Expression of β-Glucosidases from the Yak Rumen in Lactic Acid Bacteria: A Genetic Engineering Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Construction of Individual Genes and the Fusion Gene
2.3. Construction and Transformation of the Recombinant Plasmid
2.4. Enzyme Assays and Protein Analysis of Recombinant L. lactis NZ900
2.5. Enzyme Assays at Different Temperatures and pH Values
2.6. Enzyme Kinetic Parameter Assays
2.7. Enzyme Assays at Different Metal Ion Concentrations
2.8. Statistical Analysis
3. Results
3.1. Identification of Recombinant Plasmids
3.2. Enzyme Expression Levels in Recombinants
3.3. Determination of Optimum Reaction Temperature and pH of the Recombinant Enzyme
3.4. Effect of Various Substrates on the Kinetic Parameters of Recombinant Enzymes
3.5. Effect of Ions and Chemical Reagents on Recombinant Enzymes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilmore, S.P.; Henske, J.K.; O’Malley, M.A. Driving biomass breakdown through engineered cellulosomes. Bioengineered 2015, 6, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Salgado, J.C.S.; Meleiro, L.P.; Carli, S.; Ward, R.J. Glucose tolerant and glucose stimulated β-glucosidases-A review. Bioresour. Technol. 2018, 267, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Nigam, P.S. Microbial enzymes with special characteristics for biotechnological applications. Biomolecules 2013, 3, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Navarro, R.R.; Otsuka, Y.; Nojiri, M.; Ishizuka, S.; Nakamura, M.; Shikinaka, K.; Matsuo, K.; Sasaki, K.; Sasaki, K.; Kimbara, K.; et al. Simultaneous enzymatic saccharification and comminution for the valorization of lignocellulosic biomass toward natural products. BMC Biotechnol. 2018, 18, 79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cai, G.; Zheng, E.; Zhang, G.; Li, Y.; Li, Z.; Yang, H.; Wu, Z. Transgenic pigs expressing β-xylanase in the parotid gland improve nutrient utilization. Transgenic. Res. 2019, 28, 189–198. [Google Scholar] [CrossRef]
- Angelotti, J.A.F.; Dias, F.F.G.; Sato, H.H.; Fernandes, P.; Nakajima, V.M.; Macedo, J. Improvement of Aglycone Content in Soy Isoflavones Extract by Free and Immobilized Β-Glucosidase and their Effects in Lipid Accumulation. Appl. Biochem. Biotechnol. 2020, 192, 734–750. [Google Scholar] [CrossRef]
- Rani, V.; Mohanram, S.; Tiwari, R.; Nain, L.; Arora, A. Beta-Glucosidase: Key Enzyme in Determining Efficiency of Cellulase and Biomass Hydrolysis. J. Bioprocess. Biotech. 2014, 5, 197. [Google Scholar]
- Martins, E.D.S.; Gomes, E.; da Silva, R.; Junior, R.B. Production of cellulases by Thermomucor indicae-seudaticae: Characterization of a thermophilic β-glucosidase. Prep. Biochem. Biotechnol. 2019, 49, 830–836. [Google Scholar] [CrossRef]
- Li, Y.; Bu, M.; Chen, P.; Li, X.; Chen, C.; Gao, G.; Feng, Y.; Han, W.; Zhang, Z. Characterization of a Thermophilic Monosaccharide Stimulated β-Glucosidase from Acidothermus cellulolyticus. Chem. Res. Chin. Univ. 2018, 34, 212–220. [Google Scholar] [CrossRef]
- Li, Y.; Liu, N.; Yang, H.; Zhao, F.; Yu, Y.; Tian, Y.; Lu, X. Cloning and characterization of a new β-glucosidase from a metagenomic library of rumen of cattle feeding with Miscanthus sinensis. BMC Biotechnol. 2014, 14, 85. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, N.; Xu, J.; Qi, Y.; Wei, X.; Fan, M. Exploring the catalytic mechanism of a novel β-glucosidase BGL0224 from Oenococcus oeni SD-2a: Kinetics, spectroscopic and molecular simulation. Enzyme Microb. Technol. 2021, 148, 109814. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Wu, Y.; Qin, X.; Ma, R.; Luo, H.; Su, X.; Yao, B. Revisiting overexpression of a heterologous β-glucosidase in Trichoderma reesei: Fusion expression of the Neosartorya fischeri Bgl3A to cbh1 enhances the overall as well as individual cellulase activities. Microb. Cell. Fact. 2016, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Acebrón, I.; Plaza-Vinuesa, L.; de Las Rivas, B.; Muñoz, R.; Cumella, J.; Sánchez-Sancho, F.; Mancheño, J.M. Structural basis of the substrate specificity and instability in solution of a glycosidase from Lactobacillus plantarum. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Tak, J.Y.; Jang, W.J.; Lee, J.M.; Suraiya, S.; Kong, I.S. Expression in Lactococcus lactis of a β-1,3-1,4-glucanase gene from Bacillus sp. SJ-10 isolated from fermented fish. Protein Expr. Purif. 2019, 162, 18–23. [Google Scholar] [CrossRef]
- Boonkaew, B.; Udompaisarn, S.; Arthan, D.; Somana, J. Expression and characterization of a recombinant stevioside hydrolyzing β-glycosidase from Enterococcus casseliflavus. Protein Expr. Purif. 2019, 163, 105449. [Google Scholar] [CrossRef]
- Xia, W.; Xu, X.; Qian, L.; Shi, P.; Bai, Y.; Luo, H.; Ma, R.; Yao, B. Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharification performance. Biotechnol. Biofuels 2016, 9, 147. [Google Scholar] [CrossRef]
- Zegers, N.D.; Kluter, E.; van Der Stap, H.; van Dura, E.; van Dalen, P.; Shaw, M.; Baillie, L. Expression of the protective antigen of Bacillus anthracis by Lactobacillus casei: Towards the development of an oral vaccine against anthrax. J. Appl. Microbiol. 1999, 87, 309–314. [Google Scholar] [CrossRef]
- Gaya, P.; Peirotén, Á.; Landete, J.M. Expression of a β-glucosidase in bacteria with biotechnological interest confers them the ability to deglycosylate lignans and flavonoids in vegetal foods. Appl. Microbiol. Biotechnol. 2020, 104, 4903–4913. [Google Scholar] [CrossRef]
- van de Guchte, M.; van der Vossen, J.M.; Kok, J.; Venema, G. Construction of a lactococcal expression vector: Expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 1989, 55, 224–228. [Google Scholar] [CrossRef]
- Pang, J.; Liu, Z.Y.; Hao, M.; Zhang, Y.F.; Qi, Q.S. An isolated cellulolytic Escherichia coli from bovine rumen produces ethanol and hydrogen from corn straw. Biotechnol. Biofuels 2017, 10, 165. [Google Scholar] [CrossRef]
- Liu, Z.L.; Li, H.N.; Song, H.T.; Xiao, W.J.; Xia, W.C.; Liu, X.P.; Jiang, Z.B. Construction of a trifunctional cellulase and expression in Saccharomyces cerevisiae using a fusion protein. BMC Biotechnol. 2018, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Kurniasih, S.D.; Alfi, A.; Natalia, D.; Radjasa, O.K.; Nurachman, Z. Construction of individual, fused, and co-expressed proteins of endoglucanase and β-glucosidase for hydrolyzing sugarcane bagasse. Microbiol. Res. 2014, 169, 725–732. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Dong, X.; Dong, Z. Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses. Anaerobe 2005, 11, 207–215. [Google Scholar] [CrossRef]
- Wei, Y.Q.; Yang, H.J.; Luan, Y.; Long, R.J.; Wu, Y.J.; Wang, Z.Y. Isolation, identification and fibrolytic characteristics of rumen fungi grown with indigenous methanogen from yaks (Bos grunniens) grazing on the Qinghai-Tibetan Plateau. J. Appl. Microbiol. 2016, 120, 571–587. [Google Scholar] [CrossRef]
- Zhao, J.; Shao, T.; Chen, S.; Tao, X.; Li, J. Characterization and identification of cellulase-producing Enterococcus species isolated from Tibetan yak (Bos grunniens) rumen and their application in various forage silages. J. Appl. Microbiol. 2021, 131, 1102–1112. [Google Scholar] [CrossRef]
- Jiang, H.; Cao, H.W.; Chai, Z.X.; Chen, X.Y.; Zhang, C.F.; Zhu, Y.; Xin, J.W. Dynamic alterations in yak (Bos grunniens) rumen microbiome in response to seasonal variations in diet. Physiol. Genomics 2022, 54, 514–525. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, L.; Ke, S.; Chen, X.; Kenéz, Á.; Xu, W.; Wang, D.; Zhang, F.; Li, Y.; Cui, Z.; et al. Yak rumen microbiome elevates fiber degradation ability and alters rumen fermentation pattern to increase feed efficiency. Anim. Nutr. 2022, 11, 201–214. [Google Scholar] [CrossRef]
- Kuipers, O.P.; de Ruyter, P.G.; Kleerebezem, M.; de Vos, W.M. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 1998, 64, 15–21. [Google Scholar] [CrossRef]
- Niu, L.; Zhang, H.; Wu, Z.; Wang, Y.; Liu, H.; Wu, X.; Wang, W. Correction: Modified TCA/acetone precipitation of plant proteins for proteomic analysis. PLoS ONE 2019, 14, e0211612. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Hong, J.; Ye, X. Cellulase assays. Methods Mol. Biol. 2009, 581, 213–231. [Google Scholar] [PubMed]
- Dashtban, M.; Maki, M.; Leung, K.T.; Mao, C.; Qin, W. Cellulase activities in biomass conversion: Measurement methods and comparison. Crit. Rev. Biotechnol. 2010, 30, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Singhvi, M.S.; Zinjarde, S.S. Production of pharmaceutically important genistein and daidzein from soybean flour extract by using β-glucosidase derived from Penicillium janthinellum NCIM 1171. Process. Biochem. 2020, 97, 183–190. [Google Scholar] [CrossRef]
- Jang, W.J.; Lee, G.H.; Lee, J.M.; Kim, T.Y.; Jeon, M.H.; Kim, Y.H.; Lee, E.W. Improving enzyme activity, thermostability and storage stability of β-1,3-1,4-glucanase with poly-γ-glutamic acid produced by Bacillus sp. SJ-10. Enzyme Microb. Technol. 2021, 143, 109703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, C.; Huang, C.Y.; Yu, Q.; Liu, H.C.; Zhang, C.W.; Pei, X.F. Construction and secretory expression of beta-galactosidase gene from Lactobacillus bulgaricus in Lactococcus lactis. Biomed. Environ. Sci. 2012, 25, 203–209. [Google Scholar]
- Liu, F.; van Heel, A.J.; Chen, J.; Kuipers, O.P. Functional production of clostridial circularin A in Lactococcus lactis NZ9000 and mutational analysis of its aromatic and cationic residues. Front. Microbiol. 2022, 13, 1026290. [Google Scholar] [CrossRef]
- King, M.S.; Boes, C.; Kunji, E.R. Membrane protein expression in Lactococcus lactis. Methods Enzymol. 2015, 556, 77–97. [Google Scholar] [PubMed]
- Sriraman, K.; Jayaraman, G. Enhancement of recombinant streptokinase production in Lactococcus lactis by suppression of acid tolerance response. Appl. Microbiol. Biotechnol. 2006, 72, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.H.; Shao, T.; Dong, Z.H.; Bai, Y.F. Solution for promoting egl3 gene of Trichoderma reesei high-efficiency secretory expression in Escherichia coli and Lactococcus lactis. Process. Biochem. 2017, 62, 135–143. [Google Scholar]
- Stephenson, D.P.; Moore, R.J.; Allison, G.E. Transformation of, and heterologous protein expression in, Lactobacillus agilis and Lactobacillus vaginalis isolates from the chicken gastrointestinal tract. Appl. Environ. Microbiol. 2011, 77, 220–228. [Google Scholar] [CrossRef]
- van Asseldonk, M.; Rutten, G.; Oteman, M.; Siezen, R.J.; de Vos, W.M.; Simons, G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 1990, 95, 155–160. [Google Scholar] [CrossRef]
- Le Loir, Y.; Nouaille, S.; Commissaire, J.; Brétigny, L.; Gruss, A.; Langella, P. Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 2001, 67, 4119–4127. [Google Scholar] [CrossRef]
- Cao, G.; Guo, W.; Wang, A.; Zhao, L.; Xu, C.; Zhao, Q.; Ren, N. Enhanced cellulosic hydrogen production from lime-treated cornstalk wastes using thermophilic anaerobic microflora. Int. J. Hydrog. Energy 2012, 37, 13161–13166. [Google Scholar] [CrossRef]
- Duedu, K.O.; French, C.E. Characterization of a Cellulomonas fimi exoglucanase/xylanase-endoglucanase gene fusion which improves microbial degradation of cellulosic biomass. Enzyme Microb. Technol. 2016, 93–94, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Viikari, L.; Alapuranen, M.; Puranen, T.; Vehmaanperä, J.; Siika-Aho, M. Thermostable enzymes in lignocellulose hydrolysis. Adv. Biochem. Eng. Biotechnol. 2007, 108, 121–145. [Google Scholar] [PubMed]
- Kiran, T.; Asad, W.; Ajaz, M.; Hanif, M.; Rasool, S.A. Industrially relevant cellulase production by indigenous thermophilic Bacillus licheniformis TLW-3 strain: Isolation-molecular identification and enzyme yield optimization. Pak. J. Pharm. Sci. 2018, 31, 2333–2340. [Google Scholar]
- Sun, J.; Wang, W.; Yao, C.; Dai, F.; Zhu, X.; Liu, J.; Hao, J. Overexpression and characterization of a novel cold-adapted and salt-tolerant GH1 β-glucosidase from the marine bacterium Alteromonas sp. L82. J. Microbiol. 2018, 56, 656–664. [Google Scholar] [CrossRef]
- Wang, H.; Zhai, L.; Geng, A. Enhanced cellulase and reducing sugar production by a new mutant strain Trichoderma harzianum EUA20. J. Biosci. Bioeng. 2020, 129, 242–249. [Google Scholar] [CrossRef]
- Haq, I.U.; Tahir, S.F.; Aftab, M.N.; Akram, F.; Rehman, A.U.; Nawaz, A.; Mukhtar, H. Purification and Characterization of a Thermostable Cellobiohydrolase from Thermotoga petrophila. Protein Pept. Lett. 2018, 25, 1003–1014. [Google Scholar] [CrossRef]
- Cheng, J.; Huang, S.; Jiang, H.; Zhang, Y.; Li, L.; Wang, J.; Fan, C. Isolation and characterization of a non-specific endoglucanase from a metagenomic library of goat rumen. World J. Microbiol. Biotechnol. 2016, 32, 12. [Google Scholar] [CrossRef]
- Pyeon, H.M.; Lee, Y.S.; Choi, Y.L. Cloning, purification, and characterization of GH3 β-glucosidase, MtBgl85, from Microbulbifer thermotolerans DAU221. PeerJ 2019, 7, e7106. [Google Scholar] [CrossRef]
- Sun, S.; Sun, S.; Cao, X.; Sun, R. The role of pretreatment in improving the enzymatic hydrolysis of lignocellulosic materials. Bioresour. Technol. 2016, 199, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, J.; Zhao, J.; Wu, J.; Shao, T. Enhancement of lignocellulosic degradation in high-moisture alfalfa via anaerobic bioprocess of engineered Lactococcus lactis with the function of secreting cellulase. Biotechnol. Biofuels 2019, 12, 88. [Google Scholar] [CrossRef] [PubMed]
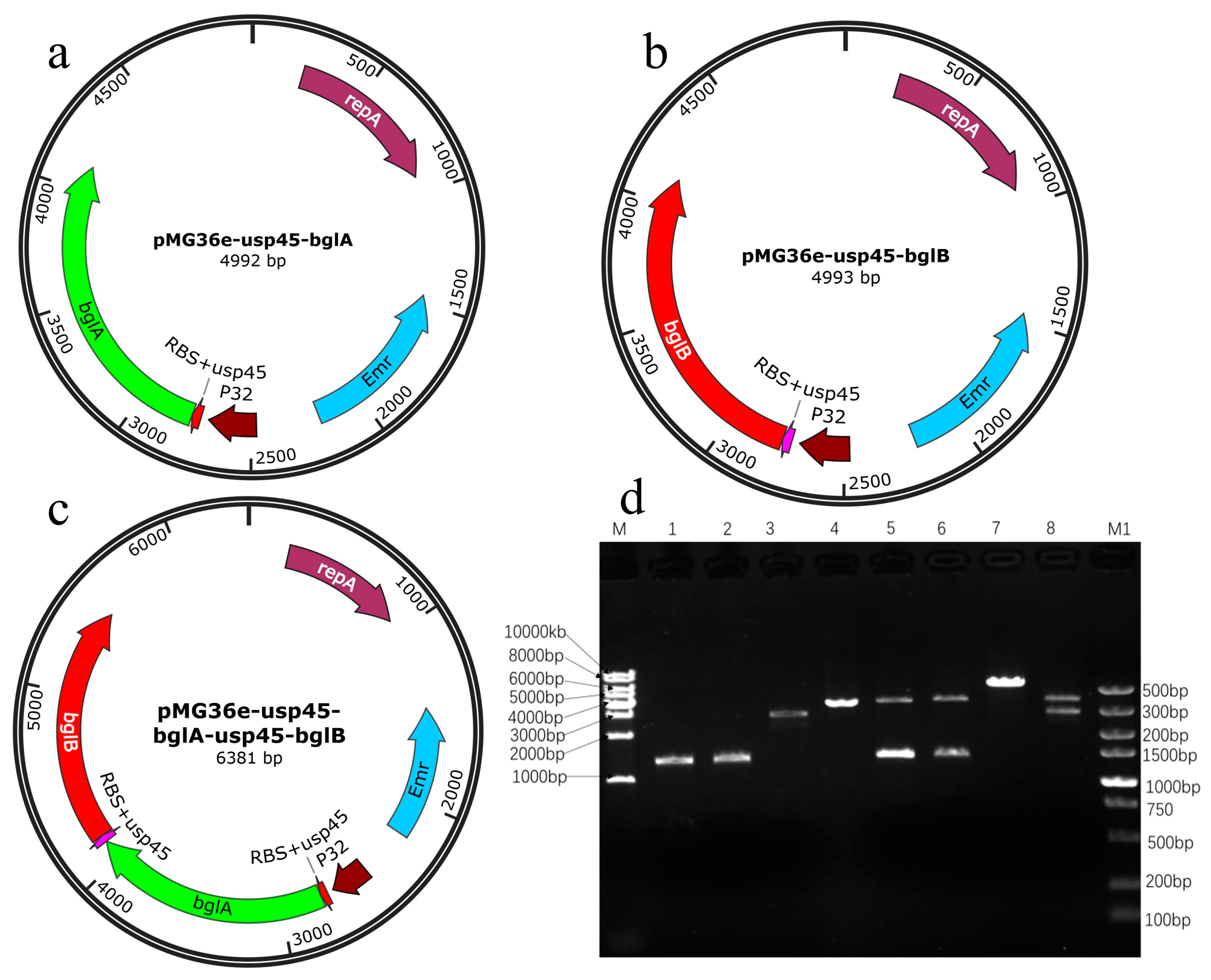

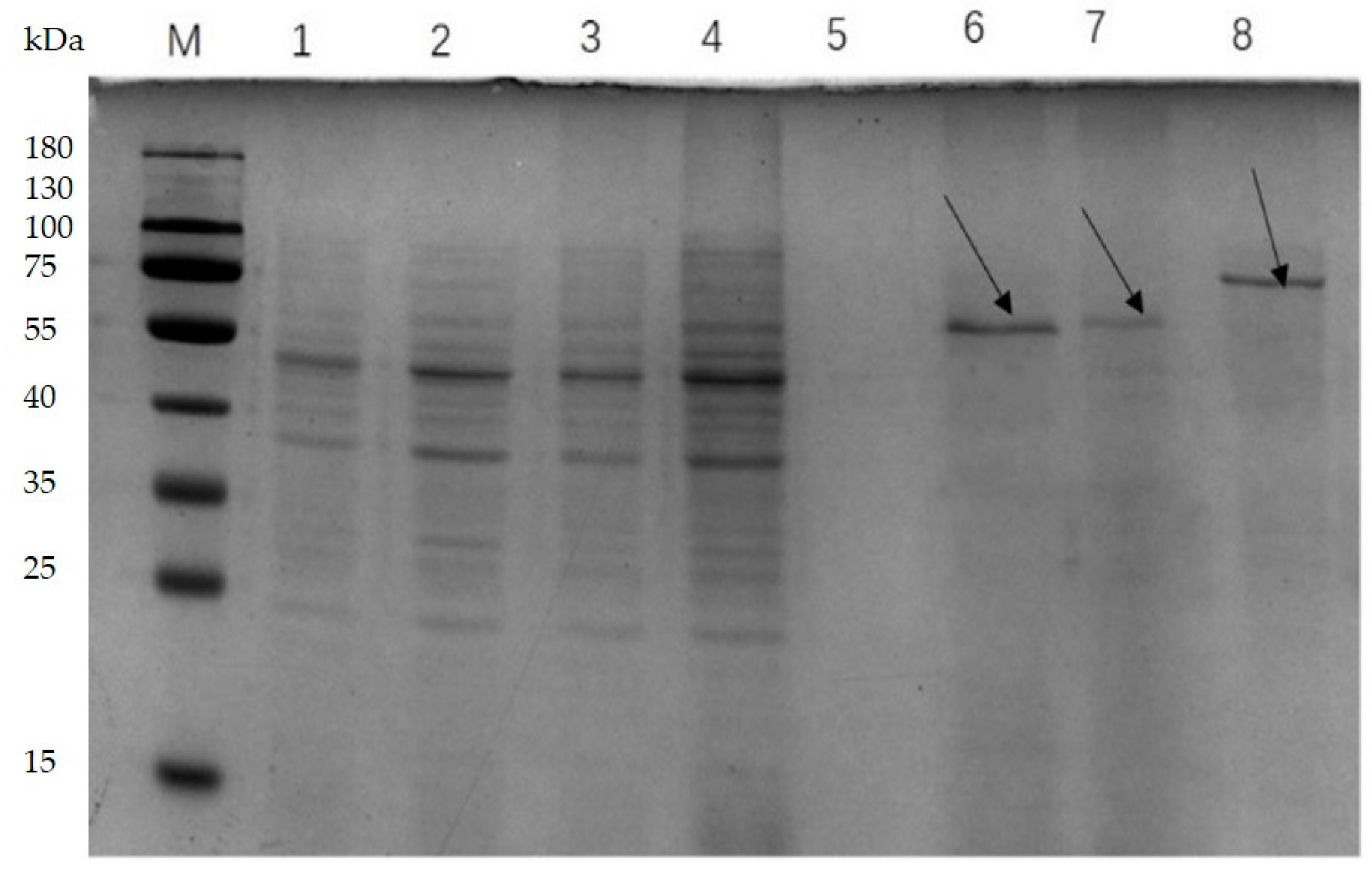
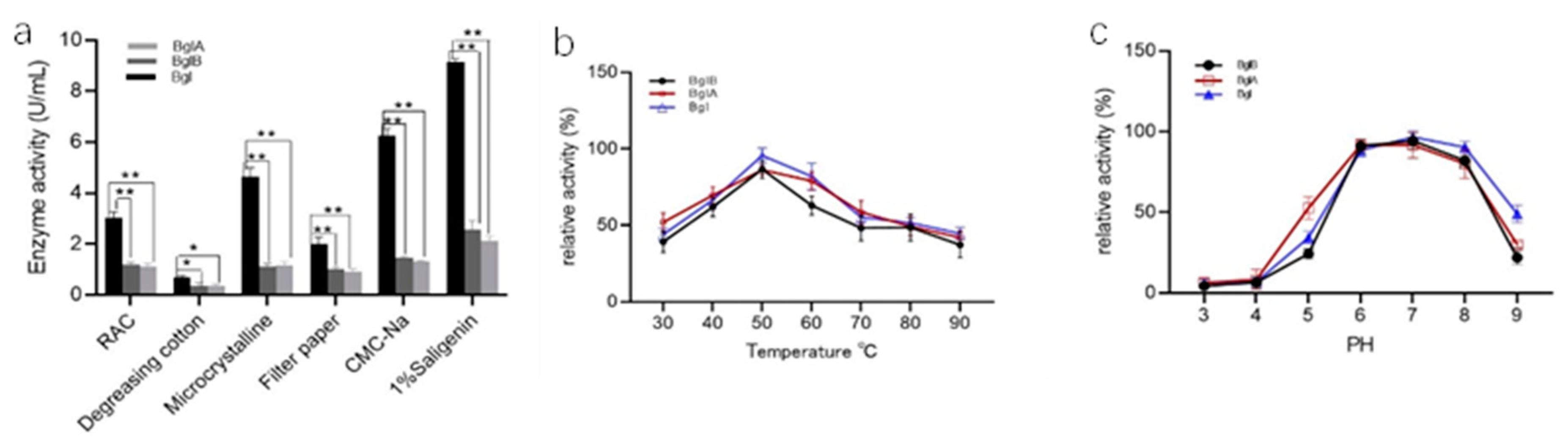
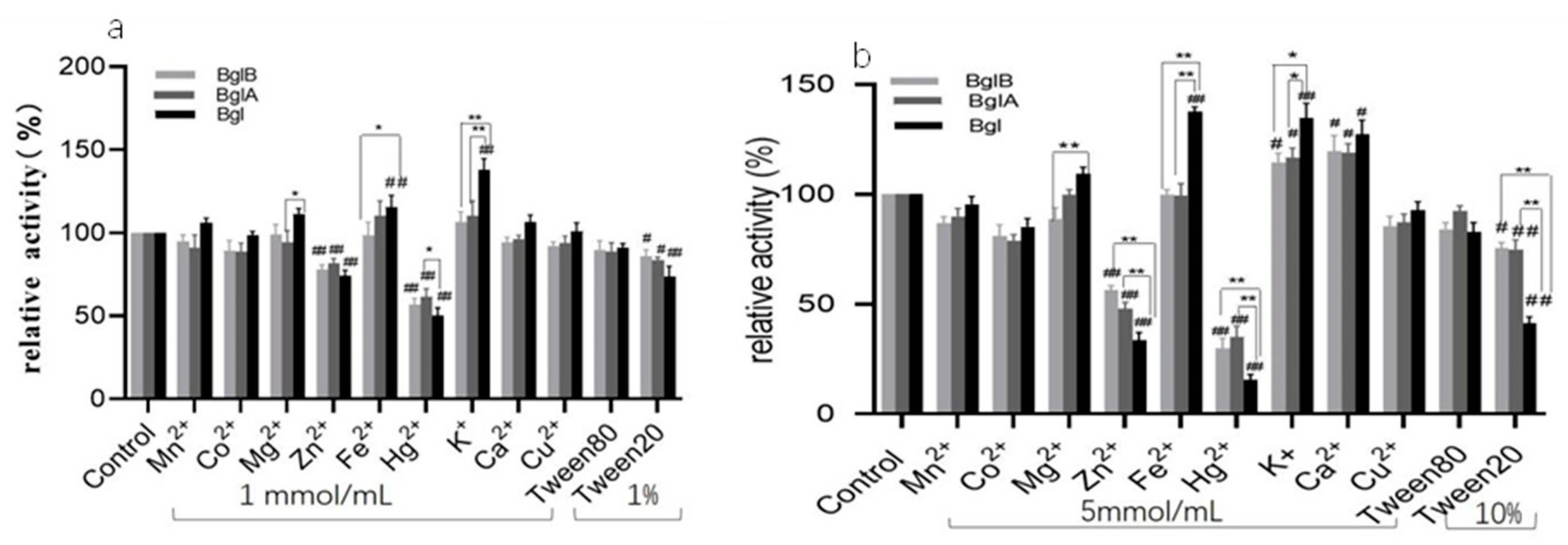
| Strain and Plasmid | Relevant Trait(s) | Source or Reference | |
|---|---|---|---|
| Strains | Escherichia coli DH5α | supE44 Δlac U169 (Φ80 lacZ ΔM15) hsdR17 recA1, endA1 gyrA96 thi-l relA1 | Our laboratory |
| L. lactis NZ9000 | Expressing recombinant plasmids | [28] | |
| E. coli DH5α/pMG36e-usp45-bglA | E. coli DH5α with pMG36e-usp45-bglA | This study | |
| E. coli DH5α/pMG36e-usp45-bglB | E. coli DH5α with pMG36e-usp45-bglB | This study | |
| E. coli DH5α/pMG36e-usp45-bglA-usp45-bglB | E. coli DH5α with pMG36e-usp45-bglA-usp45-bglB | This study | |
| L. lactis NZ9000/pMG36e | L. lactis NZ9000 with pMG36e | This study | |
| L. lactis NZ9000/pMG36e-usp45-bglA | L. lactis NZ9000 with secretory expression BglA | This study | |
| L. lactis NZ9000/pMG36e-usp45-bglB | L. lactis NZ9000 with secretory expression BglB | This study | |
| L. lactis NZ9000/pMG36e-usp45-bglA-usp45-bglB | L. lactis NZ9000 with secretory expression Bgl | This study | |
| Plasmids | pMG36e | Emr; expression vector with the P32 promoter, multiple cloning sites (MCFs), and prtP translational terminator | [19] |
| pMG36e-usp45-bglA | Emr; secretory expression of BglA | This study | |
| pMG36e-usp45-bglB | Emr; secretory expression of BglB | This study | |
| pMG36e-usp45-bglA-usp45-bglB | Emr; secretory expression of fusion Bgl | This study | |
| Primer | Sequence (5′-3′) | Restriction Enzyme |
|---|---|---|
| bglA-F | AACTGCAGAGAGCGCAAAAAAAAGATTATCTCAGCTAATGACTATTTTTCAATTTCCGC | Pst I |
| bglA-R | TCAAGCTTCTTTGTTTAGCGTC | Hind III |
| bglB-F | ATGCTGCAGAATACCTTTATATTTC | Pst I |
| bglB-R | CCCAAGCTTGCGCAAAAAAAAGATTATCTCAGCTACTTTTCTATTTAAAACCCG | Hind III |
| bglA-R1 | AACTGCAGAAGAAGGAGATATACATGCAAAAAAAAGATTATCTCAGCTAATGACTATTTTTCAATTTCCGC | Pst I |
| bglB-F1 | CCCAAGCTTCCCTTTTCTATTTAAAACCCG | Hind III |
| Strain | Vmax (μmol∙min−1mg−1) | Km (μmol∙L−1) | Kcat (s−1) | Kcat/Km (L∙s−1∙μmol−1) |
|---|---|---|---|---|
| L. lactis NZ9000/pMG36e-usp45-bglA | 1343 | 38.61 | 434.8 | 11.26 |
| L. lactis NZ9000/pMG36e-usp45-bglB | 1254 | 43.99 | 348.7 | 7.93 |
| L. lactis NZ9000/pMG36e-usp45-bglA-usp45-bglB | 1347 | 34.8 | 1016 | 29.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Yang, Y.; Ma, C.; Sunkang, Y.; Tang, S.; Zhang, Z.; Wan, X.; Wei, Y. Expression of β-Glucosidases from the Yak Rumen in Lactic Acid Bacteria: A Genetic Engineering Approach. Microorganisms 2023, 11, 1387. https://doi.org/10.3390/microorganisms11061387
Wang C, Yang Y, Ma C, Sunkang Y, Tang S, Zhang Z, Wan X, Wei Y. Expression of β-Glucosidases from the Yak Rumen in Lactic Acid Bacteria: A Genetic Engineering Approach. Microorganisms. 2023; 11(6):1387. https://doi.org/10.3390/microorganisms11061387
Chicago/Turabian StyleWang, Chuan, Yuze Yang, Chunjuan Ma, Yongjie Sunkang, Shaoqing Tang, Zhao Zhang, Xuerui Wan, and Yaqin Wei. 2023. "Expression of β-Glucosidases from the Yak Rumen in Lactic Acid Bacteria: A Genetic Engineering Approach" Microorganisms 11, no. 6: 1387. https://doi.org/10.3390/microorganisms11061387
APA StyleWang, C., Yang, Y., Ma, C., Sunkang, Y., Tang, S., Zhang, Z., Wan, X., & Wei, Y. (2023). Expression of β-Glucosidases from the Yak Rumen in Lactic Acid Bacteria: A Genetic Engineering Approach. Microorganisms, 11(6), 1387. https://doi.org/10.3390/microorganisms11061387






