Abstract
Shotgun proteomics has proven to be an attractive alternative for identifying a pathogen and characterizing the antimicrobial resistance genes it produces. Because of its performance, proteotyping of microorganisms by tandem mass spectrometry is expected to become an essential tool in modern healthcare. Proteotyping microorganisms that have been isolated from the environment by culturomics is also a cornerstone for the development of new biotechnological applications. Phylopeptidomics is a new strategy that estimates the phylogenetic distances between the organisms present in the sample and calculates the ratio of their shared peptides, thus improving the quantification of their contributions to the biomass. Here, we established the limit of detection of tandem mass spectrometry proteotyping based on MS/MS data recorded for several bacteria. The limit of detection for Salmonella bongori with our experimental set-up is 4 × 104 colony-forming units from a sample volume of 1 mL. This limit of detection is directly related to the amount of protein per cell and therefore depends on the shape and size of the microorganism. We have demonstrated that identification of bacteria by phylopeptidomics is independent of their growth stage and that the limit of detection of the method is not degraded in presence of additional bacteria in the same proportion.
1. Introduction
Rapid detection and identification of microorganisms is of utmost importance for the clinical microbiology laboratory as well as for the food industry, water suppliers, environmental applications, and microbial biotechnology. It is also an important goal to rapidly respond to bioterrorism threats and prevent the risk of dissemination of novel biological agents. Over the past decade, mass spectrometry has become a widespread technique for pathogen identification [1,2]. Indeed, whole-cell MALDI-TOF mass spectrometry, based on a comparison between experimental m/z profiles of basic, low-molecular-weight proteins of bacteria and a comprehensive database of spectra acquired under the same conditions, has revolutionized the clinical microbiology laboratory [3]. This approach can be refined with defining specific biomarkers [4,5]. For example, Francisella tularensis subspecies can be identified using specific biomarkers detected by MALDI-TOF mass spectrometry and previously defined by proteogenomics [6]. Furthermore, this approach can even be used to identify environmental bacteria for which the database is generally still poorly documented and to screen new isolates [7,8].
However, in the whole-cell MALDI-TOF mass spectrometry methodology, a single colony must first be obtained on an agar plate, which implies an overnight culture in most cases. In addition, the approach is not applicable to mixtures of microorganisms and it will be difficult to identify new environmental species that have not been previously recorded in the database. Proteotyping bacteria by tandem mass spectrometry analysis of trypsin-generated peptides from extracted proteins has proven to be a possible alternative for classification and identification of microorganisms [9]. To this end, several methodologies for interpreting MS/MS spectra to identify microorganisms have been proposed [10]. The pioneers in this field have tested a database restricted to the annotated genomes of 87 bacteria to interpret MS/MS datasets obtained with model bacteria. In this case, identification is based on unique peptides that are specific for a single organism [11]. A more complex database of 170 fully sequenced bacterial genomes was also proven successful [12], but this approach is not suitable for giant databases meant to represent the real microbial diversity, which will inevitably decrease the number of theoretical specific peptides per microorganism [13]. Alternatives have been proposed, such as the TCUP method that relies on specific peptides tailored to characterize taxonomic composition and capable of typing multiple clinically relevant species [14]. Peptide matching against a reference database derived from metagenomics sequencing is also used for bacterial identification and quantification of biomass contributions in metaproteomics [15]. In this methodology, the analysis is either restricted to a set of representative species that do not have significant overlap or is performed by allocating peptide results to the lowest common ancestor [16]. Peptide-intensity-weighted proteome abundance similarity correction based on the calculation of a matrix containing the pairwise similarities of the reference proteomes from the identified bacteria has been proposed to improve the quantification of biomass contributions in metaproteomics [17]. Recently, a new strategy has been shown to give very reliable quantification of biomass contributions after estimating the phylogenetic distances between the organisms present in the sample and calculating their ratio of shared peptides [18]. Phylopeptidomics has been able to distinguish with high accuracy the ratio of two closely related pathogens of clinical interest, Salmonella bongori and Shigella flexneri. This approach also performs well in identifying taxa present in the sample by combining unique taxon-specific peptide sequences with shared peptides that can be inferred by deconvolution of the interpreted signals from tandem mass spectrometry [18].
Rapid detection and identification of microorganisms by tandem mass spectrometry is gaining momentum [19,20], as the methodology has been shown to be high-throughput [21] and is important for clinical diagnostics. The potential of the methodology was recently illustrated by studies on ancient samples [22,23]. For such type of samples, the method was renamed paleoproteotyping. Many alternatives have been proposed to interpret the data [24,25,26,27,28]. Unfortunately, little data are available to date on the detection limit of clinically relevant bacteria by tandem mass spectrometry. The detection limit by MALDI-TOF has been documented for culture-independent detection of pathogens from urine samples [29]. In this case, 1 × 105 to 1 × 106 colony-forming units per mL were required. For the identification of yeast from positive blood cultures, the limit of detection was established at 5.9 × 105 cfu per mL [30]. The use of specific surfactants improved the detection limit of MALDI-TOF to 1 × 103 to 1 × 104 cfu per mL of bacterial suspension [31]. Targeted proteomics coupled with immunopurification of bacteria has proven to be an effective method when targeting a specific bacterium [32]. The detection limit for identification of Yersinia pestis based on specific peptides monitored by selected reaction monitoring was 2 × 104 cfu per mL in tap water or milk. To the best of our knowledge, such a detection limit for various microorganisms is not yet available for proteotyping by tandem mass spectrometry based on the shotgun proteomics principle.
Here, we established the detection limit of bacteria using phylopeptidomics to interpret MS/MS data recorded by a high-resolution LTQ-Orbitrap XL instrument (Thermo) over a 60 min gradient. We also demonstrated that the identification of bacteria by phylopeptidomics is independent of their growth stage and that the detection limit of the method is not degraded in the presence of additional bacteria.
2. Materials and Methods
2.1. Microorganisms and Microbial Cultures
The strains Salmonella bongori NCTC 12419, Deinococcus proteolyticus MRP DSM 20540, and Bacillus thuringiensis ATCC 10792 were obtained from the Pasteur collection, the DSM collection, and the ATCC collection, respectively. S. bongori was grown overnight in liquid TSB medium under aerobic conditions with vigorous shaking (140 rpm) at 30 °C in a BSL2 safety laboratory. D. proteolyticus and B. thuringiensis were grown in the same conditions except that liquid LB medium was used. Cell densities were evaluated by means of optical density (OD) measured at 600 nm. Liquid cultures of 100 mL were inoculated with overnight cultures to obtain an initial OD at 600nm of 0.008. These liquid cultures were incubated at 30 °C until OD600nm reached 0.1. Then, a serial dilution from 10 to 10 was performed and volumes of 100 µL of each of these dilutions were spread on LB medium agar plate. The plates were incubated overnight at 30 °C and the colonies were then counted to establish the relationship between number of colony forming units and OD600nm. Several samples of 1 mL of cell suspension at OD600nm = 0.1 (exponential phase) were harvested and immediately centrifuged at 16,000× g for 5 min at room temperature. The resulting supernatants were removed, and the cell pellets were subjected to another round of centrifugation for 2 min to remove any residual liquid. Each wet bacterial pellet was dissolved in 1 mL of sterile phosphate-buffered saline (PBS) buffered at pH 7.4. Each of these cell suspensions was further diluted in order to obtain tubes of 1.0 mL of suspension at 4 × 103 cfu/mL, 8 × 103 cfu/mL, 1 × 104 cfu/mL, 2 × 104 cfu/mL, 4 × 104 cfu/mL, and 8 × 104 cfu/mL. A series of 1:1 mixtures of S. bongori and D. proteolyticus was prepared by mixing the corresponding number of bacteria prepared in PBS to get a final concentration of 2 × 104 cfu/mL, and 2 × 105 cfu/mL. For S. bongori, cells harvested at the stationary phase (overnight culture) were also prepared in similar conditions.
2.2. Sample Preparation for Shotgun Proteomics
The bacterial samples were precipitated with tricholoroacetic acid by adding to the 1 mL cell suspension a volume of 250 µL of trichloroacetic acid (Sigma Aldrich, Saint-Quentin-Fallavier, France) prepared at 50% (w/vol) and centrifuged at 16,000× g for 5 min at room temperature. The supernatants were removed and the pellets were again centrifuged for 2 min to remove any residual liquid. Each pellet was dissolved in 60 µL of LDS1X sample buffer (Invitrogen, Villebon sur Yvette, France) consisting of 106 mM Tris/HCl, 141 mM Tris base, 2% lithium dodecyl sulfate, 10% glycerol, 0.51 mM EDTA, 0.22 mM SERVA Blue G250, 0.175 mM phenol red, buffered at pH 8.5, and supplemented with 2.5% beta-mercaptoethanol. Samples were heated at 99 °C for 5 min (Eppendorf, Montesson, France) and then subjected to sonication in an ultrasonic bath (VWR ultrasonic cleaner, VWR, Rosny-sous-Bois, France) for 5 min to dissolve all the biological aggregates. The samples were transferred to tubes containing 50 mg of silica beads and subjected to bead-beating with a Precellys instrument (Bertin technology, Montigny-le-Bretonneux, France) operated at room temperature at 7800 rpm for 10 cycles of 20 s separated by 30 s pauses. After cell disruption, the tubes were centrifuged at 16,000× g for 40 s. The resulting supernatants were transferred into new tubes and heated at 99 °C for 5 min. A volume of 25 µL of protein extract was loaded onto a NuPAGE 4–12% Bis-Tris gel (Invitrogen). The proteins were subjected to a short denaturing electrophoresis migration of 5 min at 200 V in MES/SDS 1X running buffer (Invitrogen) and then stained with Simply Blue Safestain (Invitrogen). Each polyacrylamide band containing the whole soluble proteome of the sample was sliced from the gel, reduced with dithiothreitol and treated with iodoacetamide to modify cysteines as described [33]. The gel pieces were then subjected to in-gel trypsin proteolysis with trypsin in the presence of 0.01% ProteaseMAX detergent (Promega, Charbonnières-les-Bains, France) as described [34].
2.3. Liquid Chromatography and Tandem Mass Spectrometry
Peptides were analyzed with an LTQ-Orbitrap XL hybrid mass spectrometer (Thermofisher, Villebon sur Yvette, France) coupled to an ultimate 3000 nanoLC system (Thermo) operated in data-dependent mode essentially as previously described [35]. The digests (50 µL) were loaded with a specific large injection loop and desalted online on a reverse phase PepMap100 C18 µ-precolumn (5 µm, 100 Å, 300 µm i.d. × 5 mm, Thermofisher, Villebon sur Yvette, France) and resolved on a nano scale PepMap 100 C18 nano LC column (3 µm, 100 Å, 75 µm i.d. × 50 cm, Thermofisher) at a flow rate of 0.3 µL.min−1 with a gradient of CH3CN, 0.1% formic acid prior to injection into the ion trap mass spectrometer. Peptides were resolved using a 60-min gradient from 2.5% to 50% solvent B (0.1% HCOOH/20% H2O/80% CH3CN) against solvent A (0.1% HCOOH/100% H2O). A top 5 strategy was used for the acquisition of MS/MS essentially consisting in a full MS scan, followed by fragmentation and MS/MS scan on the 5 most abundant ion precursors. Full scan mass spectra were measured from m/z 300 to 1800 in the Orbitrap analyzer at 30,000 resolution. The MS/MS scans were triggered in the linear ion trap at a resolution of 10,000 with a minimum signal required set at 10,000, potential charge states of 2+ and 3+, and with a 10 s dynamic exclusion of previously selected ions.
2.4. MS/MS Data Analysis
MS/MS spectra were assigned in proteomics mode with dedicated protein sequence databases corresponding to the annotated genomes: S. bongori NCTC 12419, D. proteolyticus MRP DSM 20540, and B. thuringiensis ATCC 10792. The MS/MS spectra from the mixture of microorganisms were queried against the S. bongori NCTC 12419 and D. proteolyticus MRP DSM 20540 merged database. MS/MS spectra were assigned with the Mascot Daemon software version 2.6.1 (Matrix Science) with full-trypsin specificity, up to 1 missed cleavage allowed, static modifications of carbamidomethylated cysteine (+57.0215), variable oxydation of methionine (+15.9949), mass tolerance of 5 ppm on the parent ions, and MS/MS mass tolerance of 0.5 Da. All peptide matches with a peptide score below a query threshold set at p ≤ 0.05 and rank 1 were parsed. A protein was considered valid when at least two different peptides were detected. The false-positive rate for protein identification was estimated by a search with a reverse decoy database to be below 0.1% using the same parameters. The number of MS/MS spectra assigned per protein (spectral counts) was extracted for each sample. The normalized spectral abundance factor (NSAF) for each protein was calculated as the total spectral count divided by its molecular mass in kDa, as described previously [36]. Tfold change for comparative proteomics between growth phases was calculated after data normalization as recommended [37]. For assigning the MS/MS spectra in phylopeptidomics mode, the NCBInr database was used as previously described [18].
3. Results and Discussion
3.1. Phylopeptidomic Identification of S. bongori Is Independent of Growth Stage
Figure 1 shows the strategy used for the analysis of S. Bongori harvested either in exponential phase or in stationary phase. Three independent biological replicates for both conditions with 1 × 106 cfu were harvested, lysed, and the extracted proteins were subjected to trypsin proteolysis. The resulting peptides were characterized by tandem mass spectrometry after their separation by reverse phase liquid chromatography. As shown in Figure 1, two types of bioinformatics analysis were performed on the six raw files. First, a proteotyping assignment of the MS/MS spectra against a generalist database (NCBInr) was carried out to assign taxon-spectrum matches (TSMs) to identify the taxon present in the sample and list the specific peptide sequences of that taxonomical branch. Then, a more classical proteomic interpretation of the MS/MS spectra was performed using only the protein sequences of the annotated genome of the identified taxon.
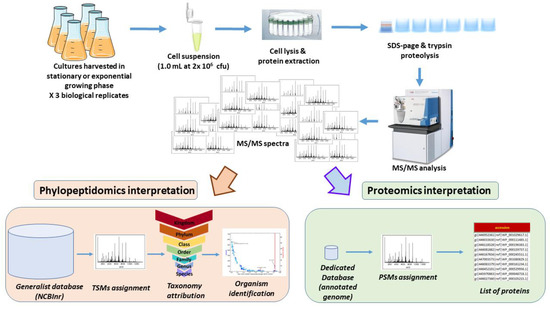
Figure 1.
Experimental and data analysis strategy.
Table 1 reports the number of MS/MS spectra recorded for each of the six samples containing a relatively small amount of biological material (1 × 106 cfu, corresponding to the equivalent of 0.4 µL of a culture of bacteria at OD of 1.0 at 600 nm). On average, 3992 MS/MS spectra were recorded, yielding 282 TSMs when interpreted against NCBInr, filtered for taxonomical links between peptide sequences and taxa, and accounting for validated taxonomical units at the various possible taxonomical ranks. This low level of TSMs is due to the small amount of sample on the one hand, and the atypically inflated database used for the MS/MS assignation on the other. For the first replicate of S. bongori cells harvested in exponential phase, 197 unique taxon-specific peptide sequences were exclusively from the Bacteria superkindgdom. Of these, the vast majority belong to a single phylum: Proteobacteria, with 368 TSMs and 83 unique taxon-specific peptide sequences. Clearly, the number of unique taxon-specific peptide sequences decreases sharply (83 instead of 197) when moving from the taxonomical rank superkingdom to phylum, whereas this decrease is relatively mild (368 instead of 372) for TSMs. Peptide sequences of proteins that are highly conserved among all bacteria are generally not considered useful in terms of taxonomy even though they contribute to the bacterial signal. Since no other daughter taxon are represented in the dataset, the vast majority of TSMs are then assigned to Proteobacteria. The same reasoning was applied to lower taxonomical ranks. Taxon-specific peptides indicate the presence of a single class, Gammaproteobacteria; an order, Enterobacterales; a family, Enterobacteriaceae; a genus, Salmonella; and a species, S. bongori. Finally, the species S. bongori is validated by the presence of four unique taxon-specific peptide sequences, and a large number of TSMs (357), which is expected for such a species.

Table 1.
Number of TSMs and specific peptides for S. bongori samples at 1 × 106 cells.
This approach based on both unique taxon-specific peptide sequences and TSMs circumvents possible false-positives when relying only on the former criterion alone. Figure 2 (Panel A) depicts the specific phylopeptidomic signature of this sample, which indicates the number of TSMs shared between S. bongori and all other organisms present in the NCBInr database and organized according to their respective phylogenetic distance from S. bongori. Logically, the shared TSMs decrease with phylogenetic distance since the average similarity between the peptide sequences of the organisms is inversely proportional to this distance. The signature fits the experimental points perfectly since only one species is present in the sample. Figure 2 (Panel B) shows the same phylopeptidomic signature for cells harvested in stationary phase. As shown in Table 1, S. bongori is the only validated species for all six samples. Although the proteomes differ between the exponential and stationary phases and, therefore the sets of peptides identified were dissimilar, enough unique taxon-specific peptide sequences and TSMs validated the species taxonomical rank, and similar phylopeptidomics signatures were obtained (Figure 2).
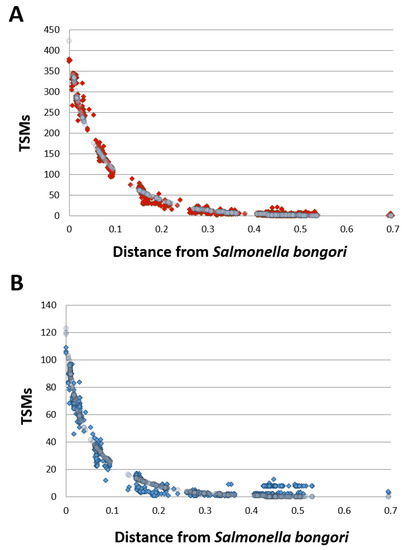
Figure 2.
Phylopeptidomic signature of S. bongori harvested in exponential and stationary phases. Panel A: Exponential phase (replicate 1). Experimental TSMs (for each taxon present in the database) are indicated by red dots and the mathematical signature is represented by grey circles. Panel B: Stationary phase (replicate 1). Experimental TSMs (for each taxon present in the database) are indicated by blue dots and the mathematical signature is represented by grey circles. The calculated distance of each taxon present in the database (x axis) is indicated from Salmonella bongori. The phylogenetic distance is expressed as the ratio of differing residues between two taxa based on a multi-alignment of conserved COGs ubiquitous across superkingdoms and considering a total of 8310 amino acid positions as previously defined (Pible et al., 2020). A phylogenetic distance of 0.1 means that 10% of the 8310 amino acids are differing between the two taxa.
Proteomic interpretation of the same dataset against the annotated S. bongori NTC 12419 genome revealed the detection of 1262 unique peptides and 6238 PSMs when the data from the 6 replicates were merged (Supplementary Materials, Table S1). These numbers are higher than those from the phylopeptidomic analysis because the database is much smaller. In total, 202 proteins could be validated with at least two different peptide sequences. As previously described for other bacteria [36], strong proteome differences are noted when comparing exponential and stationary phases. Indeed, only 55 proteins are consistently detected for the three exponential phase replicates and the three stationary phase replicates, whereas 92 and 18 are systematically only found in exponential phase and stationary phase, respectively (Supplementary Materials, Table S2). Moreover, the abundances of these proteins may differ considerably due to divergence in their metabolism. Figure 3 shows the volcano plot obtained when comparing the proteomes of the two growth stages. The most differentially abundant proteins in stationary phase are the DNA starvation/stationary phase protection protein [WP_000100806.1], the peptidoglycan-binding protein LysM [WP_015702978.1], and the outer membrane protein W [WP_000714794.1] with Tfold changes of 24.7, 6.7, and 5.0, respectively. The most differentially abundant proteins in the exponential phase are the 50S ribosomal protein L20 [WP_000124850.1], the DNA-directed RNA polymerase subunit beta [WP_000263100.1], the molecular chaperone DnaK [WP_000516126.1], and the lysine decarboxylase CadA [WP_001100654.1], with Tfold changes of −16.3, −14.8, −11.3, and −11.3, respectively. In conclusion, this analysis shows that phylopeptidomics is independent of bacterial growth stage and delivers the expected species if sufficient MS/MS were recorded.
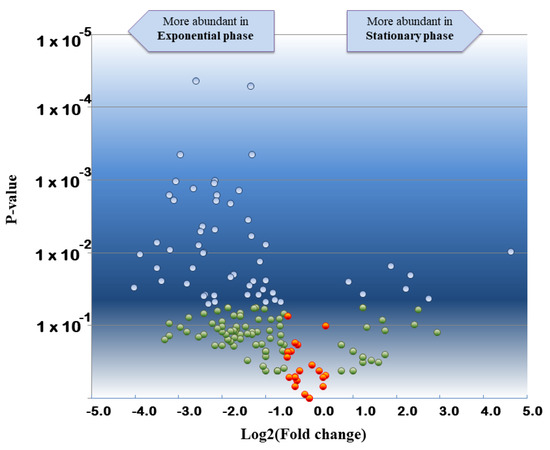
Figure 3.
Volcano plot showing the comparative proteomics results of S. bongori stationary phase versus exponential phase. Tfold change was calculated taking into consideration the three biological replicates of both conditions. Proteins are indicated with blue circles if they satisfied both, the Tfold change (≥1.5 or ≤−1.5) and statistical criteria (p-value ≤ 0.05), orange circles for identifications that did not meet the fold criterion but have low p-values, green circles if they satisfied the fold criteria but, most likely, this happened by chance (p-value > 0.05), and red circles if they did not meet the fold and p-value criteria. The Benjamini–Hochberg test indicates that the FDR is less than 10% for this dataset.
3.2. Detection Limit of Proteotyping for Three Pure Microorganism Samples
The detection limit of tandem mass spectrometry proteotyping was assessed using three different species: S. bongori, B. thuringiensis, and D. proteolyticus. These bacteria belong to three distinct phyla, namely Proteobacteria, Firmicutes, and Deinococcus-Thermus, respectively. They exhibit distinct Gram staining due to their specific membranes and walls. For this evaluation, we recorded MS/MS spectra for a range of samples normalized to 1.0 mL volume and containing 4 × 103 cfu, 8 × 103 cfu, 1 × 104 cfu, 2 × 104 cfu, 4 × 104 cfu, and 8 × 104 cfu with independent biological triplicates. Table 2 reports the interpretation of proteotyping at genus and species taxonomical ranks with the corresponding number of TSMs and unique taxon-specific peptide sequences at the corresponding limit of detection. The datasets recorded for the three S. bongori replicates at 4 × 104 cfu identify the correct species, whereas at 2 × 104 cfu the mass spectrometry signal was not sufficient to certify the correct species. At this low concentration, the interpreted signal is rather weak with only 76, 76, and 49 TSMs for the three replicates. It only certifies the presence of Enterobacteriaceae at the taxonomical rank of the family with 22, 23, and 21 TSMs and 4, 5, and 6 unique taxon-specific peptide sequences.

Table 2.
Number of TSMs and specific peptides for S. bongori, B. thuringiensis, and D. proteolyticus samples at their respective limit of detection (4 × 104, 8 × 104, 8 × 104 cfu, respectively).
The datasets recorded for the three replicates of B. thuringiensis at 8 × 104 cfu identifies the correct species, but a weaker signal at lower bacterial amounts prevents identification of the correct species. The same is true for D. proteolyticus, although only two of the three replicates at 8 × 104 cells gave the correct species. As expected, the limit of detection of the phylopeptidomics method depends on the protein biomass analyzed by tandem mass spectrometry and the recorded signal. Here, we analyzed three different bacteria with relatively different characteristics in terms of size and shape. First, S. bongori, formerly Salmonella choleraesuis subsp. bongori subsp. nov., is a Gram negative, motile and non-spore forming rod that measures 0.7–1.5 µm in diameter by 2.0–5.0 µm in length [38,39], thus its average volume is 3.3 µm3. The vegetative cells of B. thuringiensis are rod-shaped, 2.0–5.0 µm long and about 1.0 µm wide [40], so its average volume is 2.7 µm3. Finally, D. proteolyticus cells are non-motile Gram-positive spheres, with an average diameter of 0.8 to 1.0 μm, occurring singly or as diplococci (pairs) and occasionally seen in tetrads [41]. Their average volume is therefore 0.38 µm3. The pair of D. proteolyticus cells that form a single cfu when sprayed on an agar plate has an equivalent volume of 0.76 µm3. If the protein biomass contribution of each type of bacterium can be roughly estimated using their volume, then the biomass contribution of 4 × 104 cfu of S. bongori is 22% larger than 4 × 104 cfu of B. thuringiensis (×1.2) and much larger than the biomass contribution of the same number of cfu of D. proteolyticus (×4.3). This biomass contribution explains why the detection limit for the three strains oscillates between 4 × 104 and 8 × 104 cfu and is revealed here to be dependent on the size and shape of the bacteria. Of course, this detection limit depends on the tandem mass spectrometer used for the analytical measurements, the parameters for the acquisition, the chromatography performances, the efficiency of the protein extraction protocol, which can vary depending on the cell type, as well as the amount of peptides that was injected. Here, only half of the protein extract was subjected to SDS-PAGE and proteolyzed.
3.3. Identification of Possible Biomarkers for Proteomic Detection of the Three Pure Microorganism Samples for Minute Loads
We interpreted the recorded datasets for the full range of cell quantities (6 conditions × 3 replicates × 3 strains) used in the experiment by a classical proteomics approach with a dedicated database. Table S3 lists the PSMs assigned in each condition for each microbial strain and Table S4 shows the detected proteins and their respective spectral counts per condition and strain. Logically, the number of proteins detected decreases with the cell biomass used for the analysis. For S. bongori, a total of 51, 36, 22, 7, 3, and 2 proteins were consistently detected for the three replicates for the samples containing 8 × 104 cfu, 4 × 104 cfu, 2 × 104 cfu, 1 × 104 cfu, 8 × 103 cfu, and 4 × 103 cfu, respectively. The three most detected proteins in these low biomass samples are different for the three bacteria. Elongation factors Tu [WP_000031748.1] and G [WP_000124707.1] and OmpA porin annotated as a membrane protein [WP_015702792.1] are consistently detected in S. bongori samples with at least two spectral counts in each replicate if the biomass was at least 2 × 104 cfu. Only the first protein is detected from a biomass of 1 × 104 cfu, but this protein is not consistently detected with at least two spectral counts for lower biomass amounts. For B. thuringiensis, the elongation factor Tu [WP_001029617.1], the 50S ribosomal protein L5 [WP_001080829.1], and the 30S ribosomal protein S2 [WP_000111485.1] were the most detected proteins. They accounted for at least two spectral counts systematically detected in each replicate with a biomass of at least 4 × 104 cfu. The three most detected proteins for D. proteolyticus are a protein annotated as hypothetical but exhibiting sequence similarities with S-layer protein [WP_041221871.1], another S-layer domain-containing protein [WP_013614653.1], and an iron ABC transporter substrate-binding protein [WP_013615085.1]. In this case, a biomass of at least 8 × 104 cfu is required to detect them with at least two spectral counts but only in two of three replicates. Based on these results, a targeted proteomics approach can be proposed to specifically quantify these most detected proteins by shotgun proteomics and most likely improve the detection limit of the three bacteria.
3.4. The Detection Limit of Phylopeptidomics Is Not Degraded in Multi-Organism Samples
Equimolar mixtures of S. bongori and D. proteolyticus were prepared at 2 × 104 cfu, 4× 104 cfu, 8 × 104 cfu, 1 × 105 cfu, and 2 × 105 cfu in a 1.0 mL volume. In this case, both bacteria were successfully identified by phylopeptidomics when at least 8 × 104 cfu were present in the sample, as shown in Table 3. At 4 × 104 cfu, both bacteria were successfully identified in two of the three replicates, but at 2 × 104 cfu, the bacteria were not detected. Noteworthy, the presence of additional peptides (those from S. bongori which is more abundant) improves the detection of D. proteolyticus peptides, probably because less loss of peptides from the tube walls occurred during extraction and chromatography of these low amounts of products. Indeed, single-cell proteomics often use an excess of peptides or proteins from unrelated species to promote a carrier effect and minimize adsorption losses on surfaces. Figure 4 shows the corresponding krona charts. As mentioned above, the biomass contributions in terms of protein amounts are different between the two types of bacteria. S. bongori is more abundant in these mixtures than D. proteolyticus due to differences in cell size and shape. Here, for the three replicates at 8 × 104 cfu, S. bongori and D. proteolyticus are detected with 91/43/73 and 6/6/14 TSMs, respectively, while unique taxon peptides are 2/2/1 and 6/6/13, respectively. For the three replicates at 4 × 105 cfu, S. bongori and D. proteolyticus are detected with 64/166/337 and 8/67/53 TSMs, respectively, while unique taxon peptides are 1/2/5 and 7/46/36, respectively. A great deal of heterogeneity is noted among the replicates, but basically, the ratio of S. bongori TSMs to D. proteolyticus TSMs is x7.9, x10.8, and x4.4 for the samples at 8 × 104 cfu, 1 × 105 cfu, and 2 × 105 cfu, respectively. When sufficient bacteria are present, i.e., 2 × 105 cfu, the observed ratio of TSMs is strikingly consistent with the theoretical biomass contributions inferred from the shape and size of both types of bacteria, i.e., ×4.3. Table S5 lists the PSMs assigned for these samples. Table S6 shows the proteins detected when using a dedicated database of limited size. Remarkably, the number of D. proteolyticus proteins validated with at least two specific peptides increased strongly in presence of S. bongori: 41 (Table S6) instead of 5 (Table S4). Indeed, the number of unique peptide sequences sharply enlarged: 278 (Table S5) instead of 78 (Table S3), confirming a decrease in peptide loss when the peptide load is higher. The most abundant proteins detected in pure samples are also found here among the most abundant entities. Figure 5 shows four representative MS/MS spectra assigned to peptide sequences that are species-specific for S. bongori and D. proteolyticus. A relatively good fragmentation is observed with most of their sequences covered by the b and y ions although the low peptide signal. Here, we choose to explore equimolar ratio of microorganisms in order to avoid any bias due to the different responsiveness of one proteome compared to others. In real life, mixed cultures may contain microorganisms at various ratios and as a result the limit of detection may be different depending on the asymmetry of the mixture.

Table 3.
Number of TSMs and specific peptides for S. bongori and D. proteolyticus mixed samples at 8 × 104 cfu.
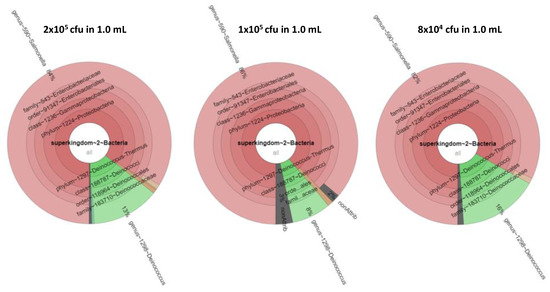
Figure 4.
Krona presentation of the results of tandem mass spectrometry proteotyping. Identification results and biomass contributions of the three concentrations 2 × 105 cfu, 1 × 105 cfu, and 8 × 104 cfu are shown from left to right. The biomass contributions of S. bongori, D. proteolyticus, and the unassigned signal are shown in red, green, and grey.
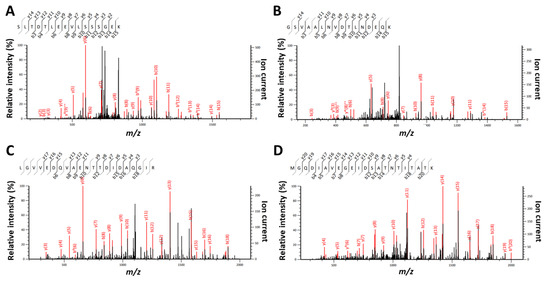
Figure 5.
Representative MS/MS spectra and their MASCOT annotation for species-specific peptides. Panel A: MS/MS spectrum assigned to peptide SLTDTLEEVLSSSGEK (m/z 847.92169; z = 2; MASCOT score = 53.33) and specific of S. bongori species. Panel B: MS/MS spectrum attributed to peptide GSVAALNVDTLNDEQK (m/z 837.42096; z = 2; MASCOT score = 45.64) and specific of S. bongori species. Panel C: MS/MS spectrum #1669 (F404441 file) corresponding to peptide LGVVEDQVAENTTDIDAQGIR (m/z 1122.0614; z = 2; MASCOT score = 102.67) and specific of D. proteolyticus species. Panel D: MS/MS spectrum attributed to peptide MGQDIAVEGEIDSATNTITATK (m/z 1133.04687; z = 2; MASCOT score = 103.94) and specific of D. proteolyticus species. Ions indicated in red are those assigned and included in the MASCOT scoring.
4. Conclusions
Phylopeptidomics is a phylogeny-based approach which is peptide-centric. This is clearly different from metaproteomics interpretation tools which are protein-centric. We illustrated this approach combining unique taxon-specific peptide sequences and taxon-spectrum-matches with several bacteria: S. bongori, B. thuringiensis, and D. proteolyticus. Identification of bacteria using phylopeptidomics was found to be independent of their growth stage. The detection limit, expressed in cfu per mL, depends on the shape and size of the microorganisms as it is strongly correlated to the protein content. It varies between 4 × 104 to 8 × 104 cfu per mL, a detection limit remarkably relevant compared to targeted proteomics approaches limited to the abundance of specific peptides. Interestingly, the method is applicable to any type of microorganisms. In addition, shotgun proteomics may help define specific, but abundant, peptide biomarkers to improve the detection limit via targeted proteomics. Last, the detection limit of D. proteolyticus was improved in the presence of S. bongori. Based on the limit of detection described in this study, tandem mass spectrometry-based proteotyping has great potential.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11051170/s1, Table S1: List of PSMs for S. bongori at 1 × 106 in exponential and stationary phases. Table S2: List of identified proteins for S. bongori at 1 × 106 in exponential and stationary phases. Table S3: List of PSMs for S. bongori, B. thuringiensis, and D. proteolyticus at 4 × 103 cfu, 8 × 103 cfu, 1 × 104 cfu, 2 × 104 cfu, 4 × 104 cfu, and 8 × 104 cfu. Table S4: List of identified proteins for S. bongori, B. thuringiensis, and D. proteolyticus at 4 × 103 cfu, 8 × 103 cfu, 1 × 104 cfu, 2 × 104 cfu, 4 × 104 cfu, and 8 × 104 cfu. Table S5: List of PSMs for S. bongori and D. proteolyticus mixtures at 2 × 104 cfu, 4 × 104 cfu, 8 × 104 cfu, 1 × 105 cfu, and 2 × 105 cfu. Table S6: List of identified proteins for S. bongori and D. proteolyticus mixtures at 2 × 104 cfu, 4 × 104 cfu, 8 × 104 cfu, 1 × 105 cfu, and 2 × 105 cfu.
Author Contributions
Conceptualization, C.M., B.A.-B. and J.A.; methodology, O.P.; validation, C.M., B.A.-B. and J.A.; investigation, C.M., B.A.-B. and J.A.; data curation, J.A.; writing—original draft preparation, J.A.; writing—review and editing, J.A.; supervision, B.A.-B. and J.A.; funding acquisition, J.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by the French joint ministerial program of R&D against CBRNE threats and the ANR program “Phylopeptidomics” (ANR-17-CE18-0023-01). CM was financially supported by DGA and CEA.
Data Availability Statement
Data are available in supplementary Tables S1–S6 provided together with the main publication.
Acknowledgments
We thank Jean-Charles Gaillard and Guylaine Miotello for invaluable technical help with the mass spectrometry platform.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Duriez, E.; Armengaud, J.; Fenaille, F.; Ezan, E. Mass spectrometry for the detection of bioterrorism agents: From environmental to clinical applications. J. Mass. Spectrom. 2016, 51, 183–199. [Google Scholar] [CrossRef]
- Ho, Y.P.; Reddy, P.M. Advances in mass spectrometry for the identification of pathogens. Mass. Spectrom. Rev. 2011, 30, 1203–1224. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, J.P.; Espinal, P.; Dunyach-Remy, C.; Messad, N.; Pantel, A.; Sotto, A. Mass spectrometry: A revolution in clinical microbiology? Clin. Chem. Lab. Med. 2013, 51, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J. Defining Diagnostic Biomarkers Using Shotgun Proteomics and MALDI-TOF Mass Spectrometry. Methods Mol. Biol. 2017, 1616, 107–120. [Google Scholar] [PubMed]
- Suarez, S.; Ferroni, A.; Lotz, A.; Jolley, K.A.; Guerin, P.; Leto, J.; Dauphin, B.; Jamet, A.; Maiden, M.C.; Nassif, X.; et al. Ribosomal proteins as biomarkers for bacterial identification by mass spectrometry in the clinical microbiology laboratory. J. Microbiol. Methods 2013, 94, 390–396. [Google Scholar] [CrossRef]
- Durighello, E.; Bellanger, L.; Ezan, E.; Armengaud, J. Proteogenomic biomarkers for identification of Francisella species and subspecies by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Anal. Chem. 2014, 86, 9394–9398. [Google Scholar] [CrossRef]
- Christie-Oleza, J.A.; Miotello, G.; Armengaud, J. Proteogenomic definition of biomarkers for the large Roseobacter clade and application for a quick screening of new environmental isolates. J. Proteome Res. 2013, 12, 5331–5339. [Google Scholar] [CrossRef]
- Christie-Oleza, J.A.; Piña-Villalonga, J.M.; Guerin, P.; Miotello, G.; Bosch, R.; Nogales, B.; Armengaud, J. Shotgun nanoLC-MS/MS proteogenomics to document MALDI-TOF biomarkers for screening new members of the Ruegeria genus. Environ. Microbiol. 2013, 15, 133–147. [Google Scholar] [CrossRef]
- Karlsson, R.; Gonzales-Siles, L.; Boulund, F.; Svensson-Stadler, L.; Skovbjerg, S.; Karlsson, A.; Davidson, M.; Hulth, S.; Kristiansson, E.; Moore, E.R. Proteotyping: Proteomic characterization, classification and identification of microorganisms--A prospectus. Syst. Appl. Microbiol. 2015, 38, 246–257. [Google Scholar] [CrossRef]
- Dworzanski, J.P.; Snyder, A.P. Classification and identification of bacteria using mass spectrometry-based proteomics. Expert. Rev. Proteom. 2005, 2, 863–878. [Google Scholar] [CrossRef]
- Dworzanski, J.P.; Snyder, A.P.; Chen, R.; Zhang, H.; Wishart, D.; Li, L. Identification of bacteria using tandem mass spectrometry combined with a proteome database and statistical scoring. Anal. Chem. 2004, 76, 2355–2366. [Google Scholar] [CrossRef] [PubMed]
- Dworzanski, J.P.; Deshpande, S.V.; Chen, R.; Jabbour, R.E.; Snyder, A.P.; Wick, C.H.; Li, L. Mass spectrometry-based proteomics combined with bioinformatic tools for bacterial classification. J. Proteome Res. 2006, 5, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Padliya, N.D.; Garrett, W.M.; Campbell, K.B.; Tabb, D.L.; Cooper, G. Tandem mass spectrometry for the detection of plant pathogenic fungi and the effects of database composition on protein inferences. Proteomics 2007, 7, 3932–3942. [Google Scholar] [CrossRef] [PubMed]
- Boulund, F.; Karlsson, R.; Gonzales-Siles, L.; Johnning, A.; Karami, N.; Al-Bayati, O.; Ahren, C.; Moore, E.R.B.; Kristiansson, E. Typing and Characterization of Bacteria Using Bottom-up Tandem Mass Spectrometry Proteomics. Mol. Cell. Proteom. 2017, 16, 1052–1063. [Google Scholar] [CrossRef]
- Kleiner, M.; Thorson, E.; Sharp, C.E.; Dong, X.; Liu, D.; Li, C.; Strous, M. Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Commun. 2017, 8, 1558. [Google Scholar] [CrossRef] [PubMed]
- Mesuere, B.; Devreese, B.; Debyser, G.; Aerts, M.; Vandamme, P.; Dawyndt, P. Unipept: Tryptic peptide-based biodiversity analysis of metaproteome samples. J. Proteome Res. 2012, 11, 5773–5780. [Google Scholar] [CrossRef]
- Penzlin, A.; Lindner, M.S.; Doellinger, J.; Dabrowski, P.W.; Nitsche, A.; Renard, B.Y. Pipasic: Similarity and expression correction for strain-level identification and quantification in metaproteomics. Bioinformatics 2014, 30, i149–i156. [Google Scholar] [CrossRef]
- Pible, O.; Allain, F.; Jouffret, V.; Culotta, K.; Miotello, G.; Armengaud, J. Estimating relative biomasses of organisms in microbiota using “phylopeptidomics”. Microbiome 2020, 8, 30. [Google Scholar] [CrossRef]
- Armengaud, J. Metaproteomics to understand how microbiota function: The crystal ball predicts a promising future. Environ. Microbiol. 2023, 25, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Grenga, L.; Pible, O.; Armengaud, J. Pathogen proteotyping: A rapidly developing application of mass spectrometry to address clinical concerns. Clin. Mass. Spectrom. 2019, 14 Pt A, 9–17. [Google Scholar] [CrossRef]
- Hayoun, K.; Gaillard, J.C.; Pible, O.; Alpha-Bazin, B.; Armengaud, J. High-throughput proteotyping of bacterial isolates by double barrel chromatography-tandem mass spectrometry based on microplate paramagnetic beads and phylopeptidomics. J. Proteom. 2020, 226, 103887. [Google Scholar] [CrossRef] [PubMed]
- Bourdin, V.; Charlier, P.; Crevat, S.; Slimani, L.; Chaussain, C.; Kielbasa, M.; Pible, O.; Armengaud, J. Deep Paleoproteotyping and Microtomography Revealed No Heart Defect nor Traces of Embalming in the Cardiac Relics of Blessed Pauline Jaricot. Int. J. Mol. Sci. 2023, 24, 3011. [Google Scholar] [CrossRef]
- Hama, H.O.; Chenal, T.; Pible, O.; Miotello, G.; Armengaud, J.; Drancourt, M. An ancient coronavirus from individuals in France, circa 16(th) century. Int. J. Infect. Dis. 2023, 131, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Alves, G.; Ogurtsov, A.; Karlsson, R.; Jaen-Luchoro, D.; Pineiro-Iglesias, B.; Salva-Serra, F.; Andersson, B.; Moore, E.R.B.; Yu, Y.K. Identification of Antibiotic Resistance Proteins via MiCId’s Augmented Workflow. A Mass Spectrometry-Based Proteomics Approach. J. Am. Soc. Mass. Spectrom. 2022, 33, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Alves, G.; Wang, G.; Ogurtsov, A.Y.; Drake, S.K.; Gucek, M.; Sacks, D.B.; Yu, Y.K. Rapid Classification and Identification of Multiple Microorganisms with Accurate Statistical Significance via High-Resolution Tandem Mass Spectrometry. J. Am. Soc. Mass. Spectrom. 2018, 29, 1721–1737. [Google Scholar] [CrossRef] [PubMed]
- Alves, G.; Yu, Y.K. Robust Accurate Identification and Biomass Estimates of Microorganisms via Tandem Mass Spectrometry. J. Am. Soc. Mass. Spectrom. 2020, 31, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Kuhring, M.; Doellinger, J.; Nitsche, A.; Muth, T.; Renard, B.Y. TaxIt: An Iterative Computational Pipeline for Untargeted Strain-Level Identification Using MS/MS Spectra from Pathogenic Single-Organism Samples. J. Proteome Res. 2020, 19, 2501–2510. [Google Scholar] [CrossRef] [PubMed]
- Mooradian, A.D.; van der Post, S.; Naegle, K.M.; Held, J.M. ProteoClade: A taxonomic toolkit for multi-species and metaproteomic analysis. PLoS Comput. Biol. 2020, 16, e1007741. [Google Scholar] [CrossRef]
- Demarco, M.L.; Burnham, C.A.D. Diafiltration MALDI-TOF mass spectrometry method for culture-independent detection and identification of pathogens directly from urine specimens. Am. J. Clin. Pathol. 2014, 141, 204–212. [Google Scholar] [CrossRef]
- Yan, Y.; He, Y.; Maier, T.; Quinn, C.; Shi, G.; Li, H.; Stratton, C.W.; Kostrzewa, M.; Tang, Y.W. Improved identification of yeast species directly from positive blood culture media by combining Sepsityper specimen processing and Microflex analysis with the matrix-assisted laser desorption ionization Biotyper system. J. Clin. Microbiol. 2011, 49, 2528–2532. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Bhaisare, M.L.; Wu, H.F. Ceria nanocubic-ultrasonication assisted dispersive liquid-liquid microextraction coupled with matrix assisted laser desorption/ionization mass spectrometry for pathogenic bacteria analysis. Talanta 2014, 120, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Chenau, J.; Fenaille, F.; Simon, S.; Filali, S.; Volland, H.; Junot, C.; Carniel, E.; Becher, F. Detection of Yersinia pestis in environmental and food samples by intact cell immunocapture and liquid chromatography-tandem mass spectrometry. Anal. Chem. 2014, 86, 6144–6152. [Google Scholar] [CrossRef]
- Hartmann, E.M.; Allain, F.; Gaillard, J.C.; Pible, O.; Armengaud, J. Taking the shortcut for high-throughput shotgun proteomic analysis of bacteria. Methods Mol. Biol. 2014, 1197, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Clair, G.; Armengaud, J.; Duport, C. Restricting fermentative potential by proteome remodeling: An adaptive strategy evidenced in Bacillus cereus. Mol. Cell. Proteom. 2012, 11, M111.013102. [Google Scholar] [CrossRef] [PubMed]
- Mappa, C.; Pible, O.; Armengaud, J.; Alpha-Bazin, B. Assessing the ratio of Bacillus spores and vegetative cells by shotgun proteomics. Environ. Sci. Pollut. Res. Int. 2021, 28, 25107–25115. [Google Scholar] [CrossRef] [PubMed]
- Christie-Oleza, J.A.; Fernandez, B.; Nogales, B.; Bosch, R.; Armengaud, J. Proteomic insights into the lifestyle of an environmentally relevant marine bacterium. ISME J. 2012, 6, 124–135. [Google Scholar] [CrossRef]
- Carvalho, P.C.; Fischer, J.S.; Chen, E.I.; Yates, J.R., 3rd; Barbosa, V.C. PatternLab for proteomics: A tool for differential shotgun proteomics. BMC Bioinform. 2008, 9, 316. [Google Scholar] [CrossRef]
- Le Minor, L.; Veron, M.; Popoff, M. [A proposal for Salmonella nomenclature]. Ann. Microbiol. (Paris) 1982, 133, 245–254. [Google Scholar]
- Reeves, M.W.; Evins, G.M.; Heiba, A.A.; Plikaytis, B.D.; Farmer, J.J., 3rd. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 1989, 27, 313–320. [Google Scholar] [CrossRef]
- Ibrahim, M.A.; Griko, N.; Junker, M.; Bulla, L.A. Bacillus thuringiensis: A genomics and proteomics perspective. Bioeng. Bugs 2010, 1, 31–50. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Watanabe, H.; Takehisa, M.; Iizuka, H. Isolation and Identification of Radiation-resistant Cocci Belonging to the Genus Deinococcus from Sewage Sludges and Animal Feeds. Agric. Biol. Chem. 1983, 47, 1239–1247. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).