The First Swedish Outbreak with VIM-2-Producing Pseudomonas aeruginosa, Occurring between 2006 and 2007, Was Probably Due to Contaminated Hospital Sinks
Abstract
1. Introduction
2. Materials and Methods
2.1. Settings and Ethics
2.2. Cases
2.3. Epidemiological Investigation
2.4. Cultures and Susceptibility Testing
2.5. Identification of MBL with PCR
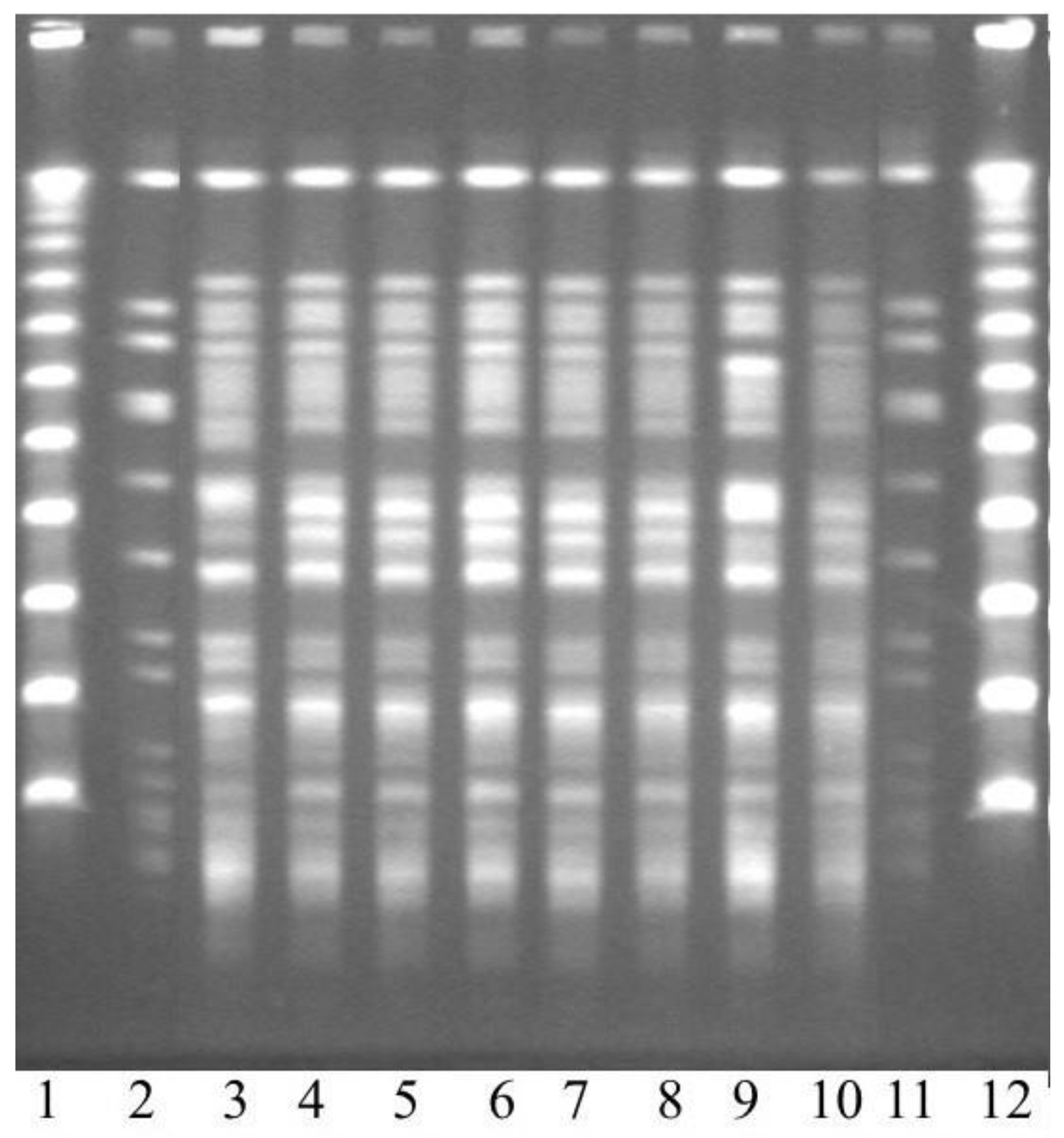
2.6. Pulsed-Field Gel Electrophoresis (PFGE)
2.7. Whole-Genome Sequencing (WGS) and Computational Analysis
3. Results
3.1. Bacterial Findings
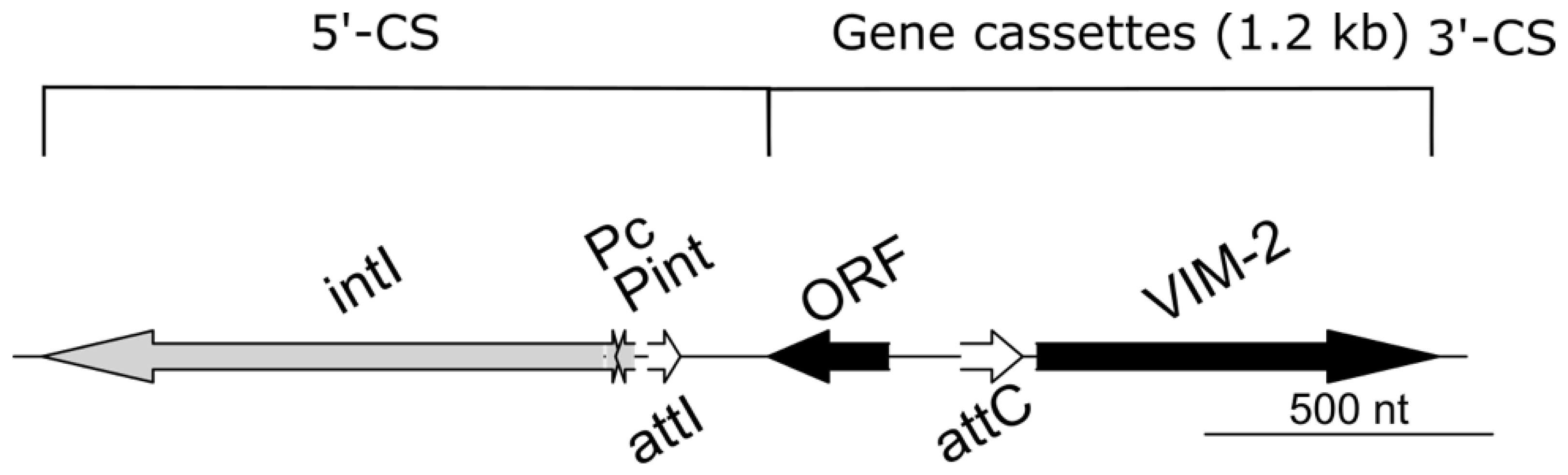
3.2. Genetic Data of the Outbreak Strain
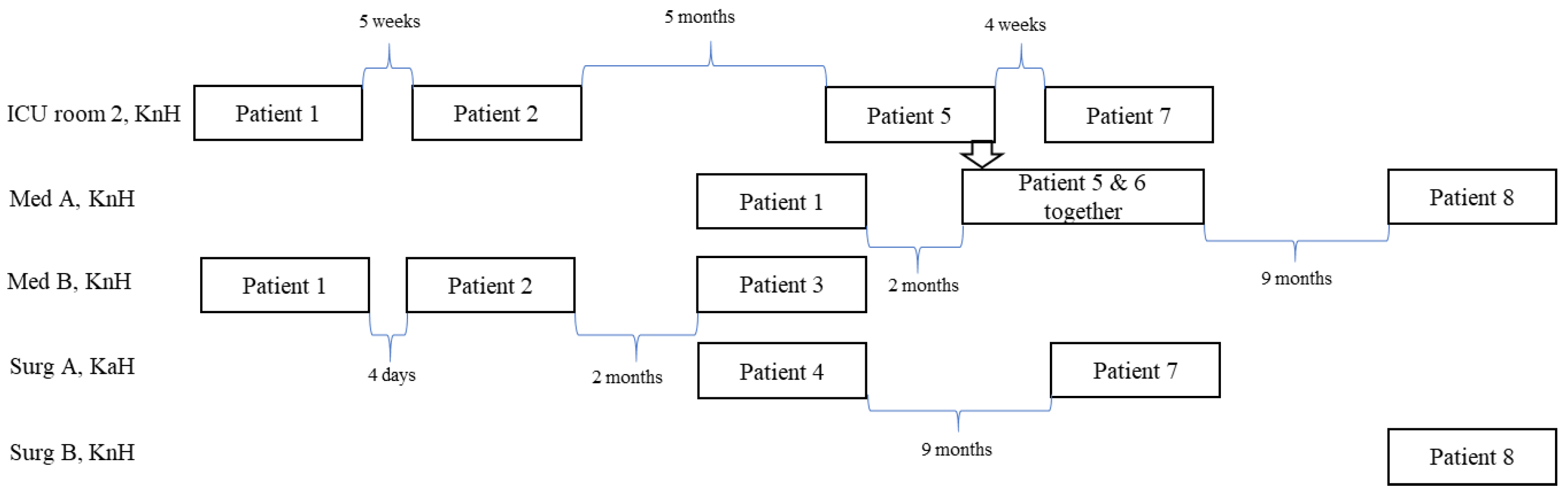
3.3. Epidemiological Investigation
3.4. Infection Control Measures
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knoester, M.; de Boer, M.G.; Maarleveld, J.J.; Claas, E.C.J.; Bernards, A.T.; de Jonge, E.; van Dissel, J.T.; Veldkamp, K.E. An integrated approach to control a prolonged outbreak of multidrug-resistant Pseudomonas aeruginosa in an intensive care unit. Clin. Microbiol. Infect. 2014, 20, O207–O215. [Google Scholar] [CrossRef] [PubMed]
- Lolans, K.; Queenan, A.M.; Bush, K.; Sahud, A.; Quinn, J.P. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-beta-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 2005, 49, 3538–3540. [Google Scholar] [CrossRef] [PubMed]
- Van der Bij, A.K.; Van Mansfeld, R.; Peirano, G.; Goessens, W.H.F.; Severin, J.A.; Pitout, J.D.D.; Willems, R.; Van Westreenen, M. First outbreak of VIM-2 metallo-beta-lactamase-producing Pseudomonas aeruginosa in The Netherlands: Microbiology, epidemiology and clinical outcomes. Int. J. Antimicrob. Agents 2011, 37, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Suarez, C.; Peña, C.; Arch, O.; Dominguez, M.A.; Tubau, F.; Juan, C.; Gavaldá, L.; Sora, M.; Oliver, A.; Pujol, M.; et al. A large sustained endemic outbreak of multiresistant Pseudomonas aeruginosa: A new epidemiological scenario for nosocomial acquisition. BMC Infect. Dis. 2011, 11, 272. [Google Scholar] [CrossRef]
- Jongerden, I.P.; Buiting, A.G.; Leverstein-van Hall, M.A.; Speelberg, B.; Zeidler, S.; Kesecioglu, J.; Bonten, M.J. Effect of open and closed endotracheal suctioning on cross-transmission with Gram-negative bacteria: A prospective crossover study. Crit. Care Med. 2011, 39, 1313–1321. [Google Scholar] [CrossRef]
- Verfaillie, C.J.; Bruno, M.J.; Holt, A.F.V.I.; Buijs, J.G.; Poley, J.-W.; Loeve, A.J.; Severin, J.A.; Abel, L.F.; Smit, B.J.; de Goeij, I.; et al. Withdrawal of a novel-design duodenoscope ends outbreak of a VIM-2-producing Pseudomonas aeruginosa. Endoscopy 2015, 47, 493–502. [Google Scholar] [CrossRef]
- Ferroni, A.; Nguyen, L.; Pron, B.; Quesne, G.; Brusset, M.C.; Berche, P. Outbreak of nosocomial urinary tract infections due to Pseudomonas aeruginosa in a paediatric surgical unit associated with tap-water contamination. J. Hosp. Infect. 1998, 39, 301–307. [Google Scholar] [CrossRef]
- Breathnach, A.S.; Cubbon, M.D.; Karunaharan, R.N.; Pope, C.F.; Planche, T.D. Multidrug-resistant Pseudomonas aeruginosa outbreaks in two hospitals: Association with contaminated hospital waste-water systems. J. Hosp. Infect. 2012, 82, 19–24. [Google Scholar] [CrossRef]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef]
- Cameron, D.R.; Pitton, M.; Oberhaensli, S.; Schlegel, K.; Prod’hom, G.; Blanc, D.S.; Jakob, S.M.; Que, Y.-A. Parallel evolution of Pseudomonas aeruginosa during a prolonged ICU-infection outbreak. Microbiol. Spectr. 2022, 10, e0274322. [Google Scholar] [CrossRef]
- Yoon, E.-J.; Jeong, S.H. Mobile carbapenemase genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef]
- Glen, K.A.; Lamont, I.L. β-lactam resistance in Pseudomonas aeruginosa: Current status, future prospects. Pathogens 2021, 10, 1638. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Doyle, T.B.; Hubler, C.M.; Collingsworth, T.D.; DeVries, S.; Mendes, R.E. The plethora of resistance mechanisms in Pseudomonas aeruginosa: Transcriptome analysis reveals a potential role of lipopolysaccharide pathway proteins to novel β-lactam/β-lactamase inhibitor combinations. J. Glob. Antimicrob. Resist. 2022, 31, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Elshamy, A.A.; Aboshanab, K.M. A review on bacterial resistance to carbapenems: Epidemiology, detection and treatment options. Future Sci. OA. 2020, 6, FSO438. [Google Scholar] [CrossRef] [PubMed]
- Del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Collet, L.; Nordmann, P. Carbapenem-hydrolyzing metallo-beta-lactamase from a nosocomial isolate of Pseudomonas aeruginosa in France. Emerg. Infect. Dis. 2000, 6, 84–85. [Google Scholar] [CrossRef]
- Pitout, J.D.; Chow, B.L.; Gregson, D.B.; Laupland, K.B.; Elsayed, S.; Church, D.L. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa in the Calgary Health Region: Emergence of VIM-2-producing isolates. J. Clin. Microbiol. 2007, 45, 294–298. [Google Scholar] [CrossRef]
- Viedma, E.; Juan, C.; Villa, J.; Barrado, L.; Orellana, M.A.; Sanz, F.; Otero, J.R.; Oliver, A.; Chaves, F. VIM-2-producing multidrug-resistant Pseudomonas aeruginosa ST175 clone, Spain. Emerg. Infect. Dis. 2012, 18, 1235–1241. [Google Scholar] [CrossRef]
- Pirzadian, J.; Persoon, M.C.; Severin, J.A.; Klaassen, C.H.W.; de Greeff, S.C.; Mennen, M.G.; Schoffelen, A.F.; Wielders, C.C.H.; Witteveen, S.; van Santen-Verheuvel, M.; et al. National surveillance pilot study unveils a multicenter, clonal outbreak of VIM-2-producing Pseudomonas aeruginosa ST111 in the Netherlands between 2015 and 2017. Sci. Rep. 2021, 11, 21015. [Google Scholar] [CrossRef]
- Samuelsen, Ö.; Toleman, M.A.; Sundsfjord, A.; Rydberg, J.; Leegaard, T.M.; Walder, M.; Lia, A.; Ranheim, T.E.; Rajendra, Y.; Hermansen, N.O.; et al. Molecular Epidemiology of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa Isolates from Norway and Sweden Shows Import of International Clones and Local Clonal Expansion. Antimicrob. Agents Chemother. 2010, 54, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, Y.; Shibata, N.; Shibayama, K.; Kurokawa, H.; Yagi, T.; H Fujiwara, H.; Goto, M. Convenient test for screening metallo-beta-lactamase-producing Gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 2000, 38, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lim, Y.S.; Yong, D.; Yum, J.H.; Chong, Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 2003, 41, 4623–4629. [Google Scholar] [CrossRef]
- Shibata, N.; Doi, Y.; Yamane, K.; Yagi, T.; Kurokawa, H.; Shibayama, K.; Kato, H.; Kai, K.; Arakawa, Y. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by Gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J. Clin. Microbiol. 2003, 41, 5407–5413. [Google Scholar] [CrossRef] [PubMed]
- Lytsy, B.; Sandegren, L.; Tano, E.; Torell, E.; Andersson, D.I.; Melhus, Å. The first major extended-spectrum beta-lactamase outbreak in Scandinavia was caused by clonal spread of a multiresistant Klebsiella pneumoniae producing CTX-M-15. APMIS 2008, 116, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Chevreux, B.; Wetter, T.; Suhai, S. Genome sequence: Assembly using trace signals and additional sequence information. Comput. Sci. Biol. Proc. Ger. Conf. Bioinform. GCB 1999, 99, 45–56. [Google Scholar]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Curran, B.; Jonas, D.; Grundmann, H.; Pitt, T.; Dowson, C.G. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 2004, 42, 5644–5649. [Google Scholar] [CrossRef]
- Cury, J.; Jove, T.; Touchon, M.; Neron, B.; Rocha, E.P. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016, 44, 4539–4550. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Stjärne Aspelund, A.; Sjöström, K.; Olsson Liljequist, B.; Mörgelin, M.; Melander, E.; Påhlman, L.I. Acetic acid as a decontamination method for sink drains in a nosocomial outbreak of metallo-beta-lactamase-producing Pseudomonas aeruginosa. J. Hosp. Infect. 2016, 94, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Constantinides, B.; Chau, K.K.; Quan, T.P.; Rodger, G.; Andersson, M.I.; Jeffery, K.; Lipworth, S.; Gweon, H.S.; Peniket, A.; Pike, G.; et al. Genomic surveillance of Escherichia coli and Klebsiella spp. in hospital sink drains and patients. Microb. Genom. 2020, 6, e000391. [Google Scholar] [CrossRef] [PubMed]
- Katzenberger, R.H.; Rösel, A.; Vonberg, R.P. Bacterial survival on inanimate surfaces: A field study. BMC Res. Notes 2021, 14, 97. [Google Scholar] [CrossRef] [PubMed]
- Lewenza, S.; Abboud, J.; Poon, K.; Kobryn, M.; Humplik, I.; Bell, J.R.; Mardan, L.; Reckseidler-Zenteno, S. Pseudomonas aeruginosa displays a dormancy phenotype during long-term survival in water. PLoS ONE 2018, 13, e0198384. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, T.A.; Johnson, P.R.; Notman, A.W.; Coia, J.E.; Hanson, M.F. Eradication of a resistant Pseudomonas aeruginosa strain after a cluster of infections in a hematology/oncology unit. Clin. Microbiol. Infect. 2000, 6, 125–130. [Google Scholar] [CrossRef]
- Hota, S.; Hirji, Z.; Stockton, K.; Lemieux, C.; Dedier, H.; Wolfaardt, G.; Gardam, M.A. Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infect. Control Hosp. Epidemiol. 2009, 30, 25–33. [Google Scholar] [CrossRef]
- Witney, A.A.; Gould, K.A.; Pope, C.F.; Bolt, F.; Stoker, N.G.; Cubbon, M.D.; Bradley, C.R.; Fraise, A.; Breathnach, A.S.; Butcher, P.D.; et al. Genome sequencing and characterization of an extensively drug-resistant sequence type 111 serotype O12 hospital outbreak strain of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2014, 20, O609–O618. [Google Scholar] [CrossRef]
- Catho, G.; Martischang, R.; Boroli, F.; Chraïti, M.N.; Martin, Y.; Tomsuk, Z.K.; Renzi, G.; Schrenzel, J.; Pugin, J.; Nordmann, P.; et al. Outbreak of Pseudomonas aeruginosa producing VIM carbapenemase in an intensive care unit and its termination by implementation of waterless patient care. Crit. Care 2021, 25, 301. [Google Scholar] [CrossRef]
- Wright, L.L.; Turton, J.F.; Livermore, D.M.; Hopkins, K.L.; Woodford, N. Dominance of international ‘high-risk clones’ among metallo-beta-lactamase-producing Pseudomonas aeruginosa in the UK. J. Antimicrob. Chemother. 2015, 70, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Mulet, X.; Lopez-Causape, C.; Juan, C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 2015, 21–22, 41–59. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Naas, T.; Nordmann, P. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
| Patient | MBL Culture + Sample | MBL Screening Prior to Admittance MBL-Induced Infection | Antibiotic Treatment 1 | Risk Factors | Outcome | Follow-Up | Possible Transmission of MBL+ P. aeruginosa 2 |
|---|---|---|---|---|---|---|---|
| 1 | 2 February 2006 Urine | Not performed Yes | GEN + CXM | Urinary catheter | Discharged on day 117 | MBL− on days 15, 21, 33, and 221 | KnH: Med A, Med B, and ICU |
| 2 | 15 February, 2006 BAL 3 | Not performed Yes | GEN + PTZ | Invasive ventilation | Dead on day 7 | - | KnH: Med B and ICU |
| 3 | 28 February 2006 Urine | Negative Yes | PTZ | Urinary catheter | Discharged on day 9 | MBL− on days 9, 111, and 154 | KnH: Med B |
| 4 | 6 March 2006 Urine | Not performed No | - | Urinary catheter | Discharged on day 4, dead of other reasons | - | KaH: Surg A |
| 5 | 7 August 2006 Sputum | Negative Probable | AZI, ERT, RIM | - | Dead on day 33 | - | KnH: Med A |
| 6 | 12 August 2006 Sputum, wound | Negative No | CTX | Leg ulcer | Discharged on day 26 | MBL− on days 46, 89, and 300 | KnH: Med A and ICU |
| 7 | 13 December 2006 Urine | Negative No | - | Urinary catheter | Discharged on day 2 | MBL+ on day 320 | KnH: Med A |
| 8 | 20 May 2007 Urine | Negative No | AMX | Urinary catheter | Dead on day 11 | - | KnH: Med A KnH: Surg B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fraenkel, C.-J.; Starlander, G.; Tano, E.; Sütterlin, S.; Melhus, Å. The First Swedish Outbreak with VIM-2-Producing Pseudomonas aeruginosa, Occurring between 2006 and 2007, Was Probably Due to Contaminated Hospital Sinks. Microorganisms 2023, 11, 974. https://doi.org/10.3390/microorganisms11040974
Fraenkel C-J, Starlander G, Tano E, Sütterlin S, Melhus Å. The First Swedish Outbreak with VIM-2-Producing Pseudomonas aeruginosa, Occurring between 2006 and 2007, Was Probably Due to Contaminated Hospital Sinks. Microorganisms. 2023; 11(4):974. https://doi.org/10.3390/microorganisms11040974
Chicago/Turabian StyleFraenkel, Carl-Johan, Gustaf Starlander, Eva Tano, Susanne Sütterlin, and Åsa Melhus. 2023. "The First Swedish Outbreak with VIM-2-Producing Pseudomonas aeruginosa, Occurring between 2006 and 2007, Was Probably Due to Contaminated Hospital Sinks" Microorganisms 11, no. 4: 974. https://doi.org/10.3390/microorganisms11040974
APA StyleFraenkel, C.-J., Starlander, G., Tano, E., Sütterlin, S., & Melhus, Å. (2023). The First Swedish Outbreak with VIM-2-Producing Pseudomonas aeruginosa, Occurring between 2006 and 2007, Was Probably Due to Contaminated Hospital Sinks. Microorganisms, 11(4), 974. https://doi.org/10.3390/microorganisms11040974





