Selecting 16S rRNA Primers for Microbiome Analysis in a Host–Microbe System: The Case of the Jellyfish Rhopilema nomadica
Abstract
1. Introduction
2. Materials and Methods
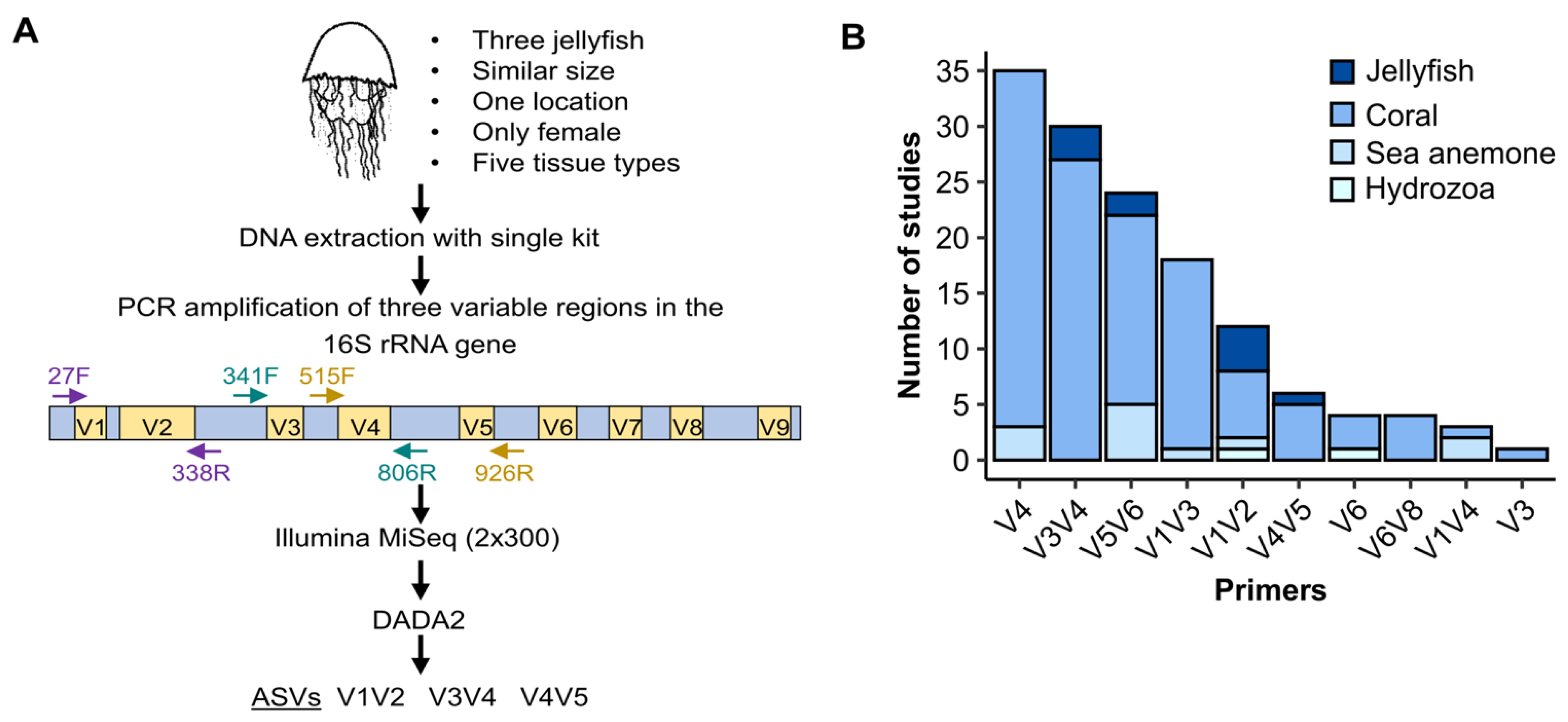
2.1. Jellyfish and Sea Water Samples Collection and Preparation
2.2. 16S rRNA Gene Library Preparation and Sequencing
2.3. Literature Survey of Microbiome Studies in Cnidaria
2.4. Data Analysis and Visualization
3. Results
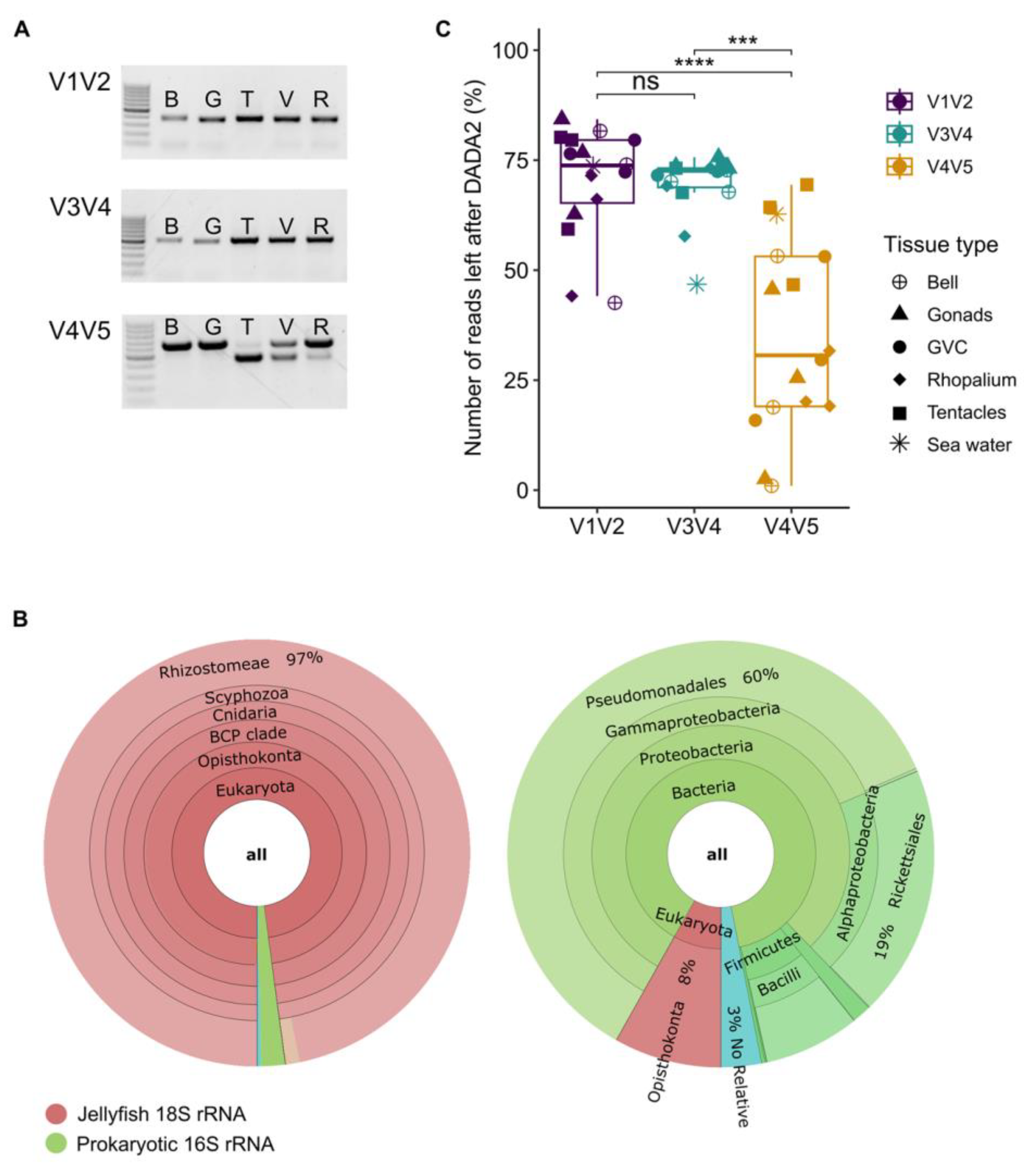
3.1. Multiple Amplicon Sequence Lengths Affect Community Coverage
3.2. Heterogeneity in Amplicon Length and ASVs Number when Examining the Mock Community Control
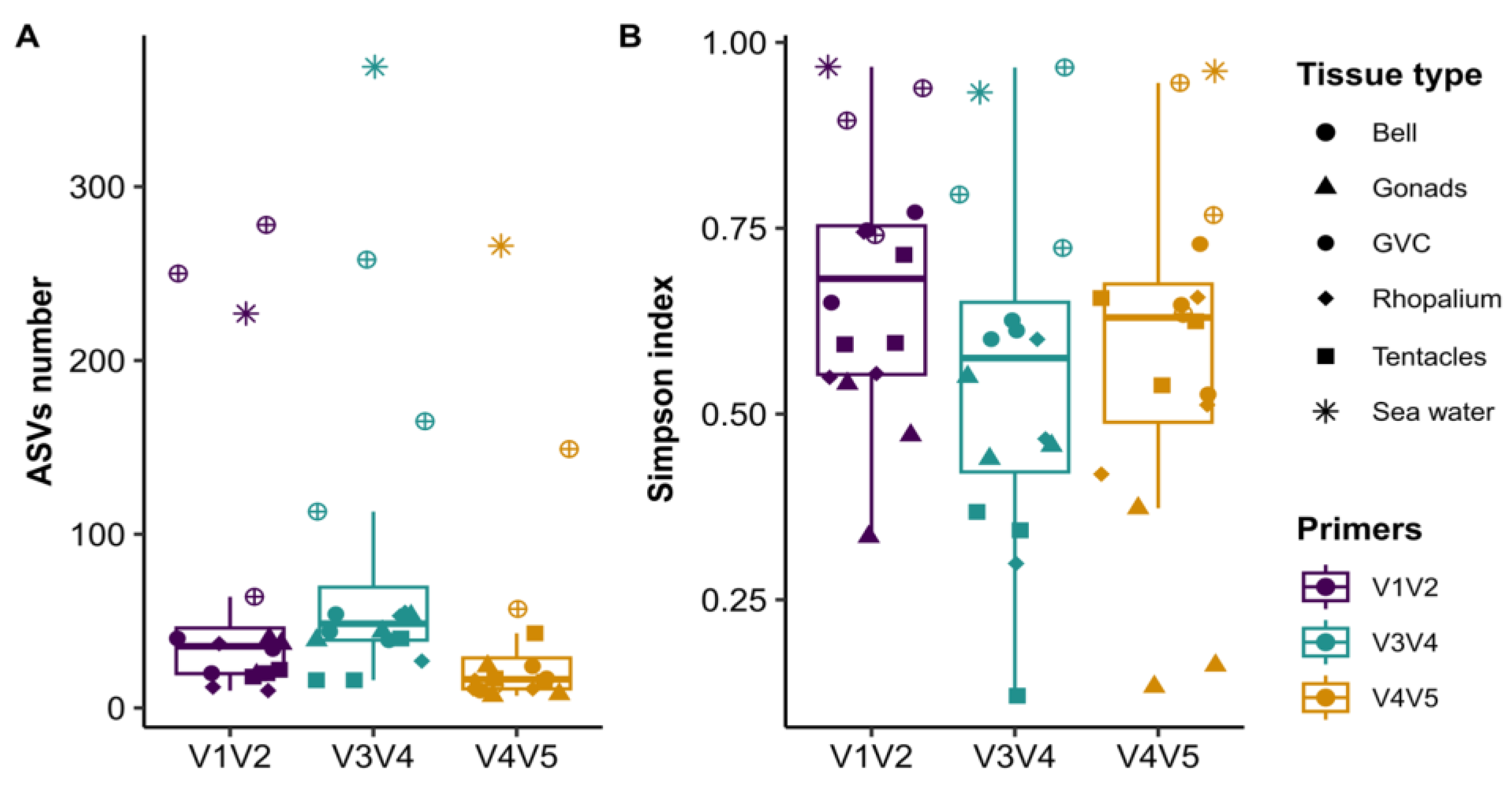
3.3. Similar Pattern of Diversity and Community Structure Revealed by All Three Primer Sets
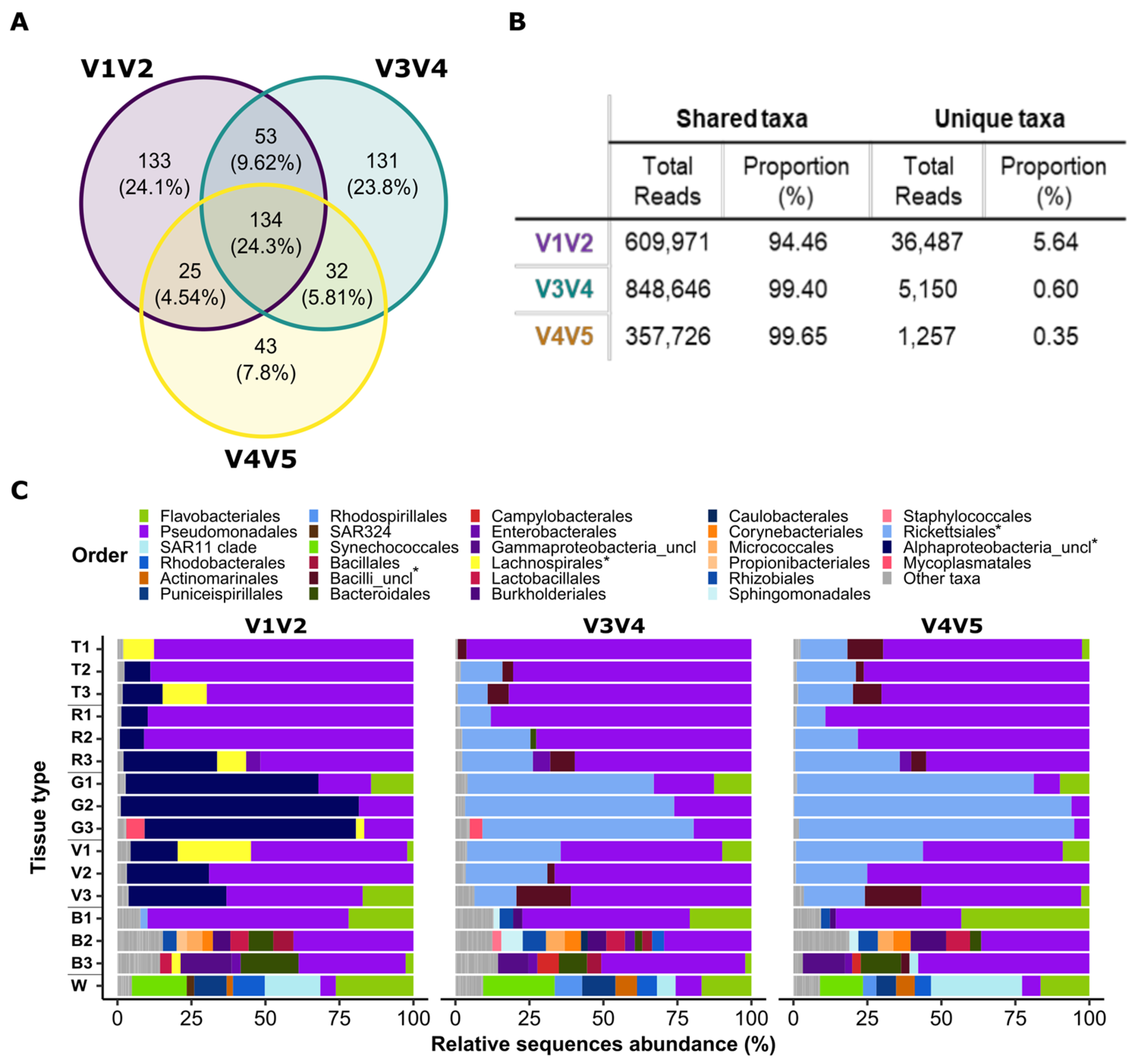
3.4. Discrepancies in Taxonomic Classifications between the Primers Affect Community Coverage
4. Discussion
4.1. Amplification of Eukaryotic 18S rRNA Gene by the V4V5 Primer Set may Affect Bacterial Diversity Observations
4.2. Sequences of the V1V2 Region May Lead to an Inaccurate Taxonomic Classification
4.3. V3V4 Primers Are Commonly Used but Not without Caveats
4.4. Can the Results of Different Primer Sets Be Compared?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bosch, T.C.G. Cnidarian-Microbe Interactions and the Origin of Innate Immunity in Metazoans. Annu. Rev. Microbiol. 2013, 67, 499–518. [Google Scholar] [CrossRef] [PubMed]
- Weiland-Bräuer, N.; Pinnow, N.; Langfeldt, D.; Roik, A.; Güllert, S.; Chibani, C.M.; Reusch, T.B.H.; Schmitz, R.A. The Native Microbiome is Crucial for Offspring Generation and Fitness of Aurelia aurita. Mbio 2020, 11, e02336-20. [Google Scholar] [CrossRef] [PubMed]
- Fraune, S.; Bosch, T.C.G. Why bacteria matter in animal development and evolution. Bioessays 2010, 32, 571–580. [Google Scholar] [CrossRef]
- Foster, K.R.; Schluter, J.; Coyte, K.Z.; Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 2017, 548, 43–51. [Google Scholar] [CrossRef]
- Schnedler-Meyer, N.A.; Kiørboe, T.; Mariani, P. Boom and Bust: Life History, Environmental Noise, and the (un)Predictability of Jellyfish Blooms. Front. Mar. Sci. 2018, 5, 257. [Google Scholar] [CrossRef]
- Rosenberg, E.; Zilber-Rosenberg, I. The hologenome concept of evolution after 10 years. Microbiome 2018, 6, 78. [Google Scholar] [CrossRef]
- Gould, A.L.; Zhang, V.; Lamberti, L.; Jones, E.W.; Obadia, B.; Korasidis, N.; Gavryushkin, A.; Carlson, J.M.; Beerenwinkel, N.; Ludington, W.B. Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. USA 2018, 115, E11951–E11960. [Google Scholar] [CrossRef]
- Baldassarre, L.; Levy, S.; Bar-Shalom, R.; Steindler, L.; Lotan, T.; Fraune, S. Contribution of Maternal and Paternal Transmission to Bacterial Colonization in Nematostella vectensis. Front. Microbiol. 2021, 12, 2892. [Google Scholar] [CrossRef]
- Stabili, L.; Parisi, M.G.; Parrinello, D.; Cammarata, M. Cnidarian Interaction with Microbial Communities: From Aid to Animal’s Health to Rejection Responses. Mar. Drugs 2018, 16, 296. [Google Scholar] [CrossRef]
- Mortzfeld, B.; Taubenheim, J.; Fraune, S.; Klimovich, A.; Bosch, T.C.G. Stem Cell Transcription Factor FoxO Controls Microbiome Resilience in Hydra. Front. Microbiol. 2018, 9, 629. [Google Scholar] [CrossRef]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Rosenberg, E.; Zilber-Rosenberg, I. Symbiosis and development: The hologenome concept. Birth Defects Res. Part C Embryo Today Rev. 2011, 93, 56–66. [Google Scholar] [CrossRef]
- Cartwright, P.; Halgedahl, S.L.; Hendricks, J.R.; Jarrard, R.D.; Marques, A.C.; Collins, A.G.; Lieberman, B.S. Excep-tionally Preserved Jellyfishes from the Middle Cambrian. PLoS ONE 2007, 2, e1121. [Google Scholar] [CrossRef]
- Van Oppen, M.J.H.; Blackall, L.L. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 2019, 17, 557–567. [Google Scholar] [CrossRef]
- Bourne, D.G.; Morrow, K.M.; Webster, N.S. Insights into the Coral Microbiome: Underpinning the Health and Resilience of Reef Ecosystems. Annu. Rev. Microbiol. 2016, 70, 317–340. [Google Scholar] [CrossRef]
- Rajasabapathy, R.; Ramasamy, K.P.; Manikandan, B.; Mohandass, C.; James, R.A. Bacterial Communities Associated with Healthy and Diseased (Skeletal Growth Anomaly) Reef Coral Acropora cytherea From Palk Bay, India. Front. Mar. Sci. 2020, 7, 92. [Google Scholar] [CrossRef]
- Tinta, T.; Kogovšek, T.; Klun, K.; Malej, A.; Herndl, G.J.; Turk, V. Jellyfish-Associated Microbiome in the Marine Environment: Exploring Its Biotechnological Potential. Mar. Drugs 2019, 17, 94. [Google Scholar] [CrossRef]
- Van De Water, J.A.J.M.; Allemand, D.; Ferrier-Pagès, C. Host-microbe interactions in octocoral holobionts–recent advances and perspectives. Microbiome 2018, 6, 64. [Google Scholar] [CrossRef] [PubMed]
- Voolstra, C.R.; Suggett, D.J.; Peixoto, R.S.; Parkinson, J.E.; Quigley, K.M.; Silveira, C.B.; Sweet, M.; Muller, E.M.; Barshis, D.J.; Bourne, D.G.; et al. Extending the natural adaptive capacity of coral holobionts. Nat. Rev. Earth Environ. 2021, 2, 747–762. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Rosado, P.M.; de Leite, D.C.A.; Rosado, A.S.; Bourne, D.G. Beneficial Microorganisms for Corals (BMC): Proposed Mechanisms for Coral Health and Resilience. Front. Microbiol. 2017, 8, 341. [Google Scholar] [CrossRef]
- Apprill, A. Marine Animal Microbiomes: Toward Understanding Host–Microbiome Interactions in a Changing Ocean. Front. Mar. Sci. 2017, 4, 222. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Methé, B.A.; Nelson, K.E.; Pop, M.; Creasy, H.H.; Giglio, M.G.; Huttenhower, C.; Gevers, D.; Petrosino, J.F.; Abubucker, S.; Badger, J.H.; et al. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef]
- Liu, P.-Y.; Wu, W.-K.; Chen, C.-C.; Panyod, S.; Sheen, L.-Y.; Wu, M.-S. Evaluation of Compatibility of 16S RRNA V3V4 and V4 Amplicon Libraries for Clinical Microbiome Profiling. bioRxiv 2020. bioRxiv:2020.08.18.256818. [Google Scholar]
- Alcon-Giner, C.; Caim, S.; Mitra, S.; Ketskemety, J.; Wegmann, U.; Wain, J.; Belteki, G.; Clarke, P.; Hall, L.J. Optimisation of 16S rRNA gut microbiota profiling of extremely low birth weight infants. BMC Genom. 2017, 18, 841. [Google Scholar] [CrossRef] [PubMed]
- Rausch, P.; Rühlemann, M.; Hermes, B.M.; Doms, S.; Dagan, T.; Dierking, K.; Domin, H.; Fraune, S.; von Frieling, J.; Hentschel, U.; et al. Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Microbiome 2019, 7, 133. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Wear, E.K.; Wilbanks, E.G.; Nelson, C.E.; Carlson, C.A. Primer selection impacts specific population abundances but not community dynamics in a monthly time-series 16S rRNA gene amplicon analysis of coastal marine bacterioplankton. Environ. Microbiol. 2018, 20, 2709–2726. [Google Scholar] [CrossRef] [PubMed]
- Willis, C.; Desai, D.; Laroche, J. Influence of 16S rRNA variable region on perceived diversity of marine microbial communities of the Northern North Atlantic. FEMS Microbiol. Lett. 2019, 366, fnz152. [Google Scholar] [CrossRef]
- Fadeev, E.; Cardozo-Mino, M.G.; Rapp, J.Z.; Bienhold, C.; Salter, I.; Salman-Carvalho, V.; Molari, M.; Tegetmeyer, H.E.; Buttigieg, P.L.; Boetius, A. Comparison of Two 16S rRNA Primers (V3–V4 and V4–V5) for Studies of Arctic Microbial Communities. Front. Microbiol. 2021, 12, 637526. [Google Scholar] [CrossRef]
- Yang, B.; Wang, Y.; Qian, P.-Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinform. 2016, 17, 135. [Google Scholar] [CrossRef]
- Angel, D.L.; Edelist, D.; Freeman, S. Local perspectives on regional challenges: Jellyfish proliferation and fish stock management along the Israeli Mediterranean coast. Reg. Environ. Chang. 2014, 16, 315–323. [Google Scholar] [CrossRef]
- Edelist, D.; Guy-Haim, T.; Kuplik, Z.; Zuckerman, N.; Nemoy, P.; Angel, D.L. Phenological shift in swarming patterns of Rhopilema nomadica in the Eastern Mediterranean Sea. J. Plankton Res. 2020, 42, 211–219. [Google Scholar] [CrossRef]
- Cruaud, P.; Vigneron, A.; Fradette, M.-S.; Charette, S.J.; Rodriguez, M.J.; Dorea, C.C.; Culley, A.I. Open the SterivexTM casing: An easy and effective way to improve DNA extraction yields. Limnol. Oceanogr. Methods 2017, 15, 1015–1020. [Google Scholar] [CrossRef]
- Naqib, A.; Poggi, S.; Wang, W.; Hyde, M.; Kunstman, K.; Green, S.J. Making and Sequencing Heavily Multiplexed. Gene Expr. Anal. Methods Protoc. 2018, 1783, 149–169. [Google Scholar]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From Raw Reads to Community Analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H.; François, R.; Henry, L.; Müller, K. Package “dplyr” Type Package Title A Grammar of Data Manipulation. 2022. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 1st ed.; Springer: New York, NY, USA, 2016; Volume 35, ISBN 978-0-387-98140-6. [Google Scholar]
- Kassambara, A. Manual Cite. 2021. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G. Vegan: Community Ecology Package. R Package Version 2.5–7. 2022. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 17 April 2022).
- Schwenk, A.J. Venn Diagram for Five Sets. Math. Mag. 1984, 57, 297. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Parada, E.A.; Fuhrman, A.J. Marine archaeal dynamics and interactions with the microbial community over 5 years from surface to seafloor. ISME J. 2017, 11, 2510–2525. [Google Scholar] [CrossRef]
- Yeh, Y.; McNichol, J.; Needham, D.M.; Fichot, E.B.; Berdjeb, L.; Fuhrman, J.A. Comprehensive single-PCR 16S and 18S rRNA community analysis validated with mock communities, and estimation of sequencing bias against 18S. Environ. Microbiol. 2021, 23, 3240–3250. [Google Scholar] [CrossRef]
- Beckers, B.; Op De Beeck, M.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Performance of 16s rDNA Primer Pairs in the Study of Rhizosphere and Endosphere Bacterial Microbiomes in Metabarcoding Studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef]
- Graspeuntner, S.; Loeper, N.; Künzel, S.; Baines, J.F.; Rupp, J. Selection of validated hypervariable regions is crucial in 16S-based microbiota studies of the female genital tract. Sci. Rep. 2018, 8, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Weiland-Bräuer, N.; Neulinger, S.C.; Pinnow, N.; Künzel, S.; Baines, J.F.; Schmitz, R.A. Composition of Bacterial Communities Associated with Aurelia aurita Changes with Compartment, Life Stage, and Population. Appl. Environ. Microbiol. 2015, 81, 6038–6052. [Google Scholar] [CrossRef]
- Haber, M.; Rosenberg, D.R.; Lalzar, M.; Burgsdorf, I.; Saurav, K.; Lionheart, R.; Lehahn, Y.; Aharonovich, D.; Gómez-Consarnau, L.; Sher, D.; et al. Spatiotemporal Variation of Microbial Communities in the Ultra-Oligotrophic Eastern Mediterranean Sea. Front. Microbiol. 2022, 13, 1129. [Google Scholar] [CrossRef]
- Mestre, M.; Ruiz-González, C.; Logares, R.; Duarte, C.M.; Gasol, J.M.; Sala, M.M. Sinking particles promote vertical connectivity in the ocean microbiome. Proc. Natl. Acad. Sci. USA 2018, 115, E6799–E6807. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Needham, D.M.; Fuhrman, J.A. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat. Microbiol. 2016, 1, 16005. [Google Scholar] [CrossRef]
- McNichol, J.; Berube, P.M.; Biller, S.J.; Fuhrman, J.A. Evaluating and Improving Small Subunit rRNA PCR Primer Coverage for Bacteria, Archaea, and Eukaryotes Using Metagenomes from Global Ocean Surveys. Msystems 2021, 6, e00565-21. [Google Scholar] [CrossRef]
- Zhang, S.L.; Bai, L.; Goel, N.; Bailey, A.; Jang, C.J.; Bushman, F.D.; Meerlo, P.; Dinges, D.F.; Sehgal, A. Human and rat gut microbiome composition is maintained following sleep restriction. Proc. Natl. Acad. Sci. USA 2017, 114, E1564–E1571. [Google Scholar] [CrossRef]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.-W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef]
- Allali, I.; Arnold, J.W.; Roach, J.; Cadenas, M.B.; Butz, N.; Hassan, H.M.; Koci, M.; Ballou, A.; Mendoza, M.; Ali, R.; et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.L.; Monteagudo-Mera, A.; Cadenas, M.B.; Lampl, M.L.; Azcarate-Peril, M.A. Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front. Cell. Infect. Microbiol. 2015, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Albores, F.; Ortiz-Suárez, L.E.; Villalpando-Canchola, E.; Martínez-Porchas, M. Size-variable zone in V3 region of 16S rRNA. RNA Biol. 2017, 14, 1514–1521. [Google Scholar] [CrossRef]
- Schön, M.E.; Martijn, J.; Vosseberg, J.; Köstlbacher, S.; Ettema, T.J.G. The evolutionary origin of host association in the Rickettsiales. Nat. Microbiol. 2022, 7, 1189–1199. [Google Scholar] [CrossRef]
- Salje, J. Cells within cells: Rickettsiales and the obligate intracellular bacterial lifestyle. Nat. Rev. Microbiol. 2021, 19, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Daley, M.C.; Urban-Rich, J.; Moisander, P.H. Bacterial associations with the hydromedusa Nemopsis bachei and scyphomedusa Aurelia aurita from the North Atlantic Ocean. Mar. Biol. Res. 2016, 12, 1088–1100. [Google Scholar] [CrossRef]
- Klinges, J.G.; Rosales, S.M.; McMinds, R.; Shaver, E.C.; Shantz, A.A.; Peters, E.C.; Eitel, M.; Wörheide, G.; Sharp, K.H.; Burkepile, D.E.; et al. Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J. 2019, 13, 2938–2953. [Google Scholar] [CrossRef]
- Gosalbes, M.J.; Durbán, A.; Pignatelli, M.; Abellan-Andres, J.J.; Hernández, N.J.; Pérez-Cobas, A.E.; Latorre, A.; Moya, A. Metatranscriptomic Approach to Analyze the Functional Human Gut Microbiota. PLoS ONE 2011, 6, e17447. [Google Scholar] [CrossRef] [PubMed]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Viver, T.; Orellana, L.H.; Hatt, J.K.; Urdiain, M.; Díaz, S.; Richter, M.; Antón, J.; Avian, M.; Amann, R.; Konstantinidis, K.T.; et al. The low diverse gastric microbiome of the jellyfish Cotylorhiza tuberculata is dominated by four novel taxa. Environ. Microbiol. 2017, 19, 3039–3058. [Google Scholar] [CrossRef] [PubMed]
- Stabili, L.; Rizzo, L.; Fanizzi, F.P.; Angilè, F.; Del Coco, L.; Girelli, C.R.; Lomartire, S.; Piraino, S.; Basso, L. The Jellyfish Rhizostoma pulmo (Cnidaria): Biochemical Composition of Ovaries and Antibacterial Lysozyme-like Activity of the Oocyte Lysate. Mar. Drugs 2019, 17, 17. [Google Scholar] [CrossRef]
- Ballinger, M.J.; Perlman, S.J. The defensive Spiroplasma. Curr. Opin. Insect Sci. 2019, 32, 36–41. [Google Scholar] [CrossRef]
- He, L.-S.; Zhang, P.-W.; Huang, J.-M.; Zhu, F.-C.; Danchin, A.; Wang, Y. The Enigmatic Genome of an Obligate Ancient Spiroplasma Symbiont in a Hadal Holothurian. Appl. Environ. Microbiol. 2018, 84, e01965-17. [Google Scholar] [CrossRef]
- Illumina 16S Metagenomic Sequencing Library. Illumina.com, 2013; 1–28.
- Fan, L.; Wang, Z.; Chen, M.; Qu, Y.; Li, J.; Zhou, A.; Xie, S.; Zeng, F.; Zou, J. Microbiota comparison of Pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Sci. Total. Environ. 2019, 657, 1194–1204. [Google Scholar] [CrossRef]
- Takagi, T.; Naito, Y.; Inoue, R.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Tsuchiya, S.; Dohi, O.; Yoshida, N.; Kamada, K.; et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J. Gastroenterol. 2019, 54, 53–63. [Google Scholar] [CrossRef]
- Dyksma, S.; Bischof, K.; Fuchs, B.; Hoffmann, K.; Meier, D.; Meyerdierks, A.; Pjevac, P.; Probandt, D.; Richter, M.; Stepanauskas, R.; et al. Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 2016, 10, 1939–1953. [Google Scholar] [CrossRef]
- Derakhshani, H.; Tun, H.M.; Khafipour, E. An extended single-index multiplexed 16S rRNA sequencing for microbial community analysis on MiSeq illumina platforms. J. Basic Microbiol. 2016, 56, 321–326. [Google Scholar] [CrossRef]
- Katiraei, S.; Anvar, Y.; Hoving, L.; Berbée, J.F.P.; van Harmelen, V.; van Dijk, K.W. Evaluation of Full-Length Versus V4-Region 16S rRNA Sequencing for Phylogenetic Analysis of Mouse Intestinal Microbiota After a Dietary Intervention. Curr. Microbiol. 2022, 79, 276. [Google Scholar] [CrossRef] [PubMed]
- García-López, R.; Cornejo-Granados, F.; Lopez-Zavala, A.A.; Sánchez-López, F.; Cota-Huízar, A.; Sotelo-Mundo, R.R.; Guerrero, A.; Mendoza-Vargas, A.; Gómez-Gil, B.; Ochoa-Leyva, A. Doing More with Less: A Comparison of 16S Hypervariable Regions in Search of Defining the Shrimp Microbiota. Microorganisms 2020, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.; Neave, M.J.; Alsheikh-Hussain, A.; Aranda, M.; Yum, L.K.; Mincer, T.; Hughen, K.; Apprill, A.; Voolstra, C.R. The Microbiome of the Red Sea Coral Stylophora pistillata Is Dominated by Tissue-Associated Endozoicomonas Bacteria. Appl. Environ. Microbiol. 2013, 79, 4759–4762. [Google Scholar] [CrossRef] [PubMed]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. Msphere 2021, 6, e01202-20. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.; Glendinning, L.; Wisedchanwet, T.; Watson, M. The Madness of Microbiome: Attempting to Find Consensus “Best Practice” for 16S Microbiome Studies. Appl. Environ. Microbiol. 2018, 84, e02627-17. [Google Scholar] [CrossRef]
- Darwish, N.; Shao, J.; Schreier, L.L.; Proszkowiec-Weglarz, M. Choice of 16S ribosomal RNA primers affects the microbiome analysis in chicken ceca. Sci. Rep. 2021, 11, 11848. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barak, N.; Fadeev, E.; Brekhman, V.; Aharonovich, D.; Lotan, T.; Sher, D. Selecting 16S rRNA Primers for Microbiome Analysis in a Host–Microbe System: The Case of the Jellyfish Rhopilema nomadica. Microorganisms 2023, 11, 955. https://doi.org/10.3390/microorganisms11040955
Barak N, Fadeev E, Brekhman V, Aharonovich D, Lotan T, Sher D. Selecting 16S rRNA Primers for Microbiome Analysis in a Host–Microbe System: The Case of the Jellyfish Rhopilema nomadica. Microorganisms. 2023; 11(4):955. https://doi.org/10.3390/microorganisms11040955
Chicago/Turabian StyleBarak, Noga, Eduard Fadeev, Vera Brekhman, Dikla Aharonovich, Tamar Lotan, and Daniel Sher. 2023. "Selecting 16S rRNA Primers for Microbiome Analysis in a Host–Microbe System: The Case of the Jellyfish Rhopilema nomadica" Microorganisms 11, no. 4: 955. https://doi.org/10.3390/microorganisms11040955
APA StyleBarak, N., Fadeev, E., Brekhman, V., Aharonovich, D., Lotan, T., & Sher, D. (2023). Selecting 16S rRNA Primers for Microbiome Analysis in a Host–Microbe System: The Case of the Jellyfish Rhopilema nomadica. Microorganisms, 11(4), 955. https://doi.org/10.3390/microorganisms11040955








