Bovine Rectoanal Junction In Vitro Organ Culture Model System to Study Shiga Toxin-Producing Escherichia coli Adherence
Abstract
1. Introduction
2. Materials and Methods
3. Results
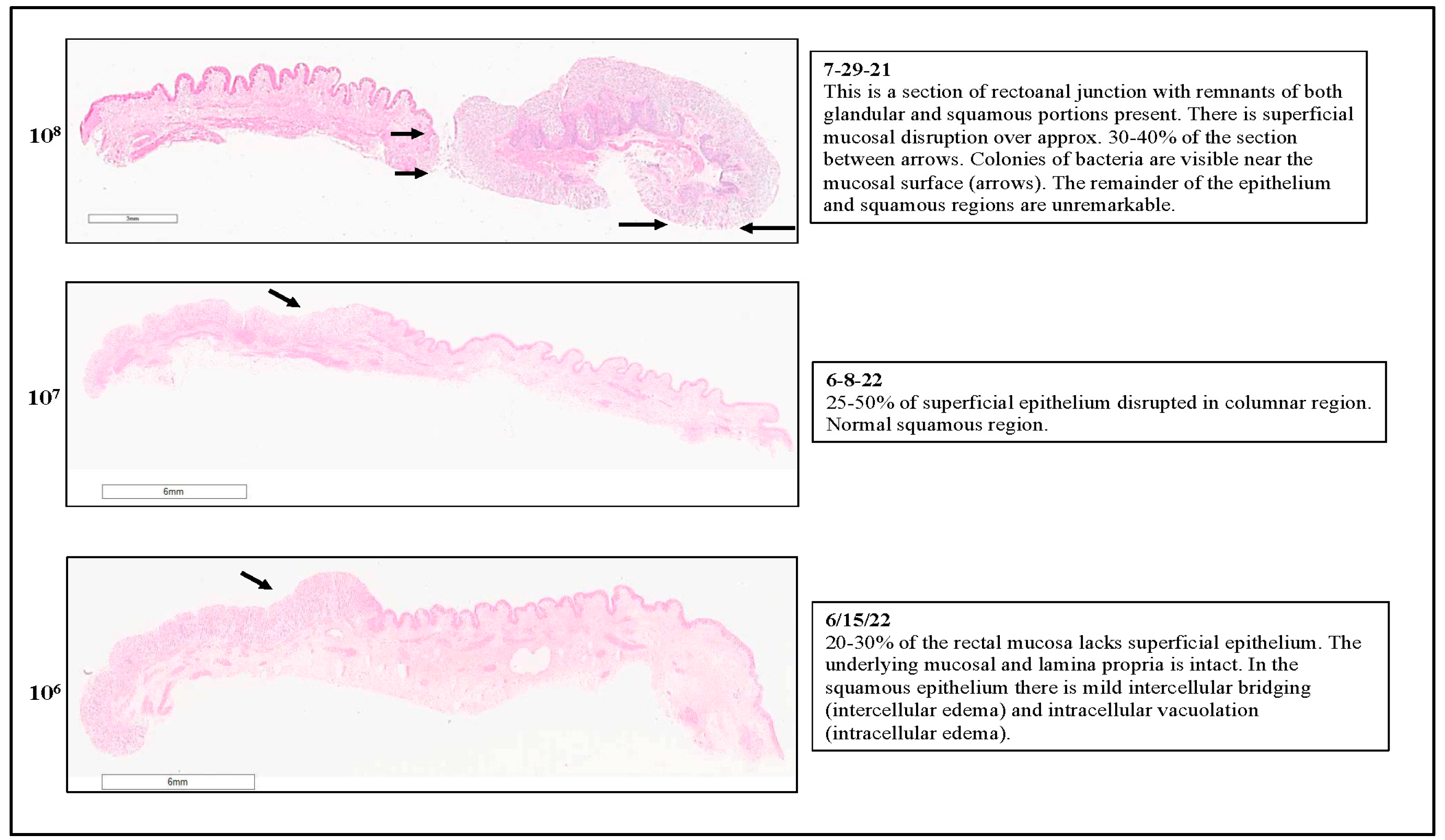
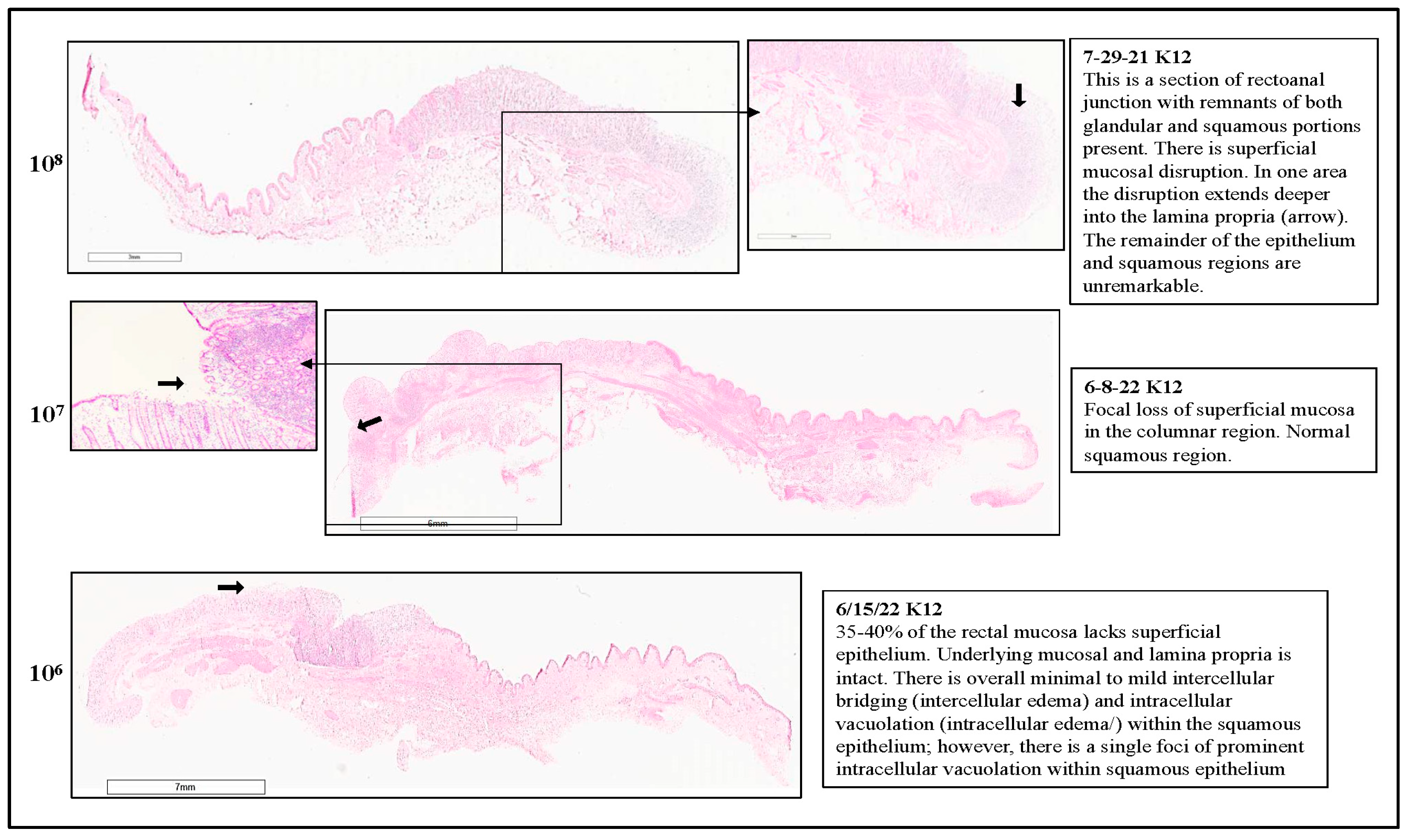
3.1. RAJ-IVOC Maintains Tissue Integrity and Viability Following Incubation for 3 h at 39 °C
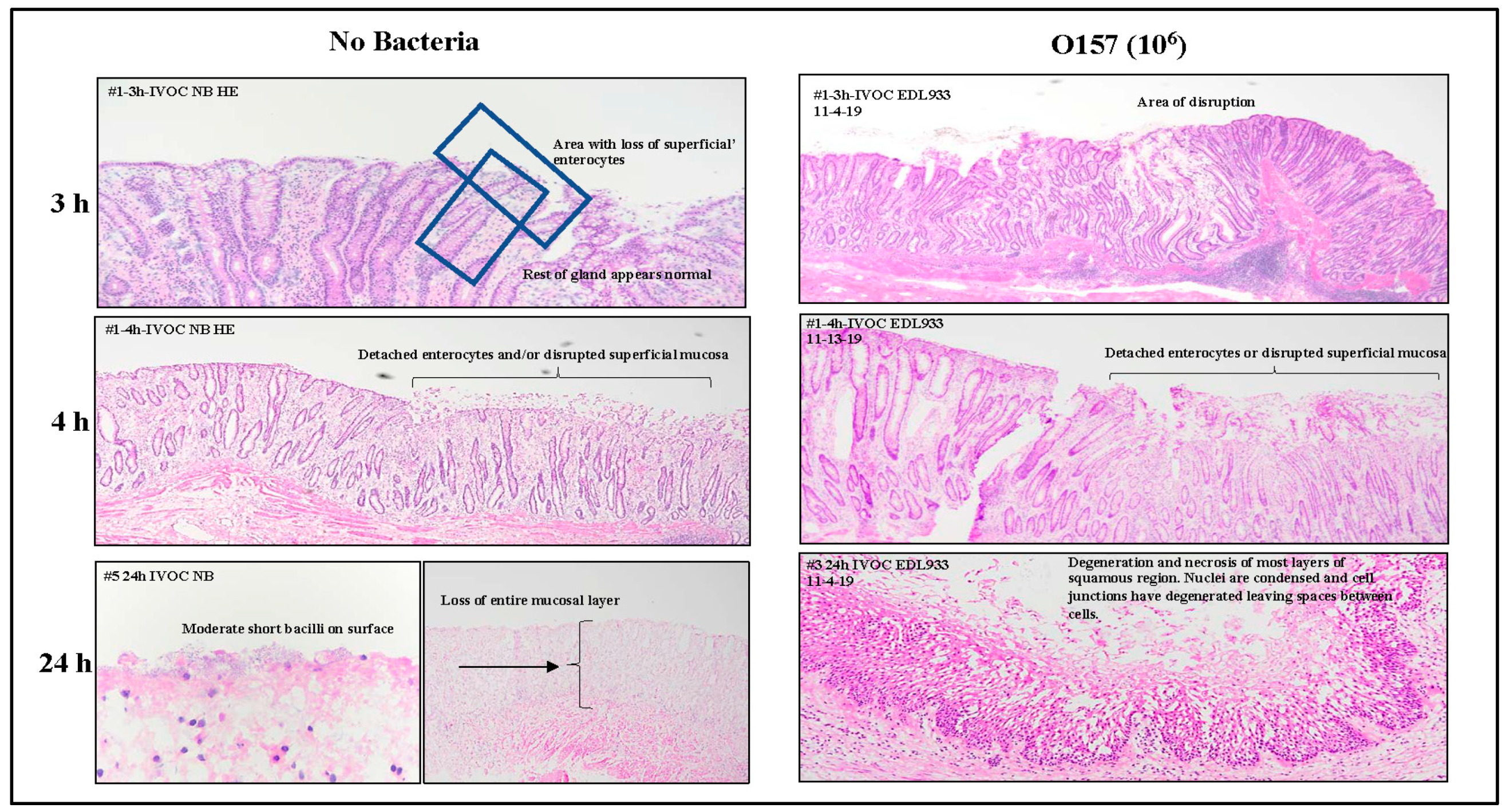
3.1.1. Histopathology
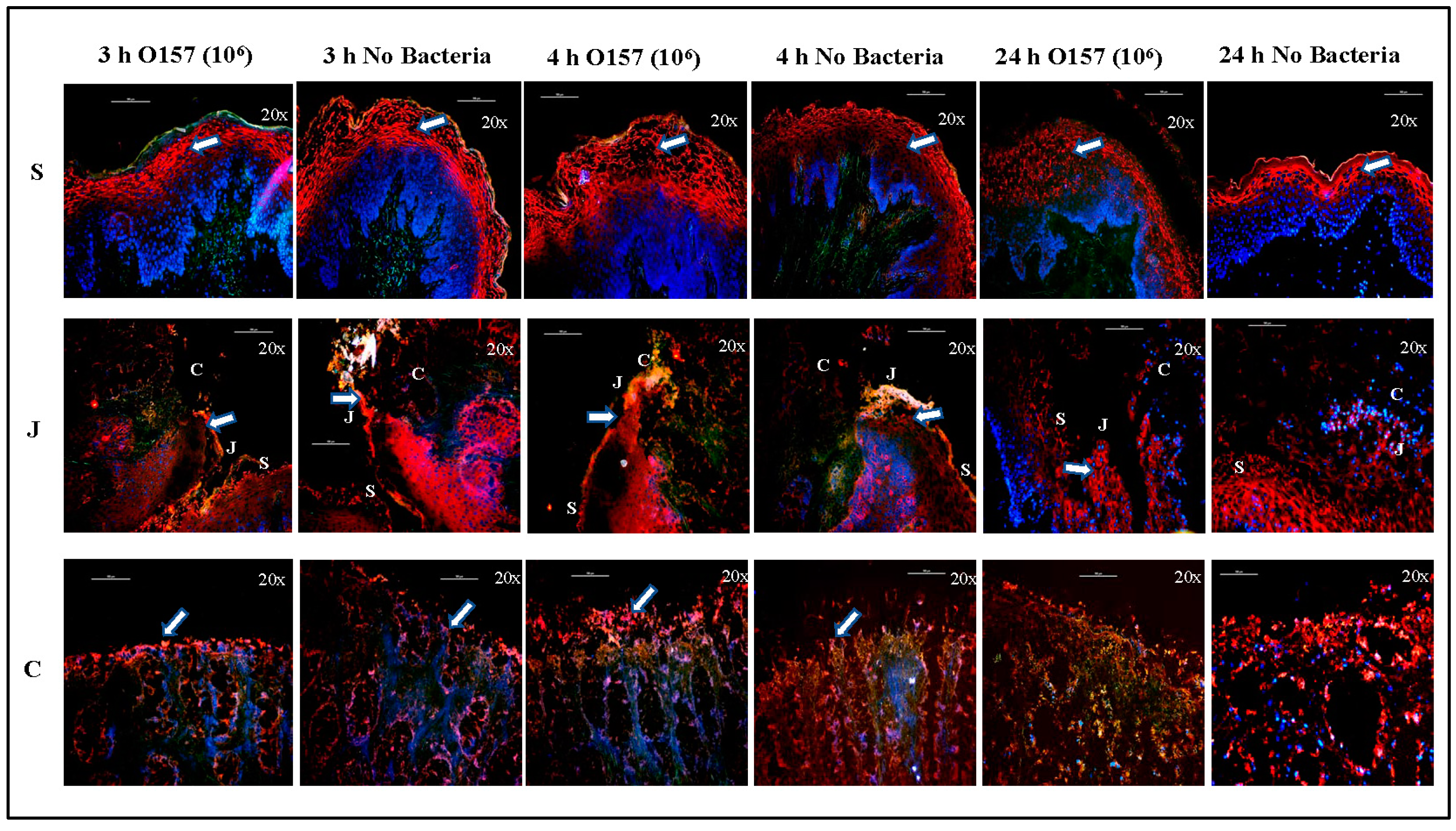
3.1.2. Cell Markers
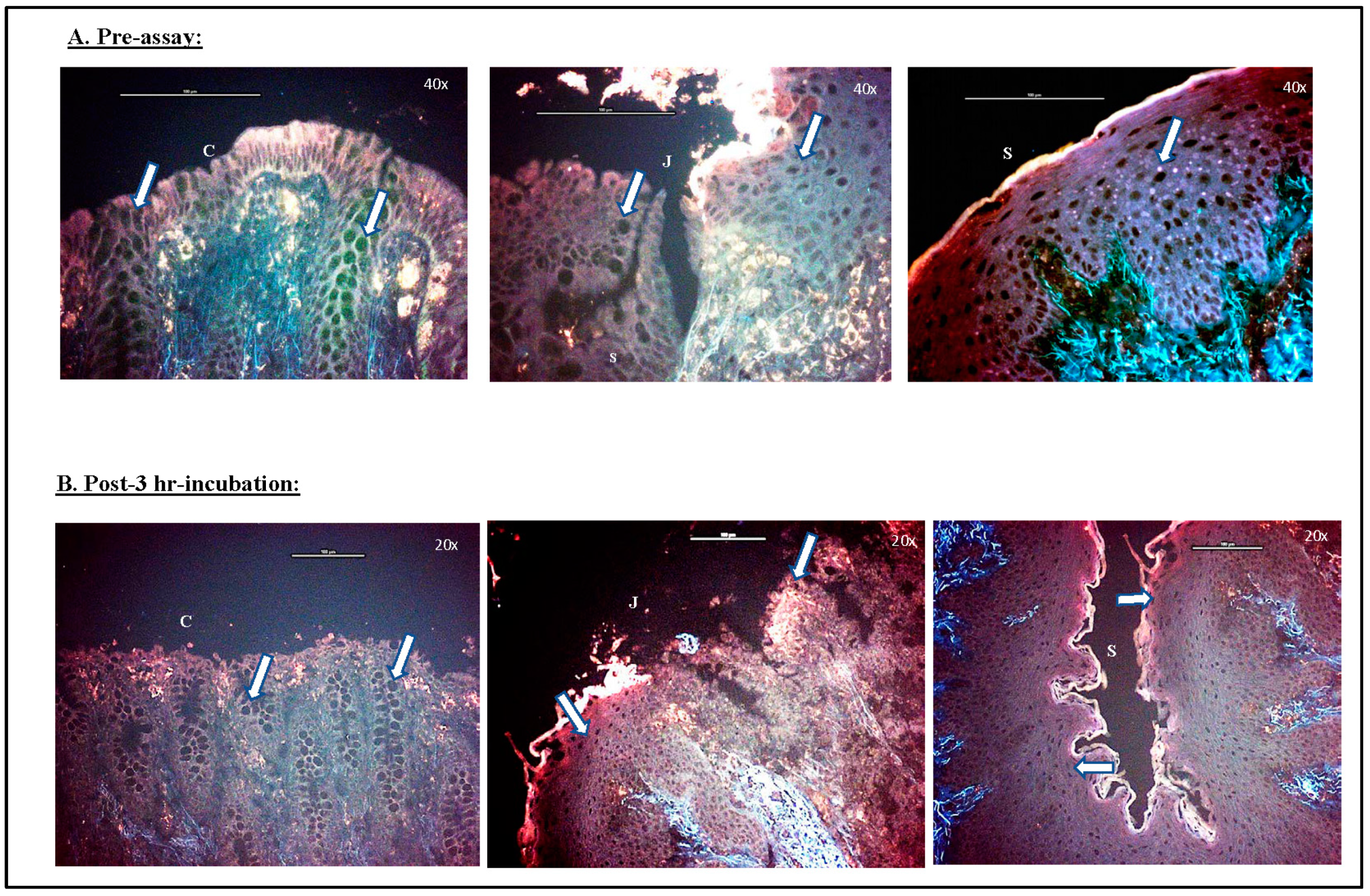
3.1.3. Viability Test
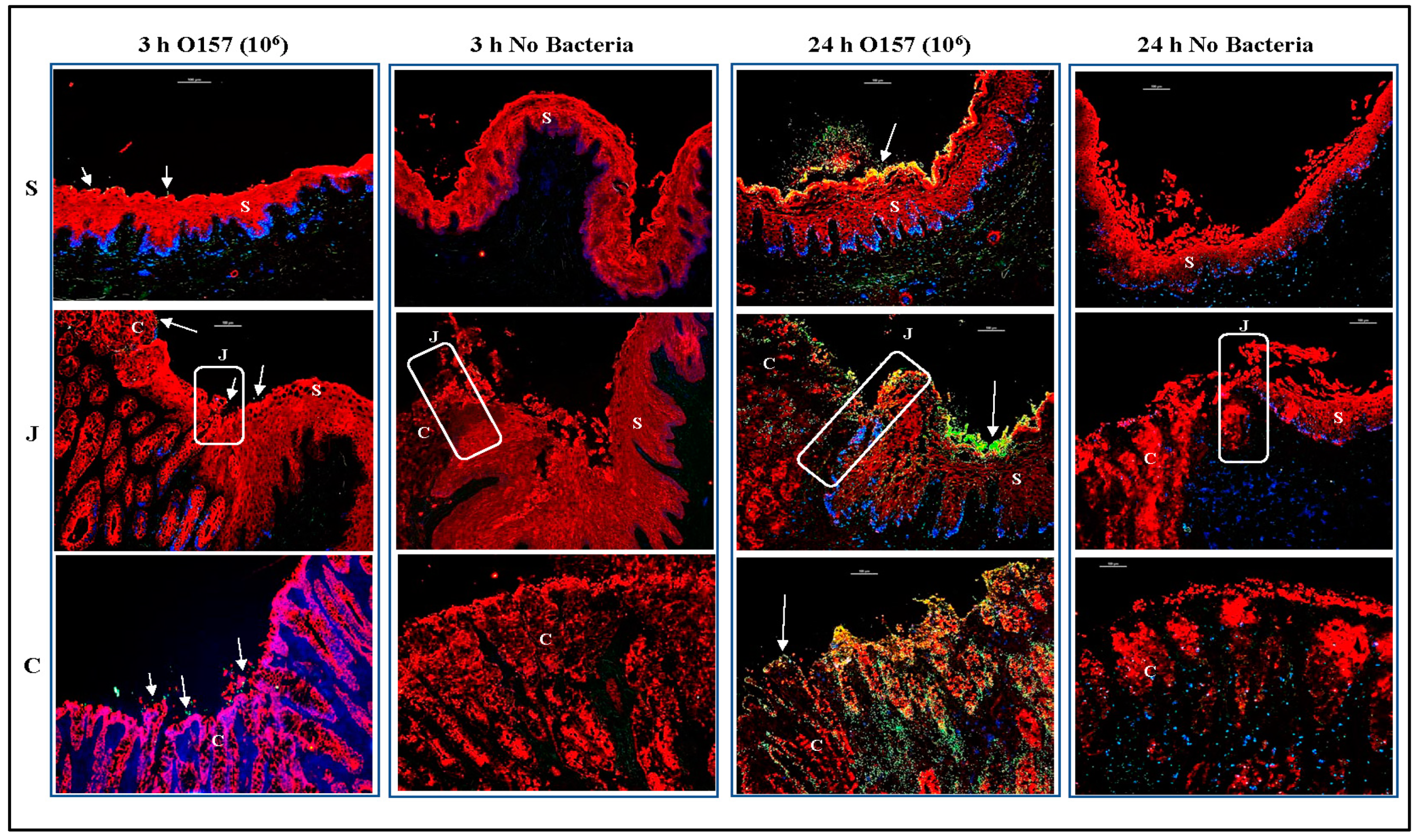
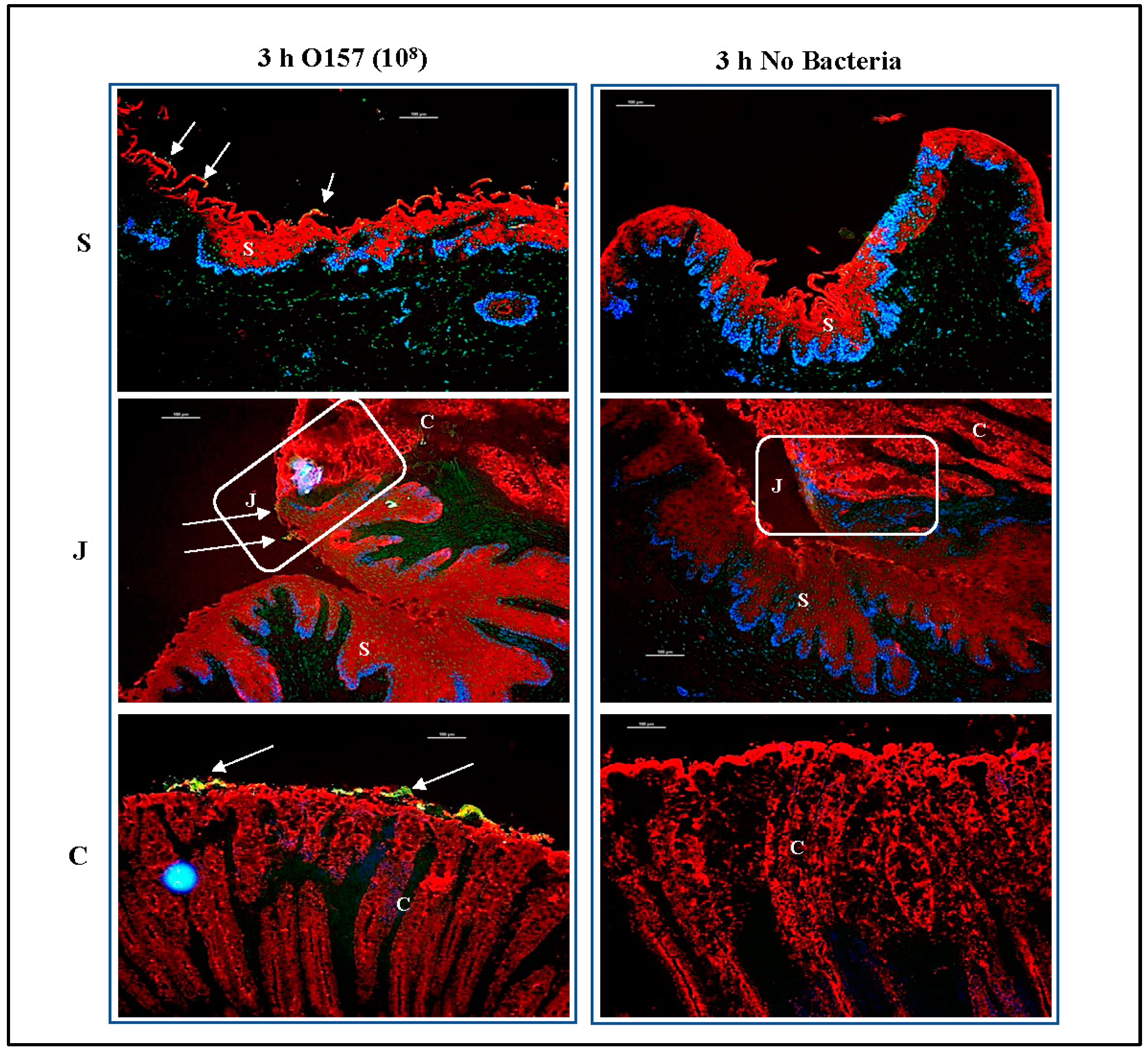
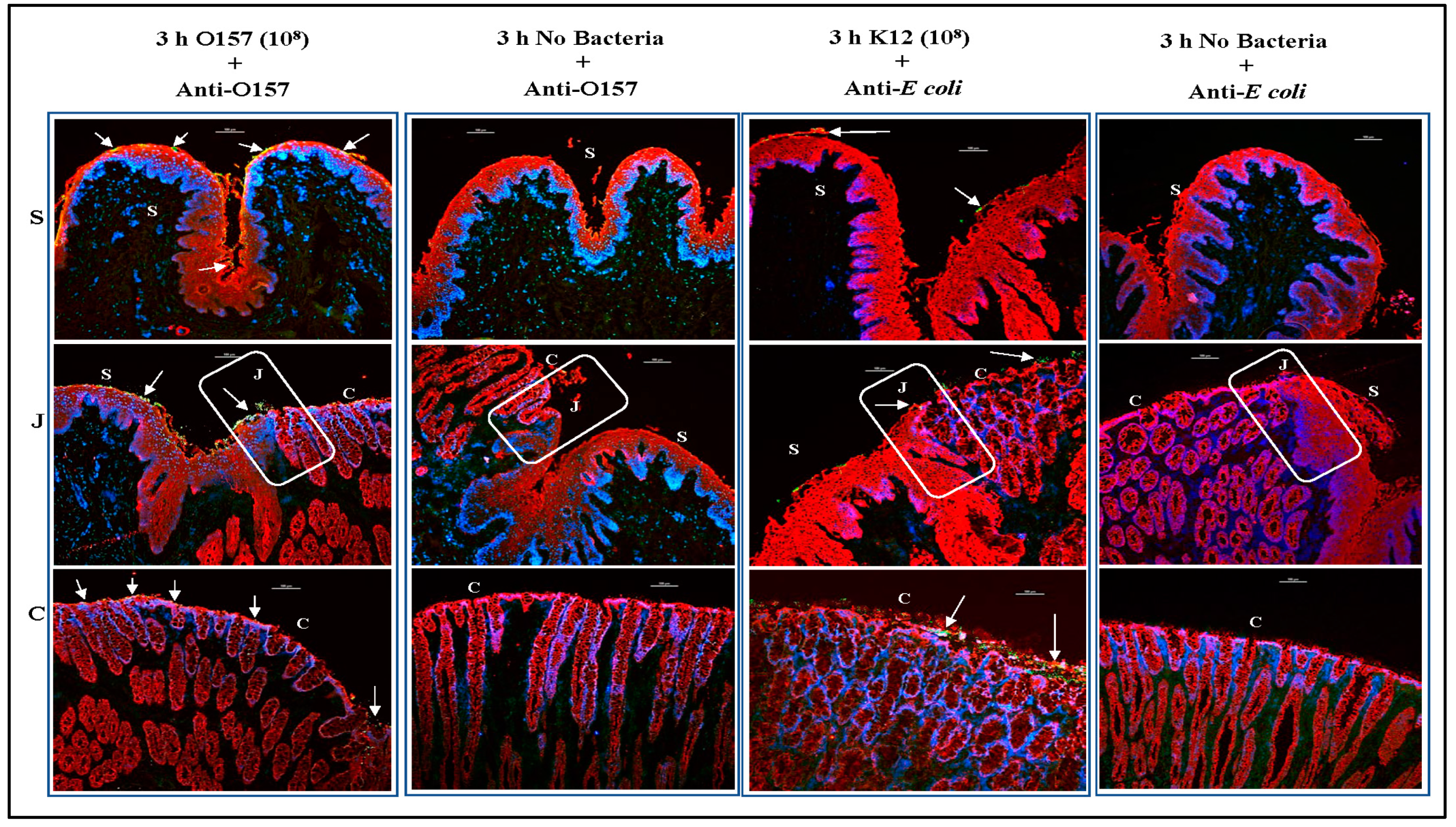
3.2. O157 Produces Distinct Adherence Patterns on the RAJ-IVOC Following 3 h Incubation at 39 °C
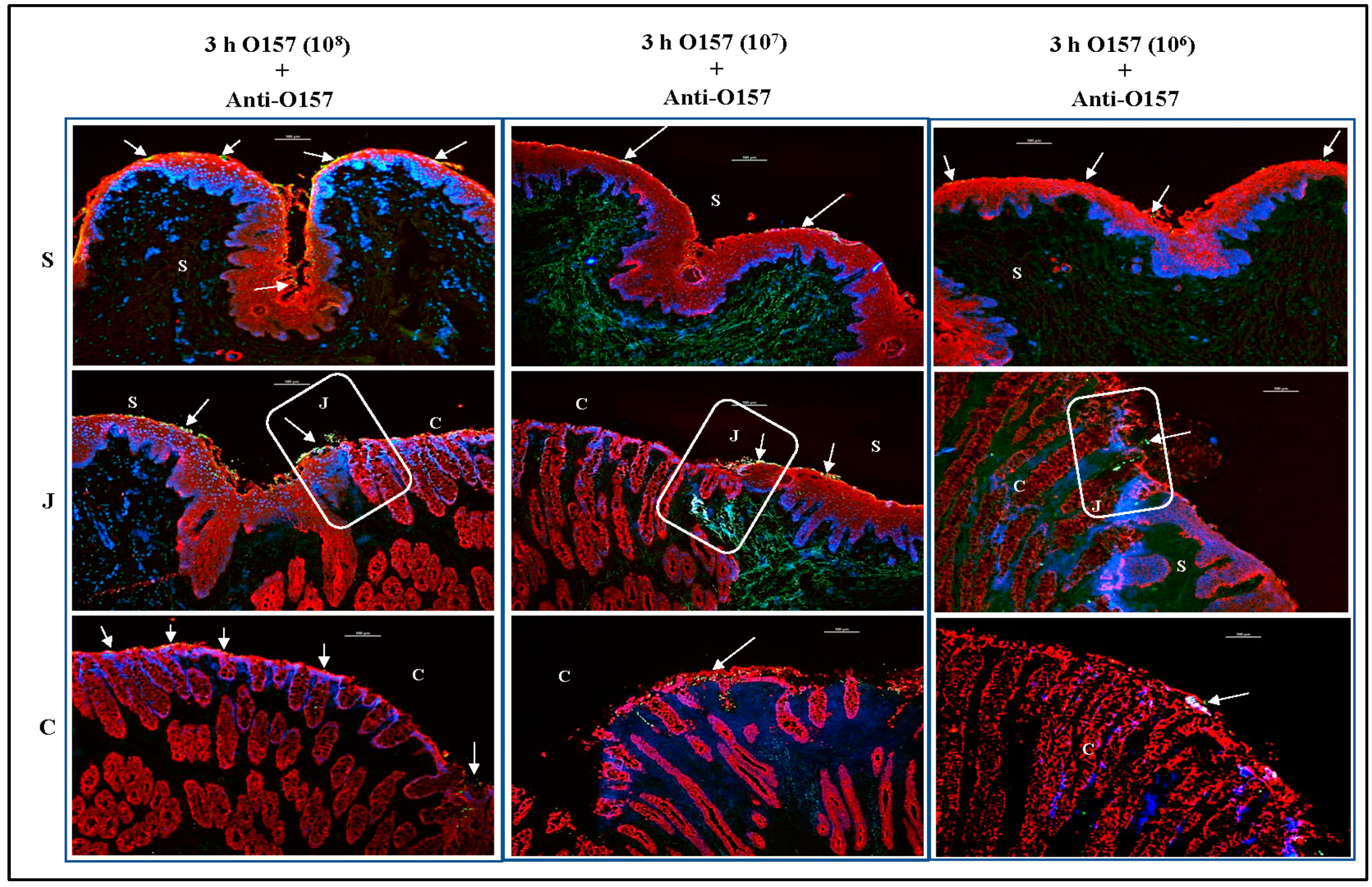
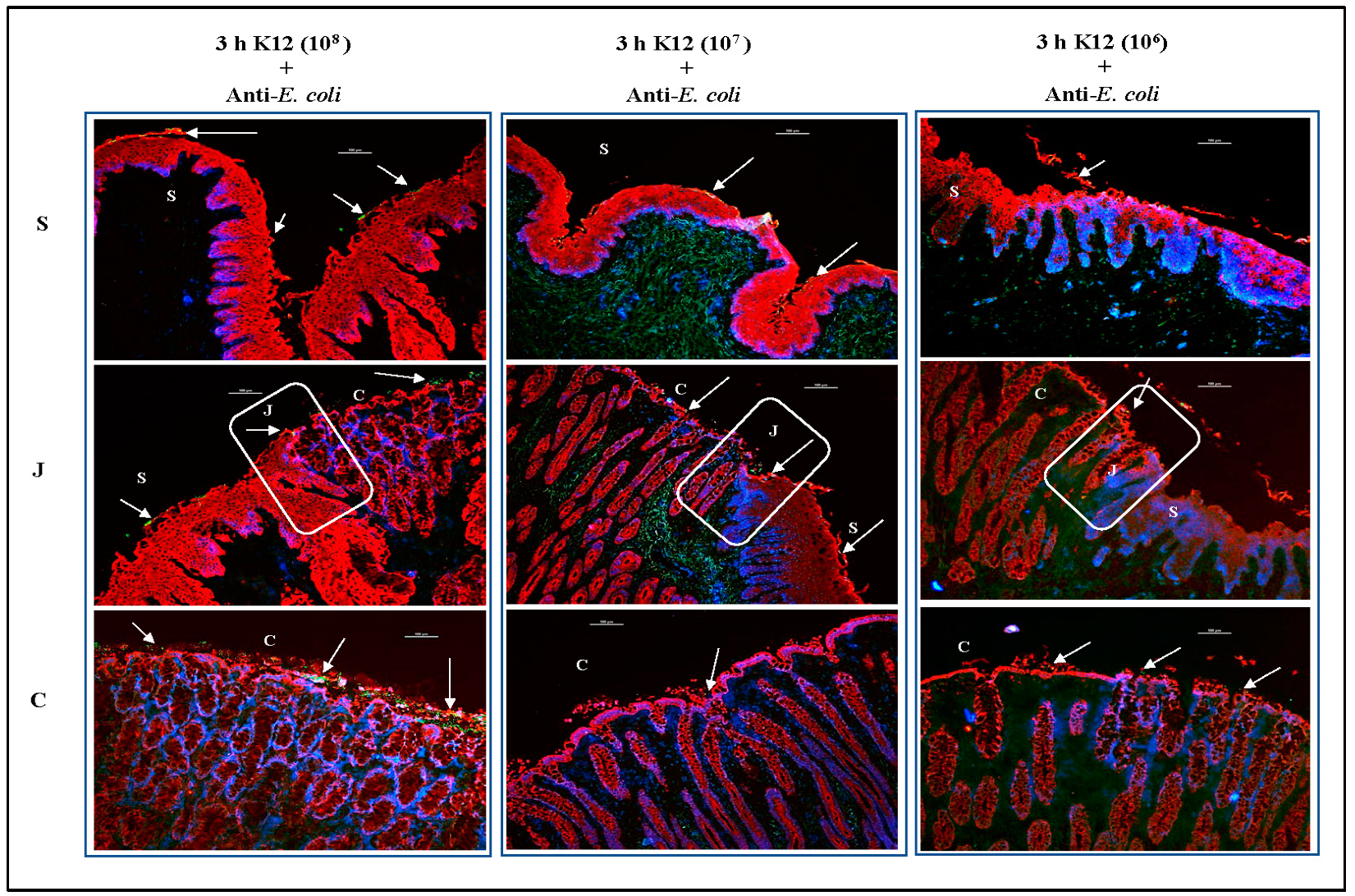
3.3. O157 Adherence Patterns Observed In Vitro and In Vivo Are Best Reproduced at the 107 Inoculum Concentration
3.3.1. For RAJ-IVOC Inoculated with 108 O157
3.3.2. For RAJ-IVOC Inoculated with 107 O157
3.3.3. For RAJ-IVOC Inoculated with 106 O157 or E. coli K12
3.4. Differences in O157 and E. coli K12 Recovered from the RAJ-IVOC Tissue Cultures Corresponded with the Adherence Patterns
3.5. PATS Verified Bacteria Recovered from the RAJ-IVOC Tissue Cultures
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wells, J.G.; Davis, B.R.; Wachsmuth, I.K.; Riley, L.W.; Remis, R.S.; Sokolow, R.; Morris, G.K. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J. Clin. Microbiol. 1983, 18, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W.; Memis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Eng. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Griffin, P.M.; Angulo, F.J.; Tauxe, R.V.; Hoekstra, R.M. Foodborne Illness Acquired in the United States—Unspecified Agents. Emerg. Infect. Dis. 2011, 17, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Foodborne Diseases Active Surveillance Network (FoodNet). Available online: https://www.cdc.gov/foodnet/index.html (accessed on 1 August 2022).
- Majowicz, S.E.; Scallan, E.; Jones-Bitton, A.; Sargeant, J.M.; Stapleton, J.; Angulo, F.J.; Yeung, D.H.; Kirk, M.D. Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: A systematic review and knowledge synthesis. Foodborne Pathog. Dis. 2014, 11, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Food Safety and Inspection Service-U.S. Department of Agriculture. Microbiological Testing Program for Escherichia coli O157:H7 and non-O157 Shiga Toxin-Producing Escherichia coli (STEC). Available online: https://www.fsis.usda.gov/science-data/data-sets-visualizations/microbiology/microbiological-testing-program-escherichia-coli (accessed on 13 August 2022).
- Rivas, M.; Chinen, I.; Miliwebsky, E.; Masana, M. Risk Factors for Shiga Toxin-Producing Escherichia coli-Associated Human Diseases. Microbiol. Spec. 2014, 2. [Google Scholar] [CrossRef]
- Kaper, J.B.; O’Brien, A.D. Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains; ASM Press: Washington, DC, USA, 1998. [Google Scholar]
- Karpman, D.; Stahl, A.L. Enterohemorrhagic Escherichia coli Pathogenesis and the Host Response. Microbiol. Spec. 2014, 2. [Google Scholar] [CrossRef]
- Pruimboom-Brees, I.M.; Morgan, T.W.; Ackermann, M.R.; Dean-Nystrom, E.A.; Samuel, J.E.; Cornick, N.A.; Moon, H.W. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc. Natl. Acad. Sci. USA 2000, 97, 10325–10329. [Google Scholar] [CrossRef]
- Persad, A.K.; LeJeune, J.T. Animal Reservoirs of Shiga Toxin-Producing Escherichia coli. Microbiol. Spec. 2014, 2. [Google Scholar] [CrossRef]
- Naylor, S.W.; Low, J.C.; Besser, T.E.; Mahajan, A.; Gunn, G.J.; Pearce, M.C.; McKendrick, I.J.; Smith, D.G.; Gally, D.L. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 2003, 71, 1505–1512. [Google Scholar] [CrossRef]
- Lim, J.Y.; Li, J.; Sheng, H.; Besser, T.E.; Potter, K.; Hovde, C.J. Escherichia coli O157:H7 colonization at the rectoanal junction of long-duration culture-positive cattle. Appl. Environ. Microbiol. 2007, 73, 1380–1382. [Google Scholar] [CrossRef]
- Keen, J.E.; Laegreid, W.W.; Chitko-McKown, C.G.; Durso, L.M.; Bono, J.L. Distribution of Shiga-toxigenic Escherichia coli O157 in the gastrointestinal tract of naturally O157-shedding cattle at necropsy. Appl. Environ. Microbiol. 2010, 76, 5278–5281. [Google Scholar] [CrossRef]
- Pohlenz, J.F.; Dean-Nystrom, E.A. Colonization of Escherichia coli O157:H7 on squamous epithelial cells at the rectal anal junction, (letter to editor). Vet. Rec. 2004, 155, 248. [Google Scholar]
- Fox, J.T.; Shi, X.; Nagaraja, T.G. Escherichia coli O157 in the rectoanal mucosal region of cattle. Foodborne Pathog. Dis. 2008, 5, 69–77. [Google Scholar] [CrossRef]
- Cobbold, R.N.; Hancock, D.D.; Rice, D.H.; Berg, J.; Stilborn, R.; Hovde, C.J.; Besser, T.E. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl. Environ. Microbiol. 2007, 73, 1563–1568. [Google Scholar] [CrossRef]
- Duffy, G.; McCabe, E. Veterinary Public Health Approach to Managing Pathogenic Verocytotoxigenic Escherichia coli in the Agri-Food Chain. Microbiol. Spec. 2014, 2. [Google Scholar] [CrossRef]
- Mahajan, A.; Naylor, S.; Mills, A.D.; Low, J.C.; Mackellar, A.; Hoey, D.E.; Currie, C.G.; Gally, D.L.; Huntley, J.; Smith, D.G. Phenotypic and functional characterisation of follicle-associated epithelium of rectal lymphoid tissue. Cell Tissue Res. 2005, 321, 365–374. [Google Scholar] [CrossRef]
- Kudva, I.T.; Dean-Nystrom, E.A. Bovine recto-anal junction squamous epithelial (RSE) cell adhesion assay for studying Escherichia coli O157 adherence. J. Appl. Microbiol. 2011, 111, 1283–1294. [Google Scholar] [CrossRef]
- Mir, R.A.; Schaut, R.G.; Allen, H.K.; Looft, T.; Loving, C.L.; Kudva, I.T.; Sharma, V.K. Cattle intestinal microbiota shifts following Escherichia coli O157:H7 vaccination and colonization. PLoS ONE 2019, 14, e0226099. [Google Scholar] [CrossRef]
- Mir, R.A.; Schaut, R.G.; Looft, T.; Allen, H.K.; Sharma, V.K.; Kudva, I.T. Recto-Anal Junction (RAJ) and Fecal Microbiomes of Cattle Experimentally Challenged with Escherichia coli O157:H7. Front. Microbiol. 2020, 11, 693. [Google Scholar] [CrossRef]
- Kudva, I.T.; Griffin, R.W.; Krastins, B.; Sarracino, D.A.; Calderwood, S.B.; John, M. Proteins other than the locus of enterocyte effacement-encoded proteins contribute to Escherichia coli O157:H7 adherence to bovine rectoanal junction stratified squamous epithelial cells. BMC Microbiol. 2012, 12, 103. [Google Scholar] [CrossRef]
- Kudva, I.T.; Hovde, C.J.; John, M. Adherence of non-O157 Shiga toxin-producing Escherichia coli to bovine recto-anal junction squamous epithelial cells appears to be mediated by mechanisms distinct from those used by O157. Foodborne Pathog. Dis. 2013, 10, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Kudva, I.T.; Krastins, B.; Torres, A.G.; Griffin, R.W.; Sheng, H.; Sarracino, D.A.; Hovde, C.J.; Calderwood, S.B.; John, M. The Escherichia coli O157:H7 cattle immunoproteome includes outer membrane protein A (OmpA), a modulator of adherence to bovine rectoanal junction squamous epithelial (RSE) cells. Proteomics 2015, 15, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Kudva, I.T.; Carter, M.Q.; Sharma, V.K.; Stasko, J.A.; Giron, J.A. Curli Temper Adherence of Escherichia coli O157:H7 to Squamous Epithelial Cells from the Bovine Recto-Anal Junction in a Strain-Dependent Manner. Appl. Environ. Microbiol. 2017, 83, e02594-16. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.A.; Brunelle, B.W.; Alt, D.P.; Arthur, T.M.; Kudva, I.T. Supershed Escherichia coli O157:H7 Has Potential for Increased Persistence on the Rectoanal Junction Squamous Epithelial Cells and Antibiotic Resistance. Int. J. Microbiol. 2020, 2020, 2368154. [Google Scholar] [CrossRef]
- Sheng, H.; Lim, J.Y.; Knecht, H.J.; Li, J.; Hovde, C.J. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun 2006, 74, 4685–4693. [Google Scholar] [CrossRef]
- Li, J.; Hovde, C.J. Expression profiles of bovine genes in the rectoanal junction mucosa during colonization with Escherichia coli O157:H7. Appl. Environ. Microbiol. 2007, 73, 2380–2385. [Google Scholar] [CrossRef]
- Sheng, H.; Wang, J.; Lim, J.Y.; Davitt, C.; Minnich, S.A.; Hovde, C.J. Internalization of Escherichia coli O157:H7 by bovine rectal epithelial cells. Front. Microbiol. 2011, 2, 32. [Google Scholar] [CrossRef]
- Wang, O.; McAllister, T.A.; Plastow, G.; Stanford, K.; Selinger, B.; Guan, L.L. Interactions of the Hindgut Mucosa-Associated Microbiome with Its Host Regulate Shedding of Escherichia coli O157:H7 by Cattle. Appl. Environ. Microbiol. 2018, 84, e01738-17. [Google Scholar] [CrossRef]
- Larzabal, M.; Da Silva, W.M.; Multani, A.; Vagnoni, L.E.; Moore, D.P.; Marin, M.S.; Riviere, N.A.; Delgado, F.O.; Vilte, D.A.; Victorica, M.R.; et al. Early immune innate hallmarks and microbiome changes across the gut during Escherichia coli O157:H7 infection in cattle. Sci. Rep. 2020, 10, 21535. [Google Scholar] [CrossRef]
- Moreau, M.R.; Kudva, I.T.; Katani, R.; Cote, R.; Li, L.; Arthur, T.M.; Kapur, V. Nonfimbrial Adhesin Mutants Reveal Divergent Escherichia coli O157:H7 Adherence Mechanisms on Human and Cattle Epithelial Cells. Int. J. Microbiol. 2021, 2021, 8868151. [Google Scholar] [CrossRef]
- Katani, R.; Kudva, I.T.; Srinivasan, S.; Stasko, J.B.; Schilling, M.; Li, L.; Cote, R.; DebRoy, C.; Arthur, T.M.; Sokurenko, E.V.; et al. Strain and host-cell dependent role of type-1 fimbriae in the adherence phenotype of super-shed Escherichia coli O157:H7. Int. J. Med. Microbiol. 2021, 311, 151511. [Google Scholar] [CrossRef]
- Kudva, I.T. In vitro adherence patterns of Shigella serogroups to bovine recto-anal junction squamous epithelial (RSE) cells are similar to those of Escherichia coli O157. Foodborne Pathog. Dis. 2012, 9, 346–351. [Google Scholar] [CrossRef]
- Kudva, I.T.; Hatfield, P.G.; Hovde, C.J. Effect of diet on the shedding of Escherichia coli O157:H7 in a sheep model. Appl. Environ. Microbiol. 1995, 61, 1363–1370. [Google Scholar] [CrossRef]
- Kudva, I.T.; Hunt, C.W.; Williams, C.J.; Nance, U.M.; Hovde, C.J. Evaluation of dietary influences on Escherichia coli O157:H7 shedding by sheep. Appl. Environ. Microbiol. 1997, 63, 3878–3886. [Google Scholar] [CrossRef]
- Kudva, I.T.; Hatfield, P.G.; Hovde, C.J. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli serotypes isolated from sheep. J. Clin. Microbiol. 1997, 35, 892–899. [Google Scholar] [CrossRef]
- Kudva, I.T.; Davis, M.A.; Griffin, R.W.; Garren, J.; Murray, M.; John, M.; Hovde, C.J.; Calderwood, S.B. Polymorphic Amplified Typing Sequences and Pulsed-Field Gel Electrophoresis Yield Comparable Results in the Strain Typing of a Diverse Set of Bovine Escherichia coli O157:H7 Isolates. Int. J. Microbiol. 2012, 2012, 140105. [Google Scholar] [CrossRef]
- Kudva, I.T.; Smole, S.; Griffin, R.W.; Garren, J.; Kalia, N.; Murray, M.; John, M.; Timperi, R.; Calderwood, S.B. Polymorphic Amplified Typing Sequences (PATS) Strain Typing System Accurately Discriminates a Set of Temporally and Spatially Disparate Escherichia coli O157 Isolates Associated with Human Infection. Open Microbiol. J. 2013, 7, 123–129. [Google Scholar] [CrossRef]
- Kudva, I.T.; Evans, P.S.; Perna, N.T.; Barrett, T.J.; Ausubel, F.M.; Blattner, F.R.; Calderwood, S.B. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 2002, 184, 1873–1879. [Google Scholar] [CrossRef]
- Kudva, I.T.; Evans, P.S.; Perna, N.T.; Barrett, T.J.; DeCastro, G.J.; Ausubel, F.M.; Blattner, F.R.; Calderwood, S.B. Polymorphic amplified typing sequences provide a novel approach to Escherichia coli O157:H7 strain typing. J. Clin. Microbiol. 2002, 40, 1152–1159. [Google Scholar] [CrossRef]
- Kudva, I.T.; Griffin, R.W.; Murray, M.; John, M.; Perna, N.T.; Barrett, T.J.; Calderwood, S.B. Insertions, deletions, and single-nucleotide polymorphisms at rare restriction enzyme sites enhance discriminatory power of polymorphic amplified typing sequences, a novel strain typing system for Escherichia coli O157:H7. J. Clin. Microbiol. 2004, 42, 2388–2397. [Google Scholar] [CrossRef]
- Kudva, I.T.; Stasko, J.A. Bison and bovine rectoanal junctions exhibit similar cellular architecture and Escherichia coli O157 adherence patterns. BMC Vet. Res. 2013, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Zihni, C.; Mills, C.; Matter, K.; Balda, M.S. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016, 17, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Angst, B.D.; Marcozzi, C.; Magee, A.I. The cadherin superfamily: Diversity in form and function. Cell Sci. 2001, 114, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.W.; Pinchuk, I.V.; Saada, J.I.; Chen, X.; Mifflin, R.C. Mesenchymal cells of the intestinal lamina propria. Annu. Rev. Physiol. 2011, 73, 213–237. [Google Scholar] [CrossRef]
- Rusu, D.; Loret, S.; Peulen, O.; Mainil, J.; Dandrifosse, G. Immunochemical, biomolecular and biochemical characterization of bovine epithelia intestinal primocultures. BMC Cell Biol. 2005, 6, 42. [Google Scholar] [CrossRef]
- Rao, R. Occludin phosphorylation in regulation of epithelial tight junctions. Ann. N. Y. Acad. Sci. 2009, 1165, 62–68. [Google Scholar] [CrossRef]
- Fedorchuk, C.; Kudva, I.T.; Kariyawasam, S. The Escherichia coli O157:H7 carbon starvation-inducible lipoprotein Slp contributes to initial adherence in vitro via the human polymeric immunoglobulin receptor. PLoS ONE 2019, 14, e0216791. [Google Scholar] [CrossRef]
- Hoefler, R.S.; Kudva, I.T. EDL933 Strains of Escherichia coli O157 can Demonstrate Genetic Diversity and Differential Adherence to Bovine Recto-Anal Junction Squamous Epithelial Cells. Open Microbiol. J. 2021, 15, 129–138. [Google Scholar] [CrossRef]
- Low, J.C.; McKendrick, I.J.; McKechnie, C.; Fenlon, D.; Naylor, S.W.; Currie, C.; Smith, D.G.; Allison, L.; Gally, D.L. Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Appl. Environ. Microbiol. 2005, 71, 93–97. [Google Scholar] [CrossRef]
- Rice, D.H.; Sheng, H.Q.; Wynia, S.A.; Hovde, C.J. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. J. Clin. Microbiol. 2003, 41, 4924–4929. [Google Scholar] [CrossRef]
- Trueba, G.; Garces, V.; Barragan, V.A.; Colman, R.E.; Seymour, M.; Vogler, A.J.; Keim, P. Escherichia coli O157:H7 in Ecuador: Animal reservoirs, yet no human disease. Vector Borne Zoonotic Dis. 2013, 13, 295–298. [Google Scholar] [CrossRef]
- Stromberg, Z.R.; Lewis, G.L.; Schneider, L.G.; Erickson, G.E.; Patel, I.R.; Smith, D.R.; Moxley, R.A. Culture-Based Quantification with Molecular Characterization of Non-O157 and O157 Enterohemorrhagic Escherichia coli Isolates from Rectoanal Mucosal Swabs of Feedlot Cattle. Foodborne Pathog. Dis. 2018, 15, 26–32. [Google Scholar] [CrossRef]
- Wells, J.E.; Berry, E.D.; Kim, M.; Bono, J.L.; Oliver, W.T.; Kalchayanand, N.; Wang, R.; Freetly, H.C.; Means, W.J. Determination of gastrointestinal tract colonization sites from feedlot cattle transiently shedding or super-shedding Escherichia coli O157:H7 at harvest. J. Appl. Microbiol. 2020, 129, 1419–1426. [Google Scholar] [CrossRef]
- Sheng, H.; Davis, M.A.; Knecht, H.J.; Hovde, C.J. Rectal administration of Escherichia coli O157:H7: Novel model for colonization of ruminants. Appl. Environ. Microbiol. 2004, 70, 4588–4595. [Google Scholar] [CrossRef]
- Carter, M.Q.; Brandl, M.T.; Kudva, I.T.; Katani, R.; Moreau, M.R.; Kapur, V. Conditional Function of Autoaggregative Protein Cah and Common cah Mutations in Shiga Toxin-Producing Escherichia coli. Appl. Environ. Microbiol. 2018, 84, e01739-17. [Google Scholar] [CrossRef]
- Cote, R.; Katani, R.; Moreau, M.R.; Kudva, I.T.; Arthur, T.M.; DebRoy, C.; Mwangi, M.M.; Albert, I.; Raygoza Garay, J.A.; Li, L.; et al. Comparative analysis of super-shedder strains of Escherichia coli O157:H7 reveals distinctive genomic features and a strongly aggregative adherent phenotype on bovine rectoanal junction squamous epithelial cells. PLoS ONE 2015, 10, e0116743. [Google Scholar] [CrossRef]
- Schaut, R.G.; Palmer, M.V.; Boggiatto, P.M.; Kudva, I.T.; Loving, C.L.; Sharma, V.K. Mucosal IFNgamma production and potential role in protection in Escherichia coli O157:H7 vaccinated and challenged cattle. Sci. Rep. 2021, 11, 9769. [Google Scholar] [CrossRef]
- Grivel, J.-C.; Margolis, L. Use of human tissue explants to study human infectious agents. Nat. Protoc. 2009, 4, 256–269. [Google Scholar] [CrossRef]
- Soto-Rivera, J.; Patterson, B.K.; Chen, Y.; Shen, C.; Ratner, D.; Ding, M.; Tumne, A.; Gupta, P. Study of HIV-1 transmission across cervical mucosa to tonsil tissue cells using an organ culture. Am. J. Reprod. Immunol. 2013, 69, 52–63. [Google Scholar] [CrossRef]
- Maher, D.; Wu, X.; Schacker, T.; Horbul, J.; Southern, P. HIV binding, penetration, and primary infection in human cervicovaginal tissue. Proc. Natl. Acad. Sci. USA 2005, 102, 11504–11509. [Google Scholar] [CrossRef]
- Maher, D.M.; Zhang, Z.Q.; Schacker, T.W.; Southern, P.J. Ex vivo modeling of oral HIV transmission in human palatine tonsil. J. Histochem. Cytochem. 2005, 53, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Middleton, A.M.; Chadwick, M.V.; Nicholson, A.G.; Wilson, R.; Thornton, D.J.; Kirkham, S.; Sheehan, J.K. Interaction between mycobacteria and mucus on a human respiratory tissue organ culture model with an air interface. Exp. Lung Res. 2004, 30, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.V.; Stasko, J.; Waters, W.R.; Thacker, T.C. Examination of the reticular epithelium of the bovine pharyngeal tonsil. Anat. Rec. 2011, 294, 1939–1950. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.D.; Frankel, G. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 2000, 181, 1496–1500. [Google Scholar] [CrossRef]
- Phillips, A.D.; Navabpour, L.; Hicks, S.; Dougan, G.; Wallis, T.; Frankel, G. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer’s patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 2000, 47, 377–381. [Google Scholar] [CrossRef]




| PATS Type | Polymorphic XbaI sites | Polymorphic AvrII sites | Virulence Genes | Strain | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IK8 | IK25 | IK114 | IK118 | IK123 | IK127 | IKB3 | IKB5 | IKNR3 | IKNR7 | IKNR10 | IKNR12 | IKNR16 | IKNR27 | IKNR33 | stx1 | stx2 | eaeA | hlyA | ||
| I | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | E. coli K12 |
| II | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | O157 strain EDL933 |
| III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | A 1 |
| IV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | B 2 |
| V | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | C 3 |
| VI | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | D 4 |
| VII | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | E 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudva, I.T.; Biernbaum, E.N.; Cassmann, E.D.; Palmer, M.V. Bovine Rectoanal Junction In Vitro Organ Culture Model System to Study Shiga Toxin-Producing Escherichia coli Adherence. Microorganisms 2023, 11, 1289. https://doi.org/10.3390/microorganisms11051289
Kudva IT, Biernbaum EN, Cassmann ED, Palmer MV. Bovine Rectoanal Junction In Vitro Organ Culture Model System to Study Shiga Toxin-Producing Escherichia coli Adherence. Microorganisms. 2023; 11(5):1289. https://doi.org/10.3390/microorganisms11051289
Chicago/Turabian StyleKudva, Indira T., Erika N. Biernbaum, Eric D. Cassmann, and Mitchell V. Palmer. 2023. "Bovine Rectoanal Junction In Vitro Organ Culture Model System to Study Shiga Toxin-Producing Escherichia coli Adherence" Microorganisms 11, no. 5: 1289. https://doi.org/10.3390/microorganisms11051289
APA StyleKudva, I. T., Biernbaum, E. N., Cassmann, E. D., & Palmer, M. V. (2023). Bovine Rectoanal Junction In Vitro Organ Culture Model System to Study Shiga Toxin-Producing Escherichia coli Adherence. Microorganisms, 11(5), 1289. https://doi.org/10.3390/microorganisms11051289








