A Review of Carbapenem Resistance in Enterobacterales and Its Detection Techniques
Abstract
1. Introduction
1.1. Development of Antibiotic Resistance and Their Mechanisms
1.2. Factors Converging Emergence and Transmission of Antibiotic Resistance
2. Urgent Threat of Infections by Antimicrobial Resistant Bacteria: Carbapenem-Resistant Bacteria
2.1. Carbapenemases
| Ambler Class | Representative Gene | No of Variants | Gene Location | Bacterial Origins |
|---|---|---|---|---|
| A | KPC (Klebsiella pneumoniae carbapenemase) | >84 | Plasmid | K. pneumoniae |
| GES (Guiana extended spectrum) | >27 | Plasmid | P. aeruginosa | |
| IMI (Imipenem-hydrolysing beta-lactamase) | >9 | Chromosome | E. cloacae | |
| SME (Serratia marcescencens enzyme) | >5 | Chromosome | S. marcescencens | |
| SFC (Serratia fonticola carbapenemase-1) | >1 | Chromosome | S. fonticola | |
| NMC-A (not metalloenzyme carbapenemase A) | >1 | Chromosome | E. cloacae | |
| B | NDM (New Delhi metallo-lactamase) | >29 | Plasmid | K. pneumoniae |
| VIM (Verona integron-encoded metallo-lactamase) | >69 | Plasmid | P. aeruginosa | |
| IMP (Imipenemase), | >85 | Plasmid | S. marcescencens | |
| GIM (German imipenemase) | >2 | Plasmid | P. aeruginosa | |
| SIM (Seoul imipenemase) | >1 | Plasmid | P. aeruginosa | |
| D | OXA (Oxacillin-hydrolyzing carbapenemase) | >40 | Plasmid | K. pneumoniae |
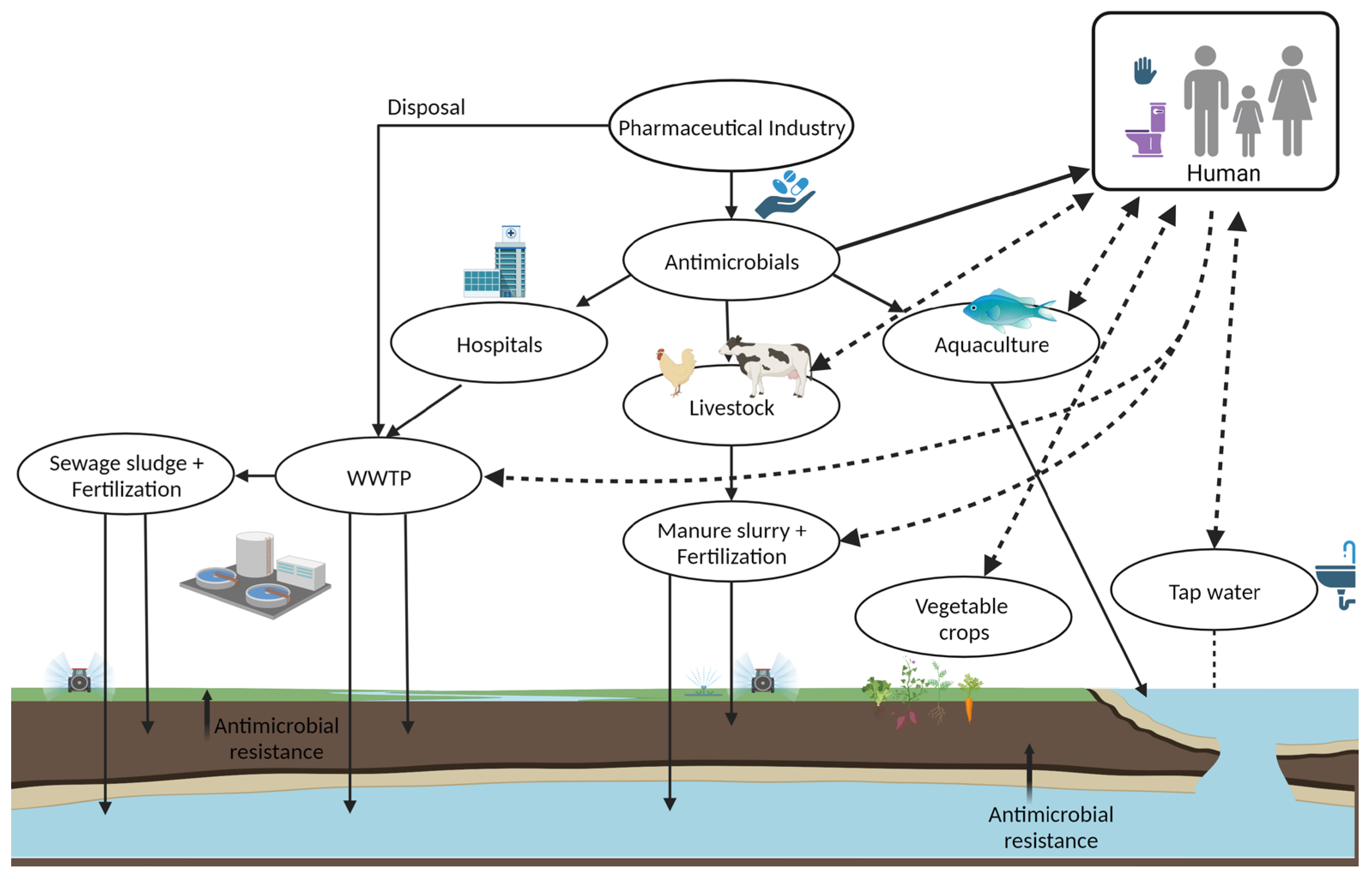
2.2. Dissemination of the Carbapenemases in Humans, Animals, Foods, and Environment
3. Current and Emerging Detection Techniques of CRE
3.1. Culture-Based Methods
3.2. Rapid Phenotypic Methods
3.3. Genotypic Methods
3.4. Rapid Serological (Immunological) Methods
3.5. Biosensing Techniques
4. Surveillance Systems for Control of Antimicrobial Resistance
5. Gaps in Detection Technology
6. Conclusions and Future Perspective
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antibiotic Resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threatsreport/2019-ar-threats-report-508.pdf (accessed on 6 September 2022).
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Morehead, M.S.; Scarbrough, C. Emergence of Global Antibiotic Resistance. Prim. Care Clin. Off. Pract. 2018, 45, 467–484. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 10 June 2022).
- Smith, R.; Coast, J. The true cost of antimicrobial resistance. BMJ 2013, 346, f1493. [Google Scholar] [CrossRef]
- Littmann, J.; Buyx, A.; Cars, O. Antibiotic resistance: An ethical challenge. Int. J. Antimicrob. Agents 2015, 46, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rojas, A.; Rodríguez-Beltrán, J.; Couce, A.; Blázquez, J. Antibiotics and antibiotic resistance: A bitter fight against evolution. Int. J. Med. Microbiol. 2013, 303, 293–297. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- CDC (Centers for Disease Control and Prevention). “Antimicrobial Resistance”, CDC. Available online: https://www.cdc.gov/drugresistance/about/how-resistance-happens.html (accessed on 10 September 2022).
- Moore, D.W. Antibiotic Classification & Mechanism, Ortho Bullets. Available online: https://www.orthobullets.com/basic-science/9059/antibiotic-classification-and-mechanism (accessed on 10 February 2023).
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Virulence Mech. Bact. Pathog. 2016, 4, 481–511. [Google Scholar] [CrossRef]
- Capita, R.; Alonso-Calleja, C. Antibiotic-Resistant Bacteria: A Challenge for the Food Industry. Crit. Rev. Food Sci. Nutr. 2013, 53, 11–48. [Google Scholar] [CrossRef]
- Holmes, A.H.; Moore, L.S.P.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Corona, F.; Martinez, J.L. Phenotypic Resistance to Antibiotics. Antibiotics 2013, 2, 237–255. [Google Scholar] [CrossRef] [PubMed]
- van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired Antibiotic Resistance Genes: An Overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-W.; Weinstein, L. Morphological Changes in Gram-Negative Bacilli Exposed To Cephalothin. J. Bacteriol. 1964, 88, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Toprak, E.; Veres, A.; Michel, J.-B.; Chait, R.; Hartl, D.L.; Kishony, R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2012, 44, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef] [PubMed]
- Sandegren, L. Selection of antibiotic resistance at very low antibiotic concentrations. Upsala J. Med. Sci. 2014, 119, 103–107. [Google Scholar] [CrossRef]
- Dankittipong, N.; Fischer, E.A.J.; Swanenburg, M.; Wagenaar, J.A.; Stegeman, A.J.; de Vos, C.J. Quantitative Risk Assessment for the Introduction of Carbapenem-Resistant Enterobacteriaceae (CPE) into Dutch Livestock Farms. Antibiotics 2022, 11, 281. [Google Scholar] [CrossRef]
- Taggar, G.; Rehman, M.A.; Boerlin, P.; Diarra, M.S. Molecular Epidemiology of Carbapenemases in Enterobacteriales from Humans, Animals, Food and the Environment. Antibiotics 2020, 9, 693. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist. Updat. 2012, 15, 162–172. [Google Scholar] [CrossRef]
- Wellington, E.M.H.; Boxall, A.B.A.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Hu, Y.; Matsui, Y.; Riley, L.W. Risk factors for fecal carriage of drug-resistant Escherichia coli: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2020, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Mahamat, O.O.; Tidjani, A.; Lounnas, M.; Hide, M.; Benavides, J.; Somasse, C.; Ouedraogo, A.-S.; Sanou, S.; Carrière, C.; Bañuls, A.-L.; et al. Fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae in hospital and community settings in Chad. Antimicrob. Resist. Infect. Control 2019, 8, 169. [Google Scholar] [CrossRef]
- Band, V.I.; Weiss, D.S. Heteroresistance: A cause of unexplained antibiotic treatment failure? PLoS Pathog. 2019, 15, e1007726. [Google Scholar] [CrossRef]
- Rebold, N.; Lagnf, A.M.; Alosaimy, S.; Holger, D.J.; Witucki, P.; Mannino, A.; Dierker, M.; Lucas, K.; Coyne, A.J.K.; El Ghali, A.; et al. Risk Factors for Carbapenem-Resistant Enterobacterales Clinical Treatment Failure. Microbiol. Spectr. 2023, 11, e02647-22. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. WHO. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 6 September 2022).
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- Smith, H.Z.; Kendall, B. Carbapenem Resistant Enterobacteriaceae; Statpearls: Tampa, FL, USA, 2021. [Google Scholar]
- Bonardi, S.; Pitino, R. Carbapenemase-producing bacteria in food-producing animals, wildlife and environment: A challenge for human health. Ital. J. Food Saf. 2019, 8, 7956. [Google Scholar] [CrossRef]
- Kopotsa, K.; Sekyere, J.O.; Mbelle, N.M. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: A review. Ann. N. Y. Acad. Sci. 2019, 1457, 61–91. [Google Scholar] [CrossRef]
- Michael, G.B.; Freitag, C.; Wendlandt, S.; Eidam, C.; Feßler, A.T.; Lopes, G.V.; Kadlec, K.; Schwarz, S. Emerging issues in antimicrobial resistance of bacteria from food-producing animals. Future Microbiol. 2015, 10, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Wareham, D.W.; Guerra, B.; Teale, C. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: An emerging public health risk of our own making? J. Antimicrob. Chemother. 2014, 69, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Mallick, A.; Roy, A.; Sarkar, S.; Mondal, K.C.; Das, S. Customized molecular diagnostics of bacterial bloodstream infections for carbapenem resistance: A convenient and affordable approach. Pathog. Glob. Health 2023, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Lu, C.-Y.; Yen, T.-Y.; Chang, L.-Y.; Chen, J.-M.; Lee, P.-I.; Huang, L.-M. Clinical characteristics and outcomes of carbapenem-resistant Enterobacterales bacteremia in pediatric patients. J. Microbiol. Immunol. Infect. 2023, 56, 84–92. [Google Scholar] [CrossRef] [PubMed]
- CDC (Centers for Disease Control and Prevention). Healthcare-Associated Infections (HAIs): CRE Technical Information. 2019. Available online: https://www.cdc.gov/hai/organisms/cre/technical-info.html (accessed on 8 June 2022).
- Liu, B.-T.; Zhang, X.-Y.; Wan, S.-W.; Hao, J.-J.; Jiang, R.-D.; Song, F.-J. Characteristics of Carbapenem-Resistant Enterobacteriaceae in Ready-to-Eat Vegetables in China. Front. Microbiol. 2018, 9, 1147. [Google Scholar] [CrossRef]
- Gordon, N.; Bawa, R.; Palmateer, G. Carbapenem-Resistant Enterobacteriaceae Testing in 45 Minutes Using an Electronic Sensor. Curr. Issues Med. Diagn. Imaging 2021, 4, 1–18. [Google Scholar]
- Rabaan, A.A.; Eljaaly, K.; Alhumaid, S.; Albayat, H.; Al-Adsani, W.; Sabour, A.A.; Alshiekheid, M.A.; Al-Jishi, J.M.; Khamis, F.; Alwarthan, S.; et al. An Overview on Phenotypic and Genotypic Characterisation of Carbapenem-Resistant Enterobacterales. Medicina 2022, 58, 1675. [Google Scholar] [CrossRef]
- Armin, S.; Azimi, L.; Shariatpanahi, G.; Shirvani, A.; Tehrani, N.A. The Prevalence of Colonization with Carbapenem-resistant Enterobacteriaceae, E. coli, Klebsiella and Enterobacter, and Related Risk Factors in Children. Arch. Pediatr. Infect. Dis. 2023, in press. [Google Scholar] [CrossRef]
- Zeng, M.; Xia, J.; Zong, Z.; Shi, Y.; Ni, Y.; Hu, F.; Chen, Y.; Zhuo, C.; Hu, B.; Lv, X.; et al. Guidelines for the diagnosis, treatment, prevention and control of infections caused by carbapenem-resistant gram-negative bacilli. J. Microbiol. Immunol. Infect. 2023; in press. [Google Scholar] [CrossRef]
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef]
- Guerra, B.; Fischer, J.; Helmuth, R. An emerging public health problem: Acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet. Microbiol. 2014, 171, 290–297. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The Versatile beta-Lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Cheruvanky, A.; Stoesser, N.; Sheppard, A.E.; Crook, D.W.; Hoffman, P.S.; Weddle, E.; Carroll, J.; Sifri, C.D.; Chai, W.; Barry, K.; et al. Enhanced Klebsiella pneumoniae Carbapenemase Expression from a Novel Tn 4401 Deletion. Antimicrob. Agents Chemother. 2017, 61, e00025-17. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, X.; Liu, C.; Zhang, Y.; Cheung, Y.C.; Chan, E.W.C.; Chen, S.; Zhang, R. Identification of a KPC Variant Conferring Resistance to Ceftazidime-Avibactam from ST11 Carbapenem-Resistant Klebsiella pneumoniae Strains. Microbiol. Spectr. 2022, 10, e02655-21. [Google Scholar] [CrossRef] [PubMed]
- Farhat, N.; Khan, A.U. Evolving trends of New Delhi Metallo-betalactamse (NDM) variants: A threat to antimicrobial resistance. Infect. Genet. Evol. 2020, 86, 104588. [Google Scholar] [CrossRef] [PubMed]
- Boyd, S.E.; Livermore, D.M.; Hooper, D.C.; Hope, W.W. Metallo-β-Lactamases: Structure, Function, Epidemiology, Treatment Options, and the Development Pipeline. Antimicrob. Agents Chemother. 2020, 64, 15–22. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef]
- Boyd, S.E.; Holmes, A.; Peck, R.; Livermore, D.M.; Hope, W. OXA-48-Like β-Lactamases: Global Epidemiology, Treatment Options, and Development Pipeline. Antimicrob. Agents Chemother. 2022, 66, e00216-22. [Google Scholar] [CrossRef]
- CDC. Tracking Antibiotic Resistance. Available online: https://www.cdc.gov/drugresistance/tracking.html (accessed on 5 May 2022).
- Nair, D.V.T.; Venkitanarayanan, K.; Kollanoor Johny, A. Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control. Foods 2018, 7, 167. [Google Scholar] [CrossRef]
- Farzana, R.; Jones, L.S.; Rahman, A.; Sands, K.; van Tonder, A.J.; Portal, E.; Criollo, J.M.; Parkhill, J.; Guest, M.F.; Watkins, W.J.; et al. Genomic Insights Into the Mechanism of Carbapenem Resistance Dissemination in Enterobacterales From a Tertiary Public Heath Setting in South Asia. Clin. Infect. Dis. 2022, 76, 119–133. [Google Scholar] [CrossRef]
- Grundmann, H.; Livermore, D.M.; Giske, C.G.; Cantón, R.; Rossolini, G.M.; Campos, J.; Vatopoulos, A.; Gniadkowski, M.; Toth, A.; Pfeifer, Y.; et al. Carbapenem-non-susceptible Enterobacteriaceae in Europe: Conclusions from a meeting of national experts. Eurosurveillance 2010, 15, 19711. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Dortet, L. Rapid Detection of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2012, 18, 1503–1507. [Google Scholar] [CrossRef]
- Souli, M.; Galani, I.; Antoniadou, A.; Papadomichelakis, E.; Poulakou, G.; Panagea, T.; Vourli, S.; Zerva, L.; Armaganidis, A.; Kanellakopoulou, K.; et al. An Outbreak of Infection due to β-Lactamase Klebsiella pneumoniae Carbapenemase 2–Producing K. pneumoniae in a Greek University Hospital: Molecular Characterization, Epidemiology, and Outcomes. Clin. Infect. Dis. 2010, 50, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Raro, O.; Da Silva, R.M.C.; Filho, E.M.R.; Sukiennik, T.C.T.; Stadnik, C.; Dias, C.A.G.; Iglesias, J.O.; Pérez-Vázquez, M. Carbapenemase-Producing Klebsiella pneumoniae From Transplanted Patients in Brazil: Phylogeny, Resistome, Virulome and Mobile Genetic Elements Harboring blaKPC–2 or blaNDM–1. Front. Microbiol. 2020, 11, 1563. [Google Scholar] [CrossRef] [PubMed]
- Manenzhe, R.I.; Zar, H.J.; Nicol, M.P.; Kaba, M. The spread of carbapenemase-producing bacteria in Africa: A systematic review. J. Antimicrob. Chemother. 2014, 70, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Tula, M.Y.; Enabulele, O.I.; Ophori, E.A.; Aziegbemhin, A.S.; Iyoha, O.; Filgona, J. A systematic review of the current status of carbapenem resistance in Nigeria: Its public health implication for national intervention. Niger. Postgrad. Med. J. 2023, 30, 1–11. [Google Scholar] [CrossRef]
- Taha, M.S.; Hagras, M.M.; Shalaby, M.M.; Zamzam, Y.A.; Elkolaly, R.M.; Abdelwahab, M.A.; Maxwell, S.Y. Genotypic Characterization of Carbapenem-Resistant Klebsiella pneumoniae Isolated from an Egyptian University Hospital. Pathogens 2023, 12, 121. [Google Scholar] [CrossRef]
- Munny, N.N.; Shamsuzzaman, S.M.; Hossain, T.; Khatun, S.; Johora, F.T. Prevalence and Phenotypic Detection of Carbapenem-Resistant Enterobacter Species from the Clinical Specimens of a Tertiary Care Hospital in Bangladesh. Updat. Dent. Coll. J. 2023, 13, 17–22. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Wang, Q.; Tang, K.; Cai, X.; Li, C. Detection of carbapenem-resistant hypervirulent Klebsiella pneumoniae ST11-K64 co-producing NDM-1 and KPC-2 in a tertiary hospital in Wuhan. J. Hosp. Infect. 2022, 131, 70–80. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Demachy, M.C.; Nordmann, P. Plasmid-Mediated Carbapenem-Hydrolyzing β-Lactamase KPC-2 in Klebsiella pneumoniae Isolate from Greece. Antimicrob. Agents Chemother. 2008, 52, 796–797. [Google Scholar] [CrossRef]
- Naas, T.; Nordmann, P.; Vedel, G.; Poyart, C. Plasmid-Mediated Carbapenem-Hydrolyzing β-Lactamase KPC in a Klebsiella pneumoniae Isolate from France. Antimicrob. Agents Chemother. 2005, 49, 4423–4424. [Google Scholar] [CrossRef]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- van der Bij, A.K.; Pitout, J.D.D. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J. Antimicrob. Chemother. 2012, 67, 2090–2100. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Bradford, P.A.; Kazmierczak, K.M.; Badal, R.E.; Hackel, M.; Hoban, D.J.; Pitout, J.D. Global Incidence of Carbapenemase-Producing Escherichia coli ST131. Emerg. Infect. Dis. 2014, 20, 1928–1931. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Nordmann, P.; Poirel, L. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Barbosa-Vasconcelos, A.; Simões, R.R.; Da Costa, P.M.; Liu, W.; Nordmann, P. Environmental KPC-Producing Escherichia coli Isolates in Portugal. Antimicrob. Agents Chemother. 2012, 56, 1662–1663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lü, X.; Zong, Z. Enterobacteriaceae producing the KPC-2 carbapenemase from hospital sewage. Diagn. Microbiol. Infect. Dis. 2012, 73, 204–206. [Google Scholar] [CrossRef]
- Isozumi, R.; Yoshimatsu, K.; Yamashiro, T.; Hasebe, F.; Nguyen, B.M.; Ngo, T.C.; Yasuda, S.P.; Koma, T.; Shimizu, K.; Arikawa, J. blaNDM-1–positive Klebsiella pneumoniae from Environment, Vietnam. Emerg. Infect. Dis. 2012, 18, 1383–1385. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Schill, S.; Stoeger, A.; Pekard-Amenitsch, S.; Huhulescu, S.; Inreiter, N.; Hartl, R.; Kerschner, H.; Sorschag, S.; Springer, B.; et al. Whole genome sequencing reveals resemblance between ESBL-producing and carbapenem resistant Klebsiella pneumoniae isolates from Austrian rivers and clinical isolates from hospitals. Sci. Total Environ. 2019, 662, 227–235. [Google Scholar] [CrossRef]
- Tanner, W.D.; VanDerslice, J.A.; Goel, R.K.; Leecaster, M.K.; Fisher, M.A.; Olstadt, J.; Gurley, C.M.; Morris, A.G.; Seely, K.A.; Chapman, L. Multi-state study of Enterobacteriaceae harboring extended-spectrum beta-lactamase and carbapenemase genes in U.S. drinking water. Sci. Rep. 2019, 9, 3938. [Google Scholar] [CrossRef]
- Hoelle, J.; Johnson, J.R.; Johnston, B.D.; Kinkle, B.; Boczek, L.; Ryu, H.; Hayes, S. Survey of US wastewater for carbapenem-resistant Enterobacteriaceae. J. Water Health 2019, 17, 219–226. [Google Scholar] [CrossRef]
- Miriagou, V.; Tzouvelekis, L.S.; Rossiter, S.; Tzelepi, E.; Angulo, F.J.; Whichard, J.M. Imipenem Resistance in a Salmonella Clinical Strain Due to Plasmid-Mediated Class A Carbapenemase KPC-2. Antimicrob. Agents Chemother. 2003, 47, 1297–1300. [Google Scholar] [CrossRef]
- Chagas, T.; Seki, L.; da Silva, D.; Asensi, M. Occurrence of KPC-2-producing Klebsiella pneumoniae strains in hospital wastewater. J. Hosp. Infect. 2011, 77, 281. [Google Scholar] [CrossRef] [PubMed]
- Hamza, D.; Dorgham, S.; Ismael, E.; El-Moez, S.I.A.; ElHariri, M.; Elhelw, R.; Hamza, E. Emergence of β-lactamase- and carbapenemase-producing Enterobacteriaceae at integrated fish farms. Antimicrob. Resist. Infect. Control 2020, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Schmoger, S.; Jahn, S.; Helmuth, R.; Guerra, B. NDM-1 carbapenemase-producing Salmonella enterica subsp. enterica serovar Corvallis isolated from a wild bird in Germany. J. Antimicrob. Chemother. 2013, 68, 2954–2956. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, B.W.; Nayak, R.; Boothe, D.M. Emergence of a New Delhi Metallo-β-Lactamase (NDM-1)-Encoding Gene in Clinical Escherichia coli Isolates Recovered from Companion Animals in the United States. Antimicrob. Agents Chemother. 2013, 57, 2902–2903. [Google Scholar] [CrossRef]
- Stolle, I.; Prenger-Berninghoff, E.; Stamm, I.; Scheufen, S.; Hassdenteufel, E.; Guenther, S.; Bethe, A.; Pfeifer, Y.; Ewers, C. Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J. Antimicrob. Chemother. 2013, 68, 2802–2808. [Google Scholar] [CrossRef]
- Fischer, J.; Rodríguez, I.; Schmoger, S.; Friese, A.; Roesler, U.; Helmuth, R.; Guerra, B. Escherichia coli producing VIM-1 carbapenemase isolated on a pig farm. J. Antimicrob. Chemother. 2012, 67, 1793–1795. [Google Scholar] [CrossRef]
- Fischer, J.; Rodríguez, I.; Schmoger, S.; Friese, A.; Roesler, U.; Helmuth, R.; Guerra, B. Salmonella enterica subsp. enterica producing VIM-1 carbapenemase isolated from livestock farms. J. Antimicrob. Chemother. 2012, 68, 478–480. [Google Scholar] [CrossRef]
- Vikram, A.; Schmidt, J.W. Functional blaKPC-2 Sequences Are Present in U.S. Beef Cattle Feces Regardless of Antibiotic Use. Foodborne Pathog. Dis. 2018, 15, 444–448. [Google Scholar] [CrossRef]
- Sugawara, Y.; Hagiya, H.; Akeda, Y.; Aye, M.M.; Win, H.P.M.; Sakamoto, N.; Shanmugakani, R.K.; Takeuchi, D.; Nishi, I.; Ueda, A.; et al. Dissemination of carbapenemase-producing Enterobacteriaceae harbouring blaNDM or blaIMI in local market foods of Yangon, Myanmar. Sci. Rep. 2019, 9, 14455. [Google Scholar] [CrossRef]
- Roschanski, N.; Guenther, S.; Vu, T.T.T.; Fischer, J.; Semmler, T.; Huehn, S.; Alter, T.; Roesler, U. VIM-1 carbapenemase-producing Escherichia coli isolated from retail seafood, Germany 2016. Eurosurveillance 2017, 22, 17-00032. [Google Scholar] [CrossRef]
- Wang, J.; Yao, X.; Luo, J.; Lv, L.; Zeng, Z.; Liu, J.H. Emergence of Escherichia coli coproducing NDM-1 and KPC-2 carbapenemases from a retail vegetable, China. J. Antimicrob. Chemother. 2018, 73, 252–254. [Google Scholar] [CrossRef] [PubMed]
- Touati, A.; Mairi, A.; Baloul, Y.; Lalaoui, R.; Bakour, S.; Thighilt, L.; Gharout, A.; Rolain, J.-M. First detection of Klebsiella pneumoniae producing OXA-48 in fresh vegetables from Béjaïa city, Algeria. J. Glob. Antimicrob. Resist. 2017, 9, 17–18. [Google Scholar] [CrossRef]
- Yao, X.; Doi, Y.; Zeng, L.; Lv, L.; Liu, J.-H. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016, 16, 288–289. [Google Scholar] [CrossRef] [PubMed]
- Chaalal, N.; Touati, A.; Bakour, S.; Aissa, M.A.; Sotto, A.; Lavigne, J.-P.; Pantel, A. Spread of OXA-48 and NDM-1-Producing Klebsiella pneumoniae ST48 and ST101 in Chicken Meat in Western Algeria. Microb. Drug Resist. 2021, 27, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Guerra, B.; Rodicio, M.R. Resistance to Carbapenems in Non-Typhoidal Salmonella enterica Serovars from Humans, Animals and Food. Vet. Sci. 2018, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Morrison, B.J.; Rubin, J.E. Carbapenemase Producing Bacteria in the Food Supply Escaping Detection. PLoS ONE 2015, 10, e0126717. [Google Scholar] [CrossRef]
- Mills, M.C.; Lee, J. The threat of carbapenem-resistant bacteria in the environment: Evidence of widespread contamination of reservoirs at a global scale. Environ. Pollut. 2019, 255, 113143. [Google Scholar] [CrossRef]
- Bedos, J.; Daikos, G.; Dodgson, A.; Pan, A.; Petrosillo, N.; Seifert, H.; Vila, J.; Ferrer, R.; Wilson, P. Early identification and optimal management of carbapenem-resistant Gram-negative infection. J. Hosp. Infect. 2020, 108, 158–167. [Google Scholar] [CrossRef]
- Sutherland, J.B.; Rafii, F.J.O.L., Jr.; Williams, A.J. Rapid Analytical Methods to Identify Antibiotic-Resistant Bacteria. Antibiot. Drug Resist. 2019, 533–566. [Google Scholar] [CrossRef]
- Alizadeh, M.; Wood, R.L.; Buchanan, C.M.; Bledsoe, C.G.; Wood, M.E.; McClellan, D.S.; Blanco, R.; Ravsten, T.V.; Husseini, G.A.; Hickey, C.L.; et al. Rapid separation of bacteria from blood—Chemical aspects. Colloids Surf. B Biointerfaces 2017, 154, 365–372. [Google Scholar] [CrossRef]
- Buehler, S.S.; Madison, B.; Snyder, S.R.; Derzon, J.; Cornish, N.E.; Saubolle, M.A.; Weissfeld, A.S.; Weinstein, M.P.; Liebow, E.B.; Wolk, D.M. Effectiveness of Practices To Increase Timeliness of Providing Targeted Therapy for Inpatients with Bloodstream Infections: A Laboratory Medicine Best Practices Systematic Review and Meta-analysis. Clin. Microbiol. Rev. 2016, 29, 59–103. [Google Scholar] [CrossRef] [PubMed]
- McLain, J.E.; Cytryn, E.; Durso, L.M.; Young, S. Culture-based Methods for Detection of Antibiotic Resistance in Agroecosystems: Advantages, Challenges, and Gaps in Knowledge. J. Environ. Qual. 2016, 45, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.; Haydel, S.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.A.; Siddiqui, M.F.; Park, S. Current and Emerging Methods of Antibiotic Susceptibility Testing. Diagnostics 2019, 9, 49. [Google Scholar] [CrossRef]
- Al-Zahrani, I.A. Routine detection of carbapenem-resistant gram-negative bacilli in clinical laboratories. Saudi Med. J. 2018, 39, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Cusack, T.; Ashley, E.; Ling, C.; Rattanavong, S.; Roberts, T.; Turner, P.; Wangrangsimakul, T.; Dance, D. Impact of CLSI and EUCAST breakpoint discrepancies on reporting of antimicrobial susceptibility and AMR surveillance. Clin. Microbiol. Infect. 2019, 25, 910–911. [Google Scholar] [CrossRef] [PubMed]
- Lutgring, J.D.; Limbago, B.M. The Problem of Carbapenemase-Producing-Carbapenem-Resistant-Enterobacteriaceae Detection. J. Clin. Microbiol. 2016, 54, 529–534. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, H.; Du, H. Carbapenemases in Enterobacteriaceae: Detection and Antimicrobial Therapy. Front. Microbiol. 2019, 10, 1823. [Google Scholar] [CrossRef]
- Takayama, Y.; Adachi, Y.; Nihonyanagi, S.; Okamoto, R. Modified Hodge test using Mueller–Hinton agar supplemented with cloxacillin improves screening for carbapenemase-producing clinical isolates of Enterobacteriaceae. J. Med. Microbiol. 2015, 64, 774–777. [Google Scholar] [CrossRef]
- Amjad, A.; Ia, M.; Sa, A.; Farwa, U.; Malik, N.; Zia, F. Modified Hodge Test: A Simple and Effective Test for Detection of Carbapenemase Production The Isolates Which Showed Intermediate or Susceptible Zones for Imipenem Were Tested for Carbapenemase Modified Hodge Test, as CL Recommends the MHT to Be Perform. Iran. J. Microbiol. 2011, 3, 189–193. [Google Scholar]
- Saito, K.; Nakano, R.; Suzuki, Y.; Nakano, A.; Ogawa, Y.; Yonekawa, S.; Endo, S.; Mizuno, F.; Kasahara, K.; Mikasa, K.; et al. Suitability of Carbapenem Inactivation Method (CIM) for Detection of IMP Metallo-β-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 1220–1222. [Google Scholar] [CrossRef][Green Version]
- Alizadeh, N.; Rezaee, M.A.; Kafil, H.S.; Barhaghi, M.H.S.; Memar, M.Y.; Milani, M.; Hasani, A.; Ghotaslou, R. Detection of carbapenem-resistant Enterobacteriaceae by chromogenic screening media. J. Microbiol. Methods 2018, 153, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2012, 68, 487–489. [Google Scholar] [CrossRef]
- Nordmann, P.; Girlich, D.; Poirel, L. Detection of Carbapenemase Producers in Enterobacteriaceae by Use of a Novel Screening Medium. J. Clin. Microbiol. 2012, 50, 2761–2766. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Poirel, L.; Nordmann, P. Comparison of the SUPERCARBA, CHROMagar KPC, and Brilliance CRE screening media for detection of Enterobacteriaceae with reduced susceptibility to carbapenems. Diagn. Microbiol. Infect. Dis. 2013, 75, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Somily, A.M.; Garaween, G.A.; Abukhalid, N.; Absar, M.M.; Senok, A.C. Comparison of Molecular and Phenotypic Methods for the Detection and Characterization of Carbapenem Resistant Enterobacteriaceae. Acta Microbiol. Immunol. Hung. 2016, 63, 69–81. [Google Scholar] [CrossRef]
- Maugeri, G.; Lychko, I.; Sobral, R.; Roque, A.C.A. Identification and Antibiotic-Susceptibility Profiling of Infectious Bacterial Agents: A Review of Current and Future Trends. Biotechnol. J. 2018, 14, e1700750. [Google Scholar] [CrossRef]
- Reynoso, E.C.; Laschi, S.; Palchetti, I.; Torres, E. Advances in Antimicrobial Resistance Monitoring Using Sensors and Biosensors: A Review. Chemosensors 2021, 9, 232. [Google Scholar] [CrossRef]
- Silva, A.P.; Faria-Ramos, I.; Ricardo, E.; Miranda, I.M.; Espinar, M.J.; Costa-De-Oliveira, S.; Cantón, R.; Rodrigues, A.G.; Pina-Vaz, C. Rapid Flow Cytometry Test for Identification of Different Carbapenemases in Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 3824–3826. [Google Scholar] [CrossRef]
- Bashir, S.; Nawaz, H.; Majeed, M.I.; Mohsin, M.; Abdullah, S.; Ali, S.; Rashid, N.; Kashif, M.; Batool, F.; Abubakar, M.; et al. Rapid and sensitive discrimination among carbapenem resistant and susceptible E. coli strains using Surface Enhanced Raman Spectroscopy combined with chemometric tools. Photodiagn. Photodyn. Ther. 2021, 34, 102280. [Google Scholar] [CrossRef]
- Bernabeu, S.; Poirel, L.; Nordmann, P. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2012, 74, 88–90. [Google Scholar] [CrossRef] [PubMed]
- van Almsick, V.; Ghebremedhin, B.; Pfennigwerth, N.; Ahmad-Nejad, P. Rapid detection of carbapenemase-producing Acinetobacter baumannii and carbapenem-resistant Enterobacteriaceae using a bioluminescence-based phenotypic method. J. Microbiol. Methods 2018, 147, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Decousser, J.-W.; Poirel, L.; Nordmann, P. Recent advances in biochemical and molecular diagnostics for the rapid detection of antibiotic-resistant Enterobacteriaceae: A focus on ß-lactam resistance. Expert. Rev. Mol. Diagn. 2017, 17, 327–350. [Google Scholar] [CrossRef] [PubMed]
- Bianco, G.; Comini, S.; Boattini, M.; Ricciardelli, G.; Guarrasi, L.; Cavallo, R.; Costa, C. MALDI-TOF MS-Based Approaches for Direct Identification of Gram-Negative Bacteria and BlaKPC-Carrying Plasmid Detection from Blood Cultures: A Three-Year Single-Centre Study and Proposal of a Diagnostic Algorithm. Microorganisms 2022, 11, 91. [Google Scholar] [CrossRef]
- Knox, J.; Jadhav, S.; Sevior, D.; Agyekum, A.; Whipp, M.; Waring, L.; Iredell, J.; Palombo, E.; Cillo, A.R.; Vagratian, D.; et al. Phenotypic Detection of Carbapenemase-Producing Enterobacteriaceae by Use of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry and the Carba NP Test. J. Clin. Microbiol. 2014, 52, 4075–4077. [Google Scholar] [CrossRef]
- Gato, E.; Anantharajah, A.; Arroyo, M.J.; Artacho, M.J.; Caballero, J.D.D.; Candela, A.; Chudějová, K.; Constanso, I.P.; Elías, C.; Fernández, J.; et al. Multicenter Performance Evaluation of MALDI-TOF MS for Rapid Detection of Carbapenemase Activity in Enterobacterales: The Future of Networking Data Analysis With Online Software. Front. Microbiol. 2022, 12, 4145. [Google Scholar] [CrossRef]
- Yu, J.; Lin, Y.-T.; Chen, W.-C.; Tseng, K.-H.; Lin, H.-H.; Tien, N.; Cho, C.-F.; Huang, J.-Y.; Liang, S.-J.; Ho, L.-C.; et al. Direct prediction of carbapenem-resistant, carbapenemase-producing, and colistin-resistant Klebsiella pneumoniae isolates from routine MALDI-TOF mass spectra using machine learning and outcome evaluation. Int. J. Antimicrob. Agents 2023, 61, 106799. [Google Scholar] [CrossRef]
- Eltahlawi, R.A.; Jiman-Fatani, A.; Gad, N.M.; Ahmed, S.H.; Al-Rabia, M.W.; Zakai, S.; Kharaba, A.; El-Hossary, D. Detection of Carbapenem-resistance in CRE by Comparative Assessment of RAPIDEC® CARBA NP and Xpert™Carba-R Assay. Infect. Drug Resist. 2023, 16, 1123–1131. [Google Scholar] [CrossRef]
- Woodford, N.; Sundsfjord, A. Molecular detection of antibiotic resistance: When and where? J. Antimicrob. Chemother. 2005, 56, 259–261. [Google Scholar] [CrossRef]
- Rijpens, N.P.; Herman, L.M.F. Molecular Methods for Identification and Detection of Bacterial Food Pathogens. J. AOAC Int. 2002, 85, 984–995. [Google Scholar] [CrossRef]
- Smiljanic, M.; Kaase, M.; Ahmad-Nejad, P.; Ghebremedhin, B. Comparison of in-house and commercial real time-PCR based carbapenemase gene detection methods in Enterobacteriaceae and non-fermenting gram-negative bacterial isolates. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 48. [Google Scholar] [CrossRef]
- Probst, K.; Boutin, S.; Späth, I.; Scherrer, M.; Henny, N.; Sahin, D.; Heininger, A.; Heeg, K.; Nurjadi, D. Direct-PCR from rectal swabs and environmental reservoirs: A fast and efficient alternative to detect blaOXA-48 carbapenemase genes in an Enterobacter cloacae outbreak setting. Environ. Res. 2021, 203, 111808. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Ergani, A.; Carrër, A.; Nordmann, P. Real-Time PCR for Detection of NDM-1 Carbapenemase Genes from Spiked Stool Samples. Antimicrob. Agents Chemother. 2011, 55, 4038–4043. [Google Scholar] [CrossRef] [PubMed]
- Hlousek, L.; Voronov, S.; Diankov, V.; Leblang, A.B.; Wells, P.J.; Ford, D.M.; Nolling, J.; Hart, K.W.; Espinoza, P.A.; Bristol, M.R.; et al. Automated high multiplex qPCR platform for simultaneous detection and quantification of multiple nucleic acid targets. Biotechniques 2012, 52, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Fusaro, M.; Naas, T. Improvement of the Xpert Carba-R Kit for the Detection of Carbapenemase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 3832–3837. [Google Scholar] [CrossRef]
- Lau, A.F.; Fahle, G.A.; Kemp, M.A.; Jassem, A.N.; Dekker, J.P.; Frank, K.M. Clinical Performance of Check-Direct CPE, a Multiplex PCR for Direct Detection of bla KPC, bla NDM and/or bla VIM, and bla OXA-48 from Perirectal Swabs. J. Clin. Microbiol. 2015, 53, 3729–3737. [Google Scholar] [CrossRef]
- Wu, S.; Hulme, J.P. Recent Advances in the Detection of Antibiotic and Multi-Drug Resistant Salmonella: An Update. Int. J. Mol. Sci. 2021, 22, 3499. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef]
- Frye, J.G.; Jesse, T.; Long, F.; Rondeau, G.; Porwollik, S.; McClelland, M.; Jackson, C.R.; Englen, M.; Cray, P. DNA microarray detection of antimicrobial resistance genes in diverse bacteria. Int. J. Antimicrob. Agents 2006, 27, 138–151. [Google Scholar] [CrossRef]
- Marimuthu, K.; Venkatachalam, I.; Koh, V.; Harbarth, S.; Perencevich, E.; Cherng, B.P.Z.; Fong, R.K.C.; Pada, S.K.; Ooi, S.T.; Smitasin, N.; et al. Whole genome sequencing reveals hidden transmission of carbapenemase-producing Enterobacterales. Nat. Commun. 2022, 13, 3052. [Google Scholar] [CrossRef] [PubMed]
- Nakano, R.; Nakano, A.; Ishii, Y.; Ubagai, T.; Kikuchi-Ueda, T.; Kikuchi, H.; Tansho-Nagakawa, S.; Kamoshida, G.; Mu, X.; Ono, Y. Rapid detection of the Klebsiella pneumoniae carbapenemase (KPC) gene by loop-mediated isothermal amplification (LAMP). J. Infect. Chemother. 2014, 21, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Tong, X.; Chen, B.; Yuan, W.; Fu, M.; Yang, X.; Chen, H.; Zhang, G.; Wu, G.; Xu, B. Development of Microfluidic Chip-Based Loop-Mediated Isothermal Amplification (LAMP) Method for Detection of Carbapenemase Producing Bacteria. Microbiol. Spectr. 2022, 10, e00322-22. [Google Scholar] [CrossRef]
- Ceyssens, P.-J.; Garcia-Graells, C.; Fux, F.; Botteldoorn, N.; Mattheus, W.; Wuyts, V.; De Keersmaecker, S.; Dierick, K.; Bertrand, S. Development of a Luminex xTAG®assay for cost-effective multiplex detection of β-lactamases in Gram-negative bacteria. J. Antimicrob. Chemother. 2016, 71, 2479–2483. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oeschger, T.; Kret, L.; Erickson, D. Multiplexed Paper-Based Assay for Personalized Antimicrobial Susceptibility Profiling of Carbapenem-Resistant Enterobacteriaceae Performed in a Rechargeable Coffee Mug. Sci. Rep. 2022, 12, 11990. [Google Scholar] [CrossRef] [PubMed]
- McEwan, A.S.; Derome, A.; Meunier, D.; Burns, P.J.; Woodford, N.; Dodgson, A.R. Evaluation of the NucliSENS EasyQ KPC Assay for Detection of Klebsiella pneumoniae Carbapenemase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2013, 51, 1948–1950. [Google Scholar] [CrossRef]
- Laffler, T.G.; Cummins, L.L.; McClain, C.M.; Quinn, C.D.; Toro, M.A.; Carolan, H.E.; Toleno, D.M.; Rounds, M.A.; Eshoo, M.W.; Stratton, C.W.; et al. Enhanced Diagnostic Yields of Bacteremia and Candidemia in Blood Specimens by PCR-Electrospray Ionization Mass Spectrometry. J. Clin. Microbiol. 2013, 51, 3535–3541. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Kaas, R.S.; Seyfarth, A.M.; Agersø, Y.; Lund, O.; Larsen, M.V.; Aarestrup, F.M. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2012, 68, 771–777. [Google Scholar] [CrossRef]
- Bohara, R.A.; Pawar, S.H. Innovative Developments in Bacterial Detection with Magnetic Nanoparticles. Appl. Biochem. Biotechnol. 2015, 176, 1044–1058. [Google Scholar] [CrossRef] [PubMed]
- Wareham, D.W.; Shah, R.; Betts, J.W.; Phee, L.M.; Momin, M.H.F.A. Evaluation of an Immunochromatographic Lateral Flow Assay (OXA-48 K -SeT) for Rapid Detection of OXA-48-Like Carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 2016, 54, 471–473. [Google Scholar] [CrossRef]
- Hu, S.; Niu, L.; Zhao, F.; Yan, L.; Nong, J.; Wang, C.; Gao, N.; Zhu, X.; Wu, L.; Bo, T.; et al. Identification of Acinetobacter baumannii and its carbapenem-resistant gene blaOXA-23-like by multiple cross displacement amplification combined with lateral flow biosensor. Sci. Rep. 2019, 9, 17888. [Google Scholar] [CrossRef] [PubMed]
- Greissl, C.; Saleh, A.; Hamprecht, A. Rapid detection of OXA-48-like, KPC, NDM, and VIM carbapenemases in Enterobacterales by a new multiplex immunochromatographic test. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 38, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Perumal, V.; Hashim, U. Advances in biosensors: Principle, architecture and applications. J. Appl. Biomed. 2014, 12, 1–15. [Google Scholar] [CrossRef]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for Whole-Cell Bacterial Detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef]
- Ahmed, M.U.; Zourob, M.; Tamiya, E. Introduction to Food Biosensors. In Food Biosens; Food Chemistry, Function and Analysis; Royal Society of Chemistry, 2017; pp. 1–21. ISBN 978-1-5231-2621-7. Available online: https://books.rsc.org/books/edited-volume/571/chapter/245460/Introduction-to-Food-Biosensors (accessed on 23 May 2023). [CrossRef]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef]
- Chen, M.; Hou, C.; Huo, D.; Bao, J.; Fa, H.; Shen, C. An electrochemical DNA biosensor based on nitrogen-doped graphene/Au nanoparticles for human multidrug resistance gene detection. Biosens. Bioelectron. 2016, 85, 684–691. [Google Scholar] [CrossRef]
- Watanabe, K.; Kuwata, N.; Sakamoto, H.; Amano, Y.; Satomura, T.; Suye, S.-I. A smart DNA sensing system for detecting methicillin-resistant Staphylococcus aureus using modified nanoparticle probes. Biosens. Bioelectron. 2015, 67, 419–423. [Google Scholar] [CrossRef]
- Nemr, C.R.; Smith, S.J.; Liu, W.; Mepham, A.H.; Mohamadi, R.M.; Labib, M.; Kelley, S.O. Nanoparticle-Mediated Capture and Electrochemical Detection of Methicillin-Resistant Staphylococcus aureus. Anal. Chem. 2019, 91, 2847–2853. [Google Scholar] [CrossRef]
- Huang, J.M.-Y.; Henihan, G.; Macdonald, D.; Michalowski, A.; Templeton, K.; Gibb, A.P.; Schulze, H.; Bachmann, T.T. Rapid Electrochemical Detection of New Delhi Metallo-beta-lactamase Genes To Enable Point-of-Care Testing of Carbapenem-Resistant Enterobacteriaceae. Anal. Chem. 2015, 87, 7738–7745. [Google Scholar] [CrossRef]
- Homola, J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef]
- Wong, Y.L.; Kang, W.C.M.; Reyes, M.; Teo, J.W.P.; Kah, J.C.Y. Rapid Detection of Carbapenemase-Producing Enterobacteriacae Based on Surface-Enhanced Raman Spectroscopy with Gold Nanostars. ACS Infect. Dis. 2020, 6, 947–953. [Google Scholar] [CrossRef]
- Li, J.; Wang, C.; Kang, H.; Shao, L.; Hu, L.; Xiao, R.; Wang, S.; Gu, B. Label-free identification carbapenem-resistant Escherichia coli based on surface-enhanced resonance Raman scattering. RSC Adv. 2018, 8, 4761–4765. [Google Scholar] [CrossRef] [PubMed]
- Dester, E.; Kao, K.; Alocilja, E.C. Detection of Unamplified E. coli O157 DNA Extracted from Large Food Samples Using a Gold Nanoparticle Colorimetric Biosensor. Biosensors 2022, 12, 274. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, S.; Kamaladini, H.; Haddadi, F.; Sharifmoghadam, M.R. Thiol-Capped Gold Nanoparticle Biosensors for Rapid and Sensitive Visual Colorimetric Detection of Klebsiella pneumoniae. J. Fluoresc. 2018, 28, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Quintela, I.A.; de los Reyes, B.G.; Lin, C.-S.; Wu, V.C.H. Simultaneous Colorimetric Detection of a Variety of Salmonella spp. in Food and Environmental Samples by Optical Biosensing Using Oligonucleotide-Gold Nanoparticles. Front. Microbiol. 2019, 10, 1138. [Google Scholar] [CrossRef]
- Santopolo, G.; Rojo-Molinero, E.; Clemente, A.; Borges, M.; Oliver, A.; de la Rica, R. Bedside Detection of Carbapenemase-Producing Pathogens with Plasmonic Nanosensors. Sens. Actuators B Chem. 2020, 329, 129059. [Google Scholar] [CrossRef]
- Bakthavathsalam, P.; Rajendran, V.K.; Mohammed, J.A.B. A direct detection of Escherichia coli genomic DNA using gold nanoprobes. J. Nanobiotechnol. 2012, 10, 8. [Google Scholar] [CrossRef]
- Sharief, S.A.; Caliskan-Aydogan, O.; Alocilja, E. Carbohydrate-coated magnetic and gold nanoparticles for point-of-use food contamination testing. Biosens. Bioelectron. X 2023, 13, 100322. [Google Scholar] [CrossRef]
- Sharief, S.A.; Caliskan-Aydogan, O.; Alocilja, E.C. Carbohydrate-coated nanoparticles for PCR-less genomic detection of Salmonella from fresh produce. Food Control 2023, 150, 109770. [Google Scholar] [CrossRef]
- Baetsen-Young, A.M.; Vasher, M.; Matta, L.L.; Colgan, P.; Alocilja, E.C.; Day, B. Direct colorimetric detection of unamplified pathogen DNA by dextrin-capped gold nanoparticles. Biosens. Bioelectron. 2018, 101, 29–36. [Google Scholar] [CrossRef]
- Caliskan-Aydogan, O.; Sharief, S.A.; Alocilja, E.C. Nanoparticle-Based Plasmonic Biosensor for the Unamplified Genomic Detection of Carbapenem-Resistant Bacteria. Diagnostics 2023, 13, 656. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Global Antimicrobial Resistance and Use Surveillance System (GLASS). Available online: https://www.who.int/initiatives/glass (accessed on 15 January 2023).
- FDA. The National Antimicrobial Resistance Monitoring System, NARMS FDA. Available online: https://www.fda.gov/animal-veterinary/antimicrobial-resistance/national-antimicrobial-resistance-monitoring-system (accessed on 20 June 2022).
- ECDC: EARS-Net, Annual Report of The European Antimicrobial Resistance Surveillance Network (EARS-Net) Surveill. Report; ECDC. 2017. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017 (accessed on 20 October 2022).
- FDA. Clinical Laboratory Improvement Amendments (CLIA). Available online: https://www.fda.gov/medical-devices/ivd-regulatory-assistance/clinical-laboratory-improvement-amendments-clia (accessed on 15 July 2022).
- CDC. Foodborne Diseases Active Surveillance Network (FoodNet). Available online: https://www.cdc.gov/foodnet/index.html (accessed on 22 May 2022).
- CDC. Culture-Independent Diagnostic Tests. Available online: https://www.cdc.gov/foodsafety/challenges/cidt.html (accessed on 13 December 2022).
- CDC. CDC’s Antibiotic Resistance (AR) Laboratory Networks. Available online: https://www.cdc.gov/drugresistance/laboratories.html (accessed on 12 March 2023).
- Dester, E.; Alocilja, E. Current Methods for Extraction and Concentration of Foodborne Bacteria with Glycan-Coated Magnetic Nanoparticles: A Review. Biosensors 2022, 12, 112. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.; Jaykus, L.-A.; Brehm-Stecher, B. Advances in Separation and Concentration of Microorganisms from Food Samples; Woodhead Publishing Ltd.: Shaston, UK, 2013. [Google Scholar] [CrossRef]
- Pitt, W.G.; Alizadeh, M.; Husseini, G.A.; McClellan, D.S.; Buchanan, C.M.; Bledsoe, C.G.; Robison, R.A.; Blanco, R.; Roeder, B.L.; Melville, M.; et al. Rapid separation of bacteria from blood-review and outlook. Biotechnol. Prog. 2016, 32, 823–839. [Google Scholar] [CrossRef]
- Li, Z.; Ma, J.; Ruan, J.; Zhuang, X. Using Positively Charged Magnetic Nanoparticles to Capture Bacteria at Ultralow Concentration. Nanoscale Res. Lett. 2019, 14, 195. [Google Scholar] [CrossRef] [PubMed]
- Kearns, H.; Goodacre, R.; Jamieson, L.E.; Graham, D.; Faulds, K. SERS Detection of Multiple Antimicrobial-Resistant Pathogens Using Nanosensors. Anal. Chem. 2017, 89, 12666–12673. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, W.; Peng, Q.; Han, D.; Kong, L.; Fan, L.; Zhao, M.; Ding, S. Rapid detection of carbapenem-resistant Enterobacteriaceae using pH response based on vancomycin-modified Fe3O4@Au nanoparticle enrichment and the carbapenemase hydrolysis reaction. Anal. Methods 2019, 12, 104–111. [Google Scholar] [CrossRef]
- Krishna, V.D.; Wu, K.; Su, D.; Cheeran, M.C.; Wang, J.-P.; Perez, A. Nanotechnology: Review of concepts and potential application of sensing platforms in food safety. Food Microbiol. 2018, 75, 47–54. [Google Scholar] [CrossRef]
- Lv, M.; Liu, Y.; Geng, J.; Kou, X.; Xin, Z.; Yang, D. Engineering nanomaterials-based biosensors for food safety detection. Biosens. Bioelectron. 2018, 106, 122–128. [Google Scholar] [CrossRef]
- Frank, J.F. Microbial Attachment to Food and Food Conctact Surfaces. In Advances Innnnnnnn Food and Nutrition Research; Academic Press: Cambridge, MA, USA, 2001; Volume 43, ISBN 0-12-016443-4. [Google Scholar]
- Payne, M.J.; Kroll, R.G. Methods for the separation and concentration of bacteria from foods. Trends Food Sci. Technol. 1991, 2, 315–319. [Google Scholar] [CrossRef]
- Wang, K.; Li, S.; Petersen, M.; Wang, S.; Lu, X. Detection and Characterization of Antibiotic-Resistant Bacteria Using Surface-Enhanced Raman Spectroscopy. Nanomaterials 2018, 8, 762. [Google Scholar] [CrossRef]
- Galvan, D.D.; Yu, Q. Surface-Enhanced Raman Scattering for Rapid Detection and Characterization of Antibiotic-Resistant Bacteria. Adv. Health Mater. 2018, 7, e1701335. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; O’Driscoll, N.H.; Lamb, A.J. Morphological and ultrastructural changes in bacterial cells as an indicator of antibacterial mechanism of action. Cell. Mol. Life Sci. 2016, 73, 4471–4492. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.C.; Blair, K.M.; Salama, N.R. Staying in Shape: The Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Nishino, M.; Matsuzaki, I.; Musangile, F.Y.; Takahashi, Y.; Iwahashi, Y.; Warigaya, K.; Kinoshita, Y.; Kojima, F.; Murata, S.-I. Measurement and visualization of cell membrane surface charge in fixed cultured cells related with cell morphology. PLoS ONE 2020, 15, e0236373. [Google Scholar] [CrossRef]
- Maillard, A.P.F.; Espeche, J.C.; Maturana, P.; Cutro, A.C.; Hollmann, A. Zeta potential beyond materials science: Applications to bacterial systems and to the development of novel antimicrobials. Biochim. et Biophys. Acta (BBA)-Biomembr. 2021, 1863, 183597. [Google Scholar] [CrossRef] [PubMed]
- Soon, R.L.; Nation, R.L.; Cockram, S.; Moffatt, J.H.; Harper, M.; Adler, B.; Boyce, J.D.; Larson, I.; Li, J. Different surface charge of colistin-susceptible and -resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J. Antimicrob. Chemother. 2010, 66, 126–133. [Google Scholar] [CrossRef]
- Wilson, W.; Wade, M.M.; Holman, S.C.; Champlin, F.R. Status of methods for assessing bacterial cell surface charge properties based on zeta potential measurements. J. Microbiol. Methods 2001, 43, 153–164. [Google Scholar] [CrossRef]
- Kumar, N.; Wang, W.; Ortiz-Marquez, J.C.; Catalano, M.; Gray, M.; Biglari, N.; Hikari, K.; Ling, X.; Gao, J.; van Opijnen, T.; et al. Dielectrophoresis assisted rapid, selective and single cell detection of antibiotic resistant bacteria with G-FETs. Biosens. Bioelectron. 2020, 156, 112123. [Google Scholar] [CrossRef]
| Techniques | Advantages | Limitations |
|---|---|---|
| Culture-based methods | Simple and cost-effective | Time-consuming (>24 h) |
| 1. Improved AST tests: E-test or disk diffusion test [32,107,108] | Detect KPC and MBLs with good sensitivity (>82%) and specificity (>95%) | Insufficient for OXA-48 Require specific reagents and pure culture |
| 2. Modified Hodge Test (MHT) * [108,110] | Detects KPC with good sensitivity (>69%) and specificity (>90%) | Insufficient for MBLs Requires pure culture |
| 3. Carbapenem-inactivation methods (CIM) * [107,108] | Detect all carbapenemases with higher sensitivity (>90%) and specificity (>95%) | Require pure culture |
| 4. Selective media: SUPERCARBA, Colorex KPC, ID Carba, CHROM agar KPC, etc. [112,113,114] | Detect carbapenemases from direct patient samples SUPERCARBA has higher sensitivity (>96.5%) | Variable sensitivity (40–96.5%) and specificity (>50%) |
| Rapid phenotypic methods | Rapid (<24 h) | Costly equipment |
| 1. Colorimetric assay: CarbaNP test and its automated kits * [60,107,108] | Detect carbapenemases with good sensitivity (>70%) and specificity (>80%) Simple, rapid (<2 h), and cost-effective No equipment requirement | Insufficient for OXA-48 Require pure culture |
| 2. MALDI-TOF MS * [123,125,126] | Rapidly (1–4 h) detects KPC and MBLs with good sensitivity (>72.5%) and specificity (>95%) Low-measurement cost and simple | Requires data analysis Insufficient for OXA-48 Requires single isolated colonies |
| 3. Emerging techniques: BCDA, FC, microfluidic techniques, and Raman spectroscopic techniques [116,119,120,122,123] | Simple and rapid (<4 h) Good sensitivity (>80%) and specificity (>90%) from pure culture | Lower applicability on specimens Insufficient work on carbapenemases |
| Genotypic methods | Rapid and highly specific (>90%) and sensitive (>90%) | Costly and complex equipment |
| 1. PCR-based methods: qPCR, RT-PCR, mPCR, automated PCR (Xpert system, Check-Direct, and Carba-R-assay) [123,131,135] * | Gold standard and rapid (<4 h) Detect and type all carbapenemases directly from specimens | High technical requirements and specific reagents High measurement cost |
| 2. Loop-mediated isothermal amplification (LAMP) [123,142] | Simple and moderate cost Applicable in low-resource settings | Specific reagents and complex primer design |
| 3. Whole genome sequencing (WGS) [123,141] * | Discovers a new resistance mechanism | Longer turn-around time Complex data management |
| 4. Emerging techniques: FISH, microarray techniques, PCR-ESI-MS, and NucliSENS EasyQKPC [116,123,143] | Rapid (<6 h) Detect carbapenemases | Require specific equipment and reagents Insufficient work on carbapenemases |
| Immunological Methods Enzyme-linked immunosorbent assay (ELISA), an Immunochromatographic assay [99,123,138,151] | Rapid and moderate cost Poor sensitivity and specificity directly from specimens | Complex and difficult antibody design due to antigenic site modification |
| Biosensors: Emerging Technology | Rapid, Simple, and Cost-effective | Specific Equipment |
| 1. Electrochemical assays: Impedimetric, potentiometric, and voltammetric [43,156,160] 2. Optical assays: Raman scattering, SPR, and SERS [118,120,138,161] | Detect carbapenemases Moderate cost | Require equipment for signal processing and data analysis Insufficient work on AMR and carbapenemase detection from pure culture and specimens |
| 2.1. Plasmonic biosensors [167,172] | Rapid, simple, and cost-effective Detect carbapenemases with good sensitivity (78%) and specificity (97%) No equipment requirement | Insufficient work on AMR and carbapenemase detection from pure culture and specimens |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caliskan-Aydogan, O.; Alocilja, E.C. A Review of Carbapenem Resistance in Enterobacterales and Its Detection Techniques. Microorganisms 2023, 11, 1491. https://doi.org/10.3390/microorganisms11061491
Caliskan-Aydogan O, Alocilja EC. A Review of Carbapenem Resistance in Enterobacterales and Its Detection Techniques. Microorganisms. 2023; 11(6):1491. https://doi.org/10.3390/microorganisms11061491
Chicago/Turabian StyleCaliskan-Aydogan, Oznur, and Evangelyn C. Alocilja. 2023. "A Review of Carbapenem Resistance in Enterobacterales and Its Detection Techniques" Microorganisms 11, no. 6: 1491. https://doi.org/10.3390/microorganisms11061491
APA StyleCaliskan-Aydogan, O., & Alocilja, E. C. (2023). A Review of Carbapenem Resistance in Enterobacterales and Its Detection Techniques. Microorganisms, 11(6), 1491. https://doi.org/10.3390/microorganisms11061491





