJ 2016, 4, e2376. [Google Scholar] [CrossRef]
- Tan, C.H.; Lee, K.W.K.; Burmølle, M.; Kjelleberg, S.; Rice, S.A. All Together Now: Experimental Multispecies Biofilm Model Systems. Environ. Microbiol. 2017, 19, 42–53. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G.M. Geomicrobiology of the Built Environment. Nat. Microbiol. 2017, 2, 16275. [Google Scholar] [CrossRef] [PubMed]
- Milanesi, C.; Cresti, M.; Costantini, L.; Gallo, M.; Gallo, G.; Crognale, S.; Faleri, C.; Gradi, A.; Franco, B. Spoilage of Oat Bran by Sporogenic Microorganisms Revived from Soil Buried 4000 Years Ago in Iranian Archaeological Site. Int. Biodeterior. Biodegrad. 2015, 104, 83–91. [Google Scholar] [CrossRef]
- Checinska, A.; Probst, A.J.; Vaishampayan, P.; White, J.R.; Kumar, D.; Stepanov, V.G.; Fox, G.E.; Nilsson, H.R.; Pierson, D.L.; Perry, J.; et al. Microbiomes of the Dust Particles Collected from the International Space Station and Spacecraft Assembly Facilities. Microbiome 2015, 3, 50. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, L.A.C.; Karp, S.G.; Vendruscolo, F.; Kanno, K.Y.F.; Zoz, L.I.C.; Carvalho, J.C. Biotechnological Production of Carotenoids and Their Applications in Food and Pharmaceutical Products. In Carotenoids; Cvetkovic, D.J., Nikolic, G.S., Eds.; InTech: London, UK, 2017; ISBN 978-953-51-3211-0. [Google Scholar]
- Alcaíno, J.; Baeza, M.; Cifuentes, V. Carotenoid Distribution in Nature. In Carotenoids in Nature: Biosynthesis, Regulation and Function; Stange, C., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–33. ISBN 978-3-319-39126-7. [Google Scholar]
- Yabuzaki, J. Carotenoids Database: Structures, Chemical Fingerprints and Distribution among Organisms. Database 2017, 2017, bax004. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.-H.; Elmadfa, I. Biological Relevance of Terpenoids. Ann. Nutr. Metab. 2003, 47, 95–106. [Google Scholar] [CrossRef]
- Galasso, C.; Corinaldesi, C.; Sansone, C. Carotenoids from Marine Organisms: Biological Functions and Industrial Applications. Antioxidants 2017, 6, 96. [Google Scholar] [CrossRef]
- Mueller, D.R.; Vincent, W.F.; Bonilla, S.; Laurion, I. Extremotrophs, Extremophiles and Broadband Pigmentation Strategies in a High Arctic Ice Shelf Ecosystem. FEMS Microbiol. Ecol. 2005, 53, 73–87. [Google Scholar] [CrossRef]
- Guan, N.; Li, J.; Shin, H.; Du, G.; Chen, J.; Liu, L. Microbial Response to Environmental Stresses: From Fundamental Mechanisms to Practical Applications. Appl. Microbiol. Biotechnol. 2017, 101, 3991–4008. [Google Scholar] [CrossRef]
- Tribelli, P.; López, N. Reporting Key Features in Cold-Adapted Bacteria. Life 2018, 8, 8. [Google Scholar] [CrossRef]
- Raymond-Bouchard, I.; Goordial, J.; Zolotarov, Y.; Ronholm, J.; Stromvik, M.; Bakermans, C.; Whyte, L.G. Conserved Genomic and Amino Acid Traits of Cold Adaptation in Subzero-Growing Arctic Permafrost Bacteria. FEMS Microbiol. Ecol. 2018, 94, fiy023. [Google Scholar] [CrossRef] [PubMed]
- Dasila, H.; Maithani, D.; Suyal, D.C.; Debbarma, P. Cold-Adapted Microorganisms: Survival Strategies and Biotechnological Significance. In Survival Strategies in Cold-Adapted Microorganisms; Goel, R., Soni, R., Suyal, D.C., Khan, M., Eds.; Springer: Singapore, 2022; pp. 357–378. ISBN 978-981-16-2625-8. [Google Scholar]
- Kanekar, P.P.; Kanekar, S.P. Psychrophilic, Psychrotrophic, and Psychrotolerant Microorganisms. In Diversity and Biotechnology of Extremophilic Microorganisms from India; Kanekar, P.P., Kanekar, S.P., Eds.; Springer Nature: Singapore, 2022; pp. 215–249. ISBN 978-981-19157-3-4. [Google Scholar]
- Laskowska, E.; Kuczyńska-Wiśnik, D. New Insight into the Mechanisms Protecting Bacteria during Desiccation. Curr. Genet. 2020, 66, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Esbelin, J.; Santos, T.; Hébraud, M. Desiccation: An Environmental and Food Industry Stress That Bacteria Commonly Face. Food Microbiol. 2018, 69, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Caggianiello, G.; Kleerebezem, M.; Spano, G. Exopolysaccharides Produced by Lactic Acid Bacteria: From Health-Promoting Benefits to Stress Tolerance Mechanisms. Appl. Microbiol. Biotechnol. 2016, 100, 3877–3886. [Google Scholar] [CrossRef]
- Siliakus, M.F.; van der Oost, J.; Kengen, S.W.M. Adaptations of Archaeal and Bacterial Membranes to Variations in Temperature, PH and Pressure. Extremophiles 2017, 21, 651–670. [Google Scholar] [CrossRef]
- Quatrini, R.; Johnson, D.B. Microbiomes in Extremely Acidic Environments: Functionalities and Interactions That Allow Survival and Growth of Prokaryotes at Low PH. Curr. Opin. Microbiol. 2018, 43, 139–147. [Google Scholar] [CrossRef]
- Wennerström, H.; Oliveberg, M. On the Osmotic Pressure of Cells. QRB Discov. 2022, 3, e12. [Google Scholar] [CrossRef]
- Wood, J.M. Bacterial Responses to Osmotic Challenges. J. Gen. Physiol. 2015, 145, 381–388. [Google Scholar] [CrossRef]
- Wood, J.M. Osmosensing by Bacteria. Sci. STKE 2006, 2006, pe43. [Google Scholar] [CrossRef]
- Muñoz, P.A.; Márquez, S.L.; González-Nilo, F.D.; Márquez-Miranda, V.; Blamey, J.M. Structure and Application of Antifreeze Proteins from Antarctic Bacteria. Microb. Cell Factories 2017, 16, 138. [Google Scholar] [CrossRef]
- Eskandari, A.; Leow, T.C.; Rahman, M.B.; Oslan, S.N. Antifreeze Proteins and Their Practical Utilization in Industry, Medicine, and Agriculture. Biomolecules 2020, 10, 1649. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Yang, X.; Ke, L.; Hu, Y. The Properties, Biotechnologies, and Applications of Antifreeze Proteins. Int. J. Biol. Macromol. 2020, 153, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.; Cho, Y.-J.; Kim, S.; Kim, O.-S.; Putonti, C. Draft Genome Sequence of the Psychrotolerant Bacterium Methylobacterium sp. Strain BTF04, Isolated from Freshwater in Antarctica. Microbiol. Resour. Announc. 2020, 9, e00171-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, H.-C.; Zhou, Y.-G.; Xin, Y.-H. Microevolution and Adaptive Strategy of Psychrophilic Species Flavobacterium Bomense sp. nov. Isolated from Glaciers. Front. Microbiol. 2019, 10, 1069. [Google Scholar] [CrossRef]
- Schultz, J.; Rosado, A.S. Extreme Environments: A Source of Biosurfactants for Biotechnological Applications. Extremophiles 2020, 24, 189–206. [Google Scholar] [CrossRef]
- Malavenda, R.; Rizzo, C.; Michaud, L.; Gerçe, B.; Bruni, V.; Syldatk, C.; Hausmann, R.; Lo Giudice, A. Biosurfactant Production by Arctic and Antarctic Bacteria Growing on Hydrocarbons. Polar Biol. 2015, 38, 1565–1574. [Google Scholar] [CrossRef]
- Perfumo, A.; Banat, I.M.; Marchant, R. Going Green and Cold: Biosurfactants from Low-Temperature Environments to Biotechnology Applications. Trends Biotechnol. 2018, 36, 277–289. [Google Scholar] [CrossRef]
- Eswari, J.S.; Dhagat, S.; Sen, R. Biosurfactants, Bioemulsifiers, and Biopolymers from Thermophilic Microorganisms. In Thermophiles for Biotech Industry: A Bioprocess Technology Perspective; Eswari, J.S., Dhagat, S., Sen, R., Eds.; Springer: Singapore, 2019; pp. 87–97. ISBN 978-981-329-919-1. [Google Scholar]
- Weil, T.; De Filippo, C.; Albanese, D.; Donati, C.; Pindo, M.; Pavarini, L.; Carotenuto, F.; Pasqui, M.; Poto, L.; Gabrieli, J.; et al. Legal Immigrants: Invasion of Alien Microbial Communities during Winter Occurring Desert Dust Storms. Microbiome 2017, 5, 32. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satari, L.; Iglesias, A.; Porcar, M. The Microbiome of Things: Appliances, Machines, and Devices Hosting Artificial Niche-Adapted Microbial Communities. Microorganisms 2023, 11, 1507. https://doi.org/10.3390/microorganisms11061507
Satari L, Iglesias A, Porcar M. The Microbiome of Things: Appliances, Machines, and Devices Hosting Artificial Niche-Adapted Microbial Communities. Microorganisms. 2023; 11(6):1507. https://doi.org/10.3390/microorganisms11061507
Chicago/Turabian StyleSatari, Leila, Alba Iglesias, and Manuel Porcar. 2023. "The Microbiome of Things: Appliances, Machines, and Devices Hosting Artificial Niche-Adapted Microbial Communities" Microorganisms 11, no. 6: 1507. https://doi.org/10.3390/microorganisms11061507
APA StyleSatari, L., Iglesias, A., & Porcar, M. (2023). The Microbiome of Things: Appliances, Machines, and Devices Hosting Artificial Niche-Adapted Microbial Communities. Microorganisms, 11(6), 1507. https://doi.org/10.3390/microorganisms11061507






