Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype
Abstract
:1. Introduction
2. Materials and Methods
2.1. WGS Data Retrieval
2.2. Genome Assembly and Annotations
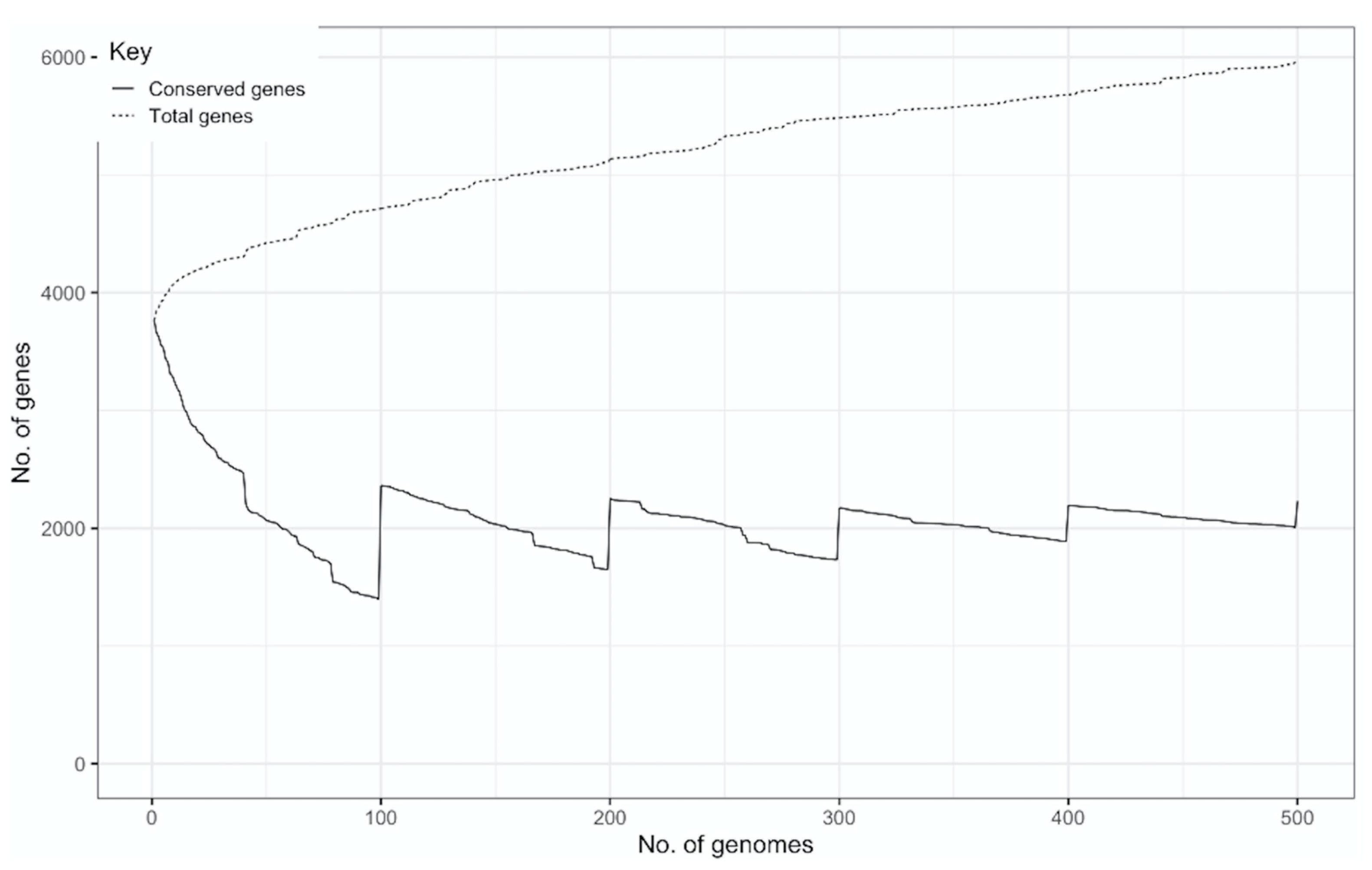
2.3. Pangenome Construction and Analysis
2.4. Phylogenetic Analysis
2.5. Statistical Analysis
3. Results
4. Discussion
| Analyzed Genomes | Core Genome | Accessory Genome Genes | Pangenome Open/Closed | Bioinformatic Tools | Reference |
|---|---|---|---|---|---|
| 36 | 3679 | 2086 | Open | PGAP pipeline | [11] |
| 47 | 3650 | 1196 | Open | BLASTP, GET_HOMOLOGUES, Perl script | [16] |
| 96 | 2066 | 6033 | Open | BLAST2GO and Perl scripts | [12] |
| 145 | 3736 | 708 | NA 4 | Roary | [38] |
| 150 | 1251 | 3758 | Nearly closed | BPGA tool | [15] |
| 183 2 | 1166 | 5870 | Closed | BLASTPBLOSUM62 | [10] |
| 1595 3 | 3419 | 7620 | Closed | CD-hit package v4.6 | [14] |
| 500 | 2231 | 3729 | Open | Roary | This study |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Tuberculosis. Available online: https://www.who.int/news-room/fact-sheets/detail/tuberculosis (accessed on 30 January 2021).
- Shah, M.; Dorman, S.E. Latent tuberculosis infection. N. Engl. J. Med. 2021, 385, 2271–2280. [Google Scholar] [CrossRef]
- Ben Ayed, H.; Koubaa, M.; Marrakchi, C.; Rekik, K.; Hammami, F.; Smaoui, F. Extrapulmonary tuberculosis: Update on the epidemiology, risk factors and prevention strategies. Int. J. Trop. Dis. 2018, 1, 1–6. [Google Scholar] [CrossRef]
- Coscolla, M. Biological and epidemiological consequences of MTBC diversity. Adv. Exp. Med. Biol. 2017, 1019, 95–116. [Google Scholar] [CrossRef]
- Saw, S.H.; Tan, J.L.; Chan, X.Y.; Chan, K.G.; Ngeow, Y.F. Chromosomal rearrangements and protein globularity changes in Mycobacterium tuberculosis isolates from cerebrospinal fluid. PeerJ 2016, 4, e2484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, K.; Verma, R.; Advani, J.; Chatterjee, O.; Solanki, H.S.; Sharma, A.; Varma, S.; Modi, M.; Ray, P.; Mukherjee, K.K.; et al. Whole genome sequencing of Mycobacterium tuberculosis isolates from extrapulmonary sites. Omics 2017, 21, 413–425. [Google Scholar] [CrossRef]
- Ruesen, C.; Chaidir, L.; van Laarhoven, A.; Dian, S.; Ganiem, A.R.; Nebenzahl-Guimaraes, H.; Huynen, M.A.; Alisjahbana, B.; Dutilh, B.E.; van Crevel, R. Large-scale genomic analysis shows association between homoplastic genetic variation in Mycobacterium tuberculosis genes and meningeal or pulmonary tuberculosis. BMC Genom. 2018, 19, 122. [Google Scholar] [CrossRef] [Green Version]
- Faksri, K.; Xia, E.; Ong, R.T.; Tan, J.H.; Nonghanphithak, D.; Makhao, N.; Thamnongdee, N.; Thanormchat, A.; Phurattanakornkul, A.; Rattanarangsee, S.; et al. Comparative whole-genome sequence analysis of Mycobacterium tuberculosis isolated from tuberculous meningitis and pulmonary tuberculosis patients. Sci. Rep. 2018, 8, 4910. [Google Scholar] [CrossRef] [Green Version]
- Laing, C.; Buchanan, C.; Taboada, E.N.; Zhang, Y.; Kropinski, A.; Villegas, A.; Thomas, J.E.; Gannon, V.P. Pan-genome sequence analysis using Panseq: An online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinform. 2010, 11, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakham, F.; Sironen, T.; Vapalahti, O.; Kant, R. Pan and core genome analysis of 183 Mycobacterium tuberculosis strains revealed a high inter-species diversity among the human adapted strains. Antibiotics 2021, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhong, J.; Zhang, J.; Li, C.; Yu, X.; Xiao, J.; Jia, X.; Ding, N.; Ma, G.; Wang, G.; et al. Pan-genomic study of Mycobacterium tuberculosis reflecting the primary/secondary genes, generality/individuality, and the interconversion through copy number variations. Front. Microbiol. 2018, 9, 1886. [Google Scholar] [CrossRef] [Green Version]
- Periwal, V.; Patowary, A.; Vellarikkal, S.K.; Gupta, A.; Singh, M.; Mittal, A.; Jeyapaul, S.; Chauhan, R.K.; Singh, A.V.; Singh, P.K.; et al. Comparative whole-genome analysis of clinical isolates reveals characteristic architecture of Mycobacterium tuberculosis pangenome. PLoS ONE 2015, 10, e0122979. [Google Scholar] [CrossRef]
- Wan, X.; Koster, K.; Qian, L.; Desmond, E.; Brostrom, R.; Hou, S.; Douglas, J.T. Genomic analyses of the ancestral Manila family of Mycobacterium tuberculosis. PLoS ONE 2017, 12, e0175330. [Google Scholar] [CrossRef] [Green Version]
- Kavvas, E.S.; Catoiu, E.; Mih, N.; Yurkovich, J.T.; Seif, Y.; Dillon, N.; Heckmann, D.; Anand, A.; Yang, L.; Nizet, V.; et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat. Commun. 2018, 9, 4306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dar, H.A.; Zaheer, T.; Ullah, N.; Bakhtiar, S.M.; Zhang, T.; Yasir, M.; Azhar, E.I.; Ali, A. Pangenome analysis of Mycobacterium tuberculosis reveals core-drug targets and screening of promising lead compounds for drug discovery. Antibiotics 2020, 9, 819. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Páez, U.; Zuluaga, N.Á.; Isaza, R.E.A.; Contreras-Moreira, B.; Rouzaud, F.; Robledo, J. Pan-genome association study of Mycobacterium tuberculosis lineage-4 revealed specific genes related to the high and low prevalence of the disease in patients from the North-Eastern area of Medellín, Colombia. Front Microbiol. 2022, 13, 1076797. [Google Scholar] [CrossRef] [PubMed]
- Negrete-Paz, A.M.; Vázquez-Marrufo, G.; Vázquez-Garcidueñas, M.S. Whole-genome comparative analysis at the lineage/sublineage level discloses relationships between Mycobacterium tuberculosis genotype and clinical phenotype. PeerJ 2021, 9, e12128. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [Green Version]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tonkin-Hill, G.; MacAlasdair, N.; Ruis, C.; Weimann, A.; Horesh, G.; Lees, J.A.; Gladstone, R.A.; Lo, S.; Beaudoin, C.; Floto, R.A.; et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020, 21, 1–21. [Google Scholar] [CrossRef]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016, 17, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Lees, J.A.; Galardini, M.; Bentley, S.D.; Weiser, J.N.; Corander, J. Pyseer: A comprehensive tool for microbial pangenome-wide association studies. Bioinformatics 2018, 34, 4310–4312. [Google Scholar] [CrossRef] [Green Version]
- Lees, J.A.; Mai, T.T.; Galardini, M.; Wheeler, N.E.; Horsfield, S.T.; Parkhill, J.; Corander, J. Improved prediction of bacterial genotype-phenotype associations using interpretable pangenome-spanning regressions. mBio 2020, 11, e01344-20. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohl, T.A.; Utpatel, C.; Schleusener, V.; De Filippo, M.R.; Beckert, P.; Cirillo, D.M.; Niemann, S. MTBseq: A comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 2018, 6, e5895. [Google Scholar] [CrossRef] [Green Version]
- Wajid, B.; Serpedin, E. Do it yourself guide to genome assembly. Brief. Funct. Genom. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillén-Nepita, A.L.; Negrete-Paz, A.M.; Vázquez-Marrufo, G.; Cruz-Hernández, A.; Fresia, P.; Naya, H.; Vázquez-Garcidueñas, M.S. Sequencing and annotation of the genome of Mycobacterium tuberculosis MYC004, a strain causing meningitis in Mexico. Genome Announ. 2018, 6, e00523-18. [Google Scholar] [CrossRef] [Green Version]
- Pepperell, C.S. Evolution of tuberculosis pathogenesis. Annu. Rev. Microbiol. 2022, 76, 661–680. [Google Scholar] [CrossRef]
- Achtman, M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 2008, 62, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Sreevatsan, S.; Pan, X.; Stockbauer, K.E.; Connell, N.D.; Kreiswirth, B.N.; Whittam, T.S.; Musser, J.M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc. Natl. Acad. Sci. USA 1997, 94, 9869–9874. [Google Scholar] [CrossRef] [Green Version]
- Gagneux, S. Genetic diversity in Mycobacterium tuberculosis. Curr. Top. Microbiol. Immunol. 2013, 374, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Warner, D.F.; Koch, A.; Mizrahi, V. Diversity and disease pathogenesis in Mycobacterium tuberculosis. Trends Microbiol. 2015, 23, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E. Genomic insights into tuberculosis. Nat. Genet. 2014, 15, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Bottai, D.; Frigui, W.; Sayes, F.; Di Luca, M.; Spadoni, D.; Pawlik, A.; Zoppo, M.; Orgeur, M.; Khanna, V.; Hardy, D.; et al. TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages. Nat. Commun. 2020, 11, 684. [Google Scholar] [CrossRef] [Green Version]
- Kayani, M.R.; Zheng, Y.C.; Xie, F.C.; Kang, K.; Li, H.Y.; Zhao, H.T. Genome sequences and comparative analysis of two extended-spectrum extensively-drug resistant Mycobacterium tuberculosis strains. Front. Pharmacol. 2018, 9, 1492. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Angala, S.K.; Pramanik, P.K.; Li, K.; Crick, D.C.; Liav, A.; Jozwiak, A.; Swiezewska, E.; Jackson, M.; Chatterjee, D. Reconstitution of functional mycobacterial arabinosyltransferase AftC proteoliposome and assessment of decaprenylphosphorylarabinose analogues as arabinofuranosyl donors. ACS Chem. Biol. 2011, 6, 819–828. [Google Scholar] [CrossRef]
- Sundararajan, S.; Muniyan, R. Latent tuberculosis: Interaction of virulence factors in Mycobacterium tuberculosis. Mol. Biol. Rep. 2021, 48, 6181–6196. [Google Scholar] [CrossRef]
- Serafini, A.; Tan, L.; Horswell, S.; Howell, S.; Greenwood, D.J.; Hunt, D.M.; Phan, M.D.; Schembri, M.; Monteleone, M.; Montague, C.R.; et al. Mycobacterium tuberculosis requires glyoxylate shunt and reverse methylcitrate cycle for lactate and pyruvate metabolism. Mol. Microbiol. 2019, 112, 1284–1307. [Google Scholar] [CrossRef] [Green Version]
- Daniel, J.; Deb, C.; Dubey, V.S.; Sirakova, T.D.; Abomoelak, B.; Morbidoni, H.R.; Kolattukudy, P.E. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 2004, 186, 5017–5030. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Hendrickson, R.C.; Meikle, V.; Lefkowitz, E.J.; Ioerger, T.R.; Niederweis, M. Comprehensive analysis of iron utilization by Mycobacterium tuberculosis. PLoS Pathog. 2020, 16, e1008337. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Olivares, C.E.; Hernández-Pando, R.; Mixcoha, E. In silico EsxG· EsxH rational epitope selection: Candidate epitopes for vaccine design against pulmonary tuberculosis. bioRxiv 2022. [Google Scholar] [CrossRef]
- Wang, L.; Asare, E.; Shetty, A.C.; Sanchez-Tumbaco, F.; Edwards, M.R.; Saranathan, R.; Weinrick, B.; Xu, J.; Chen, B.; Bénard, A.; et al. Multiple genetic paths including massive gene amplification allow Mycobacterium tuberculosis to overcome loss of ESX-3 secretion system substrates. Proc. Natl. Acad. Sci. USA 2022, 119, e2112608119. [Google Scholar] [CrossRef] [PubMed]
- Tufariello, J.M.; Chapman, J.R.; Kerantzas, C.A.; Wong, K.W.; Vilchèze, C.; Jones, C.M.; Cole, L.E.; Tinaztepe, E.; Thompson, V.; Fenyö, D.; et al. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc. Natl. Acad. Sci. USA 2016, 113, E348–E357. [Google Scholar] [CrossRef] [Green Version]
- Martini, M.C.; Hicks, N.D.; Xiao, J.; Alonso, M.N.; Barbier, T.; Sixsmith, J.; Fortune, S.M.; Shell, S.S. Loss of RNase J leads to multi-drug tolerance and accumulation of highly structured mRNA fragments in Mycobacterium tuberculosis. PLoS Pathog. 2022, 18, e1010705. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Chaiprasert, A.; Ngamphiw, C.; Tongsima, S.; Regmi, S.M.; Clark, T.G.; Ong, R.T.; Teo, Y.Y.; Prammananan, T.; Palittapongarnpim, P. Genetic signatures of Mycobacterium tuberculosis Nonthaburi genotype revealed by whole genome analysis of isolates from tuberculous meningitis patients in Thailand. PeerJ 2016, 4, e1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, G.R.; Wernisch, L.; Stabler, R.; Mangan, J.A.; Hinds, J.; Laing, K.G.; Young, D.B.; Butcher, P.D. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 2002, 148, 3129–3138. [Google Scholar] [CrossRef] [Green Version]
- Hakiem, O.R.; Batra, J.K. Role of HrcA in stress management in Mycobacterium tuberculosis. J. Appl. Microbiol. 2022, 132, 3315–3326. [Google Scholar] [CrossRef]
- Deep, A.; Kaundal, S.; Agarwal, S.; Singh, R.; Thakur, K.G. Crystal structure of Mycobacterium tuberculosis VapC20 toxin and its interactions with cognate antitoxin, VapB20, suggest a model for toxin–antitoxin assembly. FEBS J. 2017, 284, 4066–4082. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Yang, D.; Kong, Y.; Zhang, L.; Marrs, C.F.; Foxman, B.; Bates, J.H.; Wilson, F.; Cave, M.D. Clinical relevance of Mycobacterium tuberculosis plcD gene mutations. Am. J. Respir. Crit. Care Med. 2005, 171, 1436–1442. [Google Scholar] [CrossRef] [Green Version]
- Shafipour, M.; Shirzad-Aski, H.; Kochaksaraii, M.B.; Sohrabi, A.; Taziki, M.; Mahghani, G.A.; Ghaemi, E.A. The prevalence of plcD gene and evaluation of IS6110 insertion status in this gene in some clinical Mycobacterium tuberculosis isolates. Mol. Genet. Microbiol. Virol. 2021, 36, 111–118. [Google Scholar] [CrossRef]
- Kohli, S.; Singh, Y.; Sharma, K.; Mittal, A.; Ehtesham, N.Z.; Hasnain, S.E. Comparative genomic and proteomic analyses of PE/PPE multigene family of Mycobacterium tuberculosis H₃₇Rv and H₃₇Ra reveal novel and interesting differences with implications in virulence. Nucleic Acids Res. 2012, 40, 7113–7122. [Google Scholar] [CrossRef]
- Ahmad, J.; Khubaib, M.; Sheikh, J.A.; Pancsa, R.; Kumar, S.; Srinivasan, A.; Babu, M.M.; Hasnain, S.E.; Ehtesham, N.Z. Disorder-to-order transition in PE–PPE proteins of Mycobacterium tuberculosis augments the pro-pathogen immune response. FEBS Open Bio. 2020, 10, 70–85. [Google Scholar] [CrossRef] [Green Version]
- Campuzano, J.; Aguilar, D.; Arriaga, K.; León, J.C.; Salas-Rangel, L.P.; González-y-Merchand, J.; Hernández-Pando, R.; Espitia, C. The PGRS domain of Mycobacterium tuberculosis PE_PGRS Rv1759c antigen is an efficient subunit vaccine to prevent reactivation in a murine model of chronic tuberculosis. Vaccine 2007, 25, 3722–3729. [Google Scholar] [CrossRef]
- De Maio, F.; Berisio, R.; Manganelli, R.; Delogu, G. PE_PGRS proteins of Mycobacterium tuberculosis: A specialized molecular task force at the forefront of host–pathogen interaction. Virulence 2020, 11, 898–915. [Google Scholar] [CrossRef]
- Sharma, T.; Grover, S.; Arora, N.; Manjunath, P.; Ehtesham, N.Z.; Hasnain, S.E. PGRS domain of Rv0297 of Mycobacterium tuberculosis is involved in modulation of macrophage functions to favor bacterial persistence. Front. Cell Infect. Microbiol. 2020, 10, 451. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zhou, Q.; Li, L.; Zhou, Y.; Sha, W. Epidemiological characteristics of extrapulmonary tuberculosis patients with or without pulmonary tuberculosis. Epidemiol. Infect. 2022, 150, e158. [Google Scholar] [CrossRef] [PubMed]
- Yates, T.A.; Khan, P.Y.; Knight, G.M.; Taylor, J.G.; McHugh, T.D.; Lipman, M.; White, R.G.; Cohen, T.; Cobelens, F.G.; Wood, R.; et al. The transmission of Mycobacterium tuberculosis in high burden settings. Lancet Infect. Dis. 2016, 16, 227–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, R.D.; Chiu, C.; Churchyard, G.J.; Esmail, H.; Lewinsohn, D.M.; Gandhi, N.R.; Fennelly, K.P. Tuberculosis infectiousness and host susceptibility. J. Infect. Dis. 2017, 216 (Suppl. S6), S636–S643. [Google Scholar] [CrossRef] [Green Version]
- Chandra, P.; Grigsby, S.J.; Philips, J.A. Immune evasion and provocation by Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2022, 20, 750–766. [Google Scholar] [CrossRef] [PubMed]
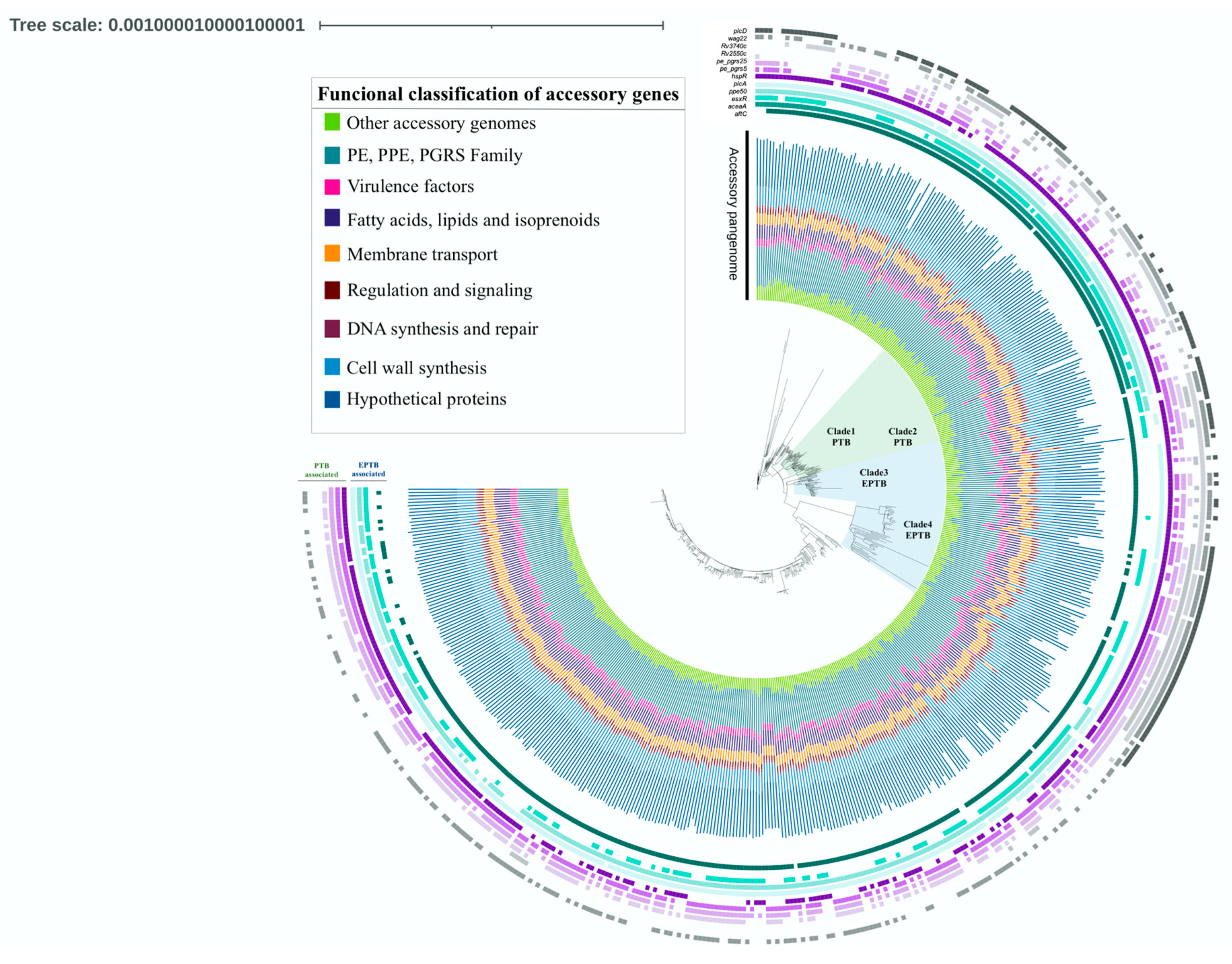

| Gene | Product | Functional Classification | p-Value | OR* | CI* 95% | Association | |
|---|---|---|---|---|---|---|---|
| Virulence | Rv0353/hspR | Heat shock protein transcriptional repressor HspR | Virulence | 0.0483 | 1.5 | 1.0034 to 2.4059 | PTB |
| Rv1755c/plcD | Phospholipase C | Virulence | 0.0006 | 2 | 1.3487 to 2.9849 | PTB | |
| Rv1915/aceA | Isocitrate lyase AceAa | Virulence | 0.0005 | 2.03 | 1.3606 to 3.0357 | EPTB | |
| Rv2351c/plcA | Phospholipase C | Virulence | 0.007 | 2.78 | 1.3136 to 5.9075 | EPTB | |
| Rv3019c/esxR | Secreted ESAT-6 like protein EsxR | Virulence | 0.0132 | 1.6 | 1.1010 to 2.2783 | EPTB | |
| Rv2550c | Antitoxin VapB20 | Virulence | 0.001 | 2.8 | 1.5405 to 5.5661 | PTB | |
| Other | Rv3135/ppe50 | PPE family protein PPE50 | Pe/ppe/pgrs | <0.0001 | 6.4 | 1.6896 to 4.5763 | EPTB |
| Rv0297/pe_pgrs5 | PE-PGRS family protein PE PGRS5 | Pe/ppe/pgrs | 0.0303 | 1.5 | 1.0419 to 2.2758 | PTB | |
| Rv1396c/pe_pgrs25 | PE-PGRS family protein PE PGRS25 | Pe/ppe/pgrs | 0.0255 | 1.6 | 1.0553 to 2.2802 | PTB | |
| Rv1759c/wag22 | PE-PGRS family protein Wag22 | Pe/ppe/pgrs | <0.0001 | 2.4 | 1.6427 to 3.4220 | PTB | |
| Rv3514/pe_pgrs57 | PE-PGRS family protein PE PGRS57 | Pe/ppe/pgrs | 0.0201 | 1.5 | 1.0679 to 2.1640 | PTB | |
| Rv3740c | Triacylglycerol synthase (diacylglycerol acyltransferase) | Lipid metabolism | 0.0266 | 1.5 | 1.0516 to 2.2586 | PTB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Negrete-Paz, A.M.; Vázquez-Marrufo, G.; Gutiérrez-Moraga, A.; Vázquez-Garcidueñas, M.S. Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype. Microorganisms 2023, 11, 1495. https://doi.org/10.3390/microorganisms11061495
Negrete-Paz AM, Vázquez-Marrufo G, Gutiérrez-Moraga A, Vázquez-Garcidueñas MS. Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype. Microorganisms. 2023; 11(6):1495. https://doi.org/10.3390/microorganisms11061495
Chicago/Turabian StyleNegrete-Paz, Andrea Monserrat, Gerardo Vázquez-Marrufo, Ana Gutiérrez-Moraga, and Ma. Soledad Vázquez-Garcidueñas. 2023. "Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype" Microorganisms 11, no. 6: 1495. https://doi.org/10.3390/microorganisms11061495
APA StyleNegrete-Paz, A. M., Vázquez-Marrufo, G., Gutiérrez-Moraga, A., & Vázquez-Garcidueñas, M. S. (2023). Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype. Microorganisms, 11(6), 1495. https://doi.org/10.3390/microorganisms11061495






