Used Nasogastric Feeding Tubes from Neonates Contain Infant-Specific Bacterial Profiles
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Ethics
2.3. Bacteriological Analyses
2.4. Bacterial Profiling with High-Throughput Sequencing
2.5. Whole Genome Sequencing (WGS) of Bacterial Isolates
2.6. Data Analysis
2.6.1. 16S rRNA Gene (V3 Region) Amplicon Sequencing
2.6.2. WGS of Bacterial Isolates
2.6.3. Data Availability
3. Results
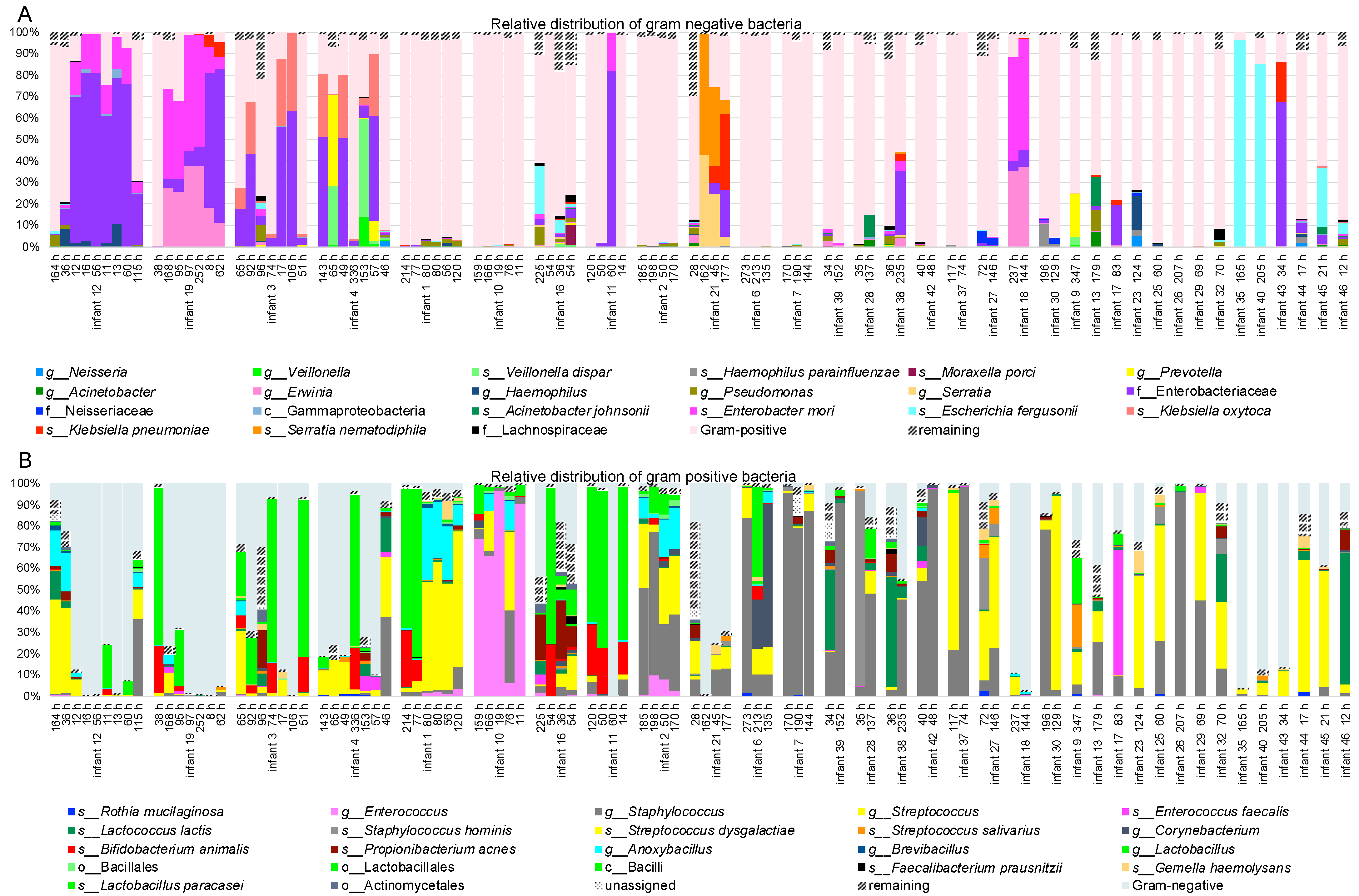
3.1. Bacterial Profiling with High-Throughput Sequencing
3.2. Whole Genome Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Brooks, B.; Firek, B.A.; Miller, C.S.; Sharon, I.; Thomas, B.C.; Baker, R.; Morowitz, M.J.; Banfield, J.F. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome 2014, 2, 1. [Google Scholar] [CrossRef]
- Hartz, L.E.; Bradshaw, W.; Brandon, D.H. Potential NICU Environmental Influences on the Neonate’s Microbiome. Adv. Neonatal Care 2015, 15, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.M.; Denning, P.W. Intestinal microbiota and its relationship with necrotizing enterocolitis. Pediatr. Res. 2015, 78, 232–238. [Google Scholar] [CrossRef]
- Warner, B.B.; Deych, E.; Zhou, Y.; Hall-Moore, C.; Weinstock, G.M.; Sodergren, E.; Shaikh, N.; Hoffmann, J.A.; Linneman, L.A.; Hamvas, A.; et al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: A prospective case-control study. Lancet 2016, 387, 1928–1936. [Google Scholar] [CrossRef] [PubMed]
- Morrow, A.L.; Lagomarcino, A.J.; Schibler, K.R.; Taft, D.H.; Yu, Z.; Wang, B.; Altaye, M.; Wagner, M.; Gevers, D.; Ward, D.V.; et al. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 2013, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.; Moles, L.; Melgar, A.; Ureta, N.; Bustos, G.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Early Gut Colonization of Preterm Infants: Effect of Enteral Feeding Tubes. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Mehall, J.; Kite, C.; Saltzman, D. Prospective study of the incidence and complications of bacterial contamination of enteral feeding in neonates. J. Pediatr. Surg. 2002, 37, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Hurrell, E.; Kucerova, E.; Loughlin, M.; Caubilla-Barron, J.; Hilton, A.; Armstrong, R.; Smith, C.; Grant, J.; Shoo, S.; Forsythe, S. Neonatal enteral feeding tubes as loci for colonisation by members of the Enterobacteriaceae. BMC Infect. Dis. 2009, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.L.; Trivedi, S.; Bhandari, N.P.; Ruf, A.; Scala, C.M.; Witowitch, G.; Chen, Y.; Renschen, C.; Meier, P.P.; Silvestri, J.M. Reducing necrotizing enterocolitis in very low birth weight infants using quality-improvement methods. J. Perinatol. 2014, 34, 850–857. [Google Scholar] [CrossRef]
- Petersen, S.M.; Greisen, G.; Krogfelt, K.A. Nasogastric feeding tubes from a neonatal department yield high concentrations of potentially pathogenic bacteria—Even 1 d after insertion. Pediatr. Res. 2016, 80, 395–400. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 2016, 1–22. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; Desantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Brister, J.R.; Bolton, E.E.; Canese, K.; Comeau, D.C.; Funk, K.; Ketter, A.; Kim, S.; Kimchi, A.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 48, D9–D16. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Costa, D.M.; Johani, K.; Melo, D.S.; Lopes, L.K.O.; Lopes Lima, L.K.O.; Tipple, A.F.V.; Hu, H.; Vickery, K. Biofilm contamination of high-touched surfaces in intensive care units: Epidemiology and potential impacts. Lett. Appl. Microbiol. 2019, 68, 269–276. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Dong, Y.; Speer, C.P. Late-onset neonatal sepsis: Recent developments. Arch. Dis. Child.-Fetal Neonatal Ed. 2015, 100, F257–F263. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.; Olm, M.; Firek, B.; Baker, R.; Geller-McGrath, D.; Reimer, S.; Soenjoyo, K.; Yip, J.; Dahan, D.; Thomas, B.; et al. The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms. Microbiome 2018, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Munkstrup, C.; Krogfelt, K.A.; Greisen, G.; Juhl, S.M. Feeding tube practices and the colonisation of the preterm stomach in the first week of life. Dan. Med. J. 2022, 69, 1–10. [Google Scholar]
- Bajorek, S.; Parker, L.; Li, N.; Winglee, K.; Weaver, M.; Johnson, J.; Sioda, M.; Gauthier, J.; Lemas, D.J.; Jobin, C.; et al. Initial microbial community of the neonatal stomach immediately after birth. Gut Microbes 2019, 10, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Reichert, F.; Piening, B.; Geffers, C.; Gastmeier, P.; Bührer, C.; Schwab, F. Pathogen-specific clustering of nosocomial blood stream infections in very preterm infants. Pediatrics 2016, 137, e20152860. [Google Scholar] [CrossRef] [PubMed]


| Patient ID | Birth Weight (g) | Gestational Age (Week) | Species Cultured in >/= 50% of NG-Tubes | Found in x of y NG-Tubes (x/y) |
|---|---|---|---|---|
| 1 | 580 | 24 + 5 | Staphylococcus epidermidis | 4/6 |
| 2 | 570 | 26 + 4 | Staphylococcus epidermidis Enterococcus faecium | 4/4 3/4 |
| 3 | 815 | 25 + 3 | Klebsiella oxytoca | 6/7 |
| 4 | 910 | 26 + 6 | Klebsiella oxytoca | 6/7 |
| 5 a | 790 | 25 + 1 | Enterococcus faecium | 5/5 |
| 6 a | 565 | 25 + 1 | Enterococcus faecium Enterobacter cloacae | 3/4 2/4 |
| 7 a | 800 | 25 + 1 | Enterobacter cloacae | 7/9 |
| 8 | 810 | 26 + 2 | None | |
| 9 | 650 | 24 + 4 | Enterobacter cloacae | 7/7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meinich Juhl, S.; Angeliki Krogfelt, K.; Kot, W.; Sandris Nielsen, D.; Krych, L. Used Nasogastric Feeding Tubes from Neonates Contain Infant-Specific Bacterial Profiles. Microorganisms 2023, 11, 1365. https://doi.org/10.3390/microorganisms11061365
Meinich Juhl S, Angeliki Krogfelt K, Kot W, Sandris Nielsen D, Krych L. Used Nasogastric Feeding Tubes from Neonates Contain Infant-Specific Bacterial Profiles. Microorganisms. 2023; 11(6):1365. https://doi.org/10.3390/microorganisms11061365
Chicago/Turabian StyleMeinich Juhl, Sandra, Karen Angeliki Krogfelt, Witold Kot, Dennis Sandris Nielsen, and Lukasz Krych. 2023. "Used Nasogastric Feeding Tubes from Neonates Contain Infant-Specific Bacterial Profiles" Microorganisms 11, no. 6: 1365. https://doi.org/10.3390/microorganisms11061365
APA StyleMeinich Juhl, S., Angeliki Krogfelt, K., Kot, W., Sandris Nielsen, D., & Krych, L. (2023). Used Nasogastric Feeding Tubes from Neonates Contain Infant-Specific Bacterial Profiles. Microorganisms, 11(6), 1365. https://doi.org/10.3390/microorganisms11061365







