Information Scale Correction for Varying Length Amplicons Improves Eukaryotic Microbiome Data Integration
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Information Scale Correction
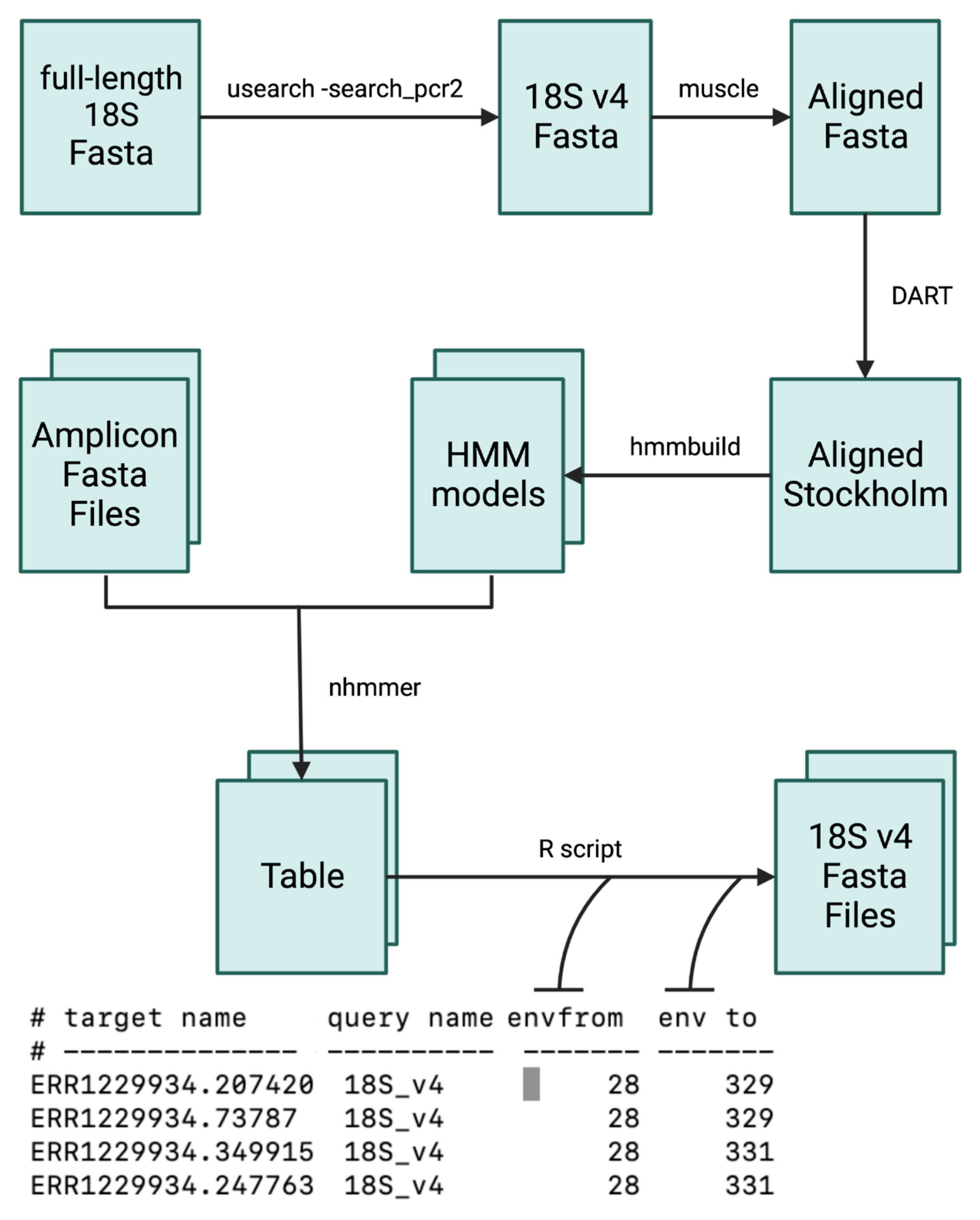
2.3. Bioinformatics Analysis
2.4. Statistical Analysis
3. Result
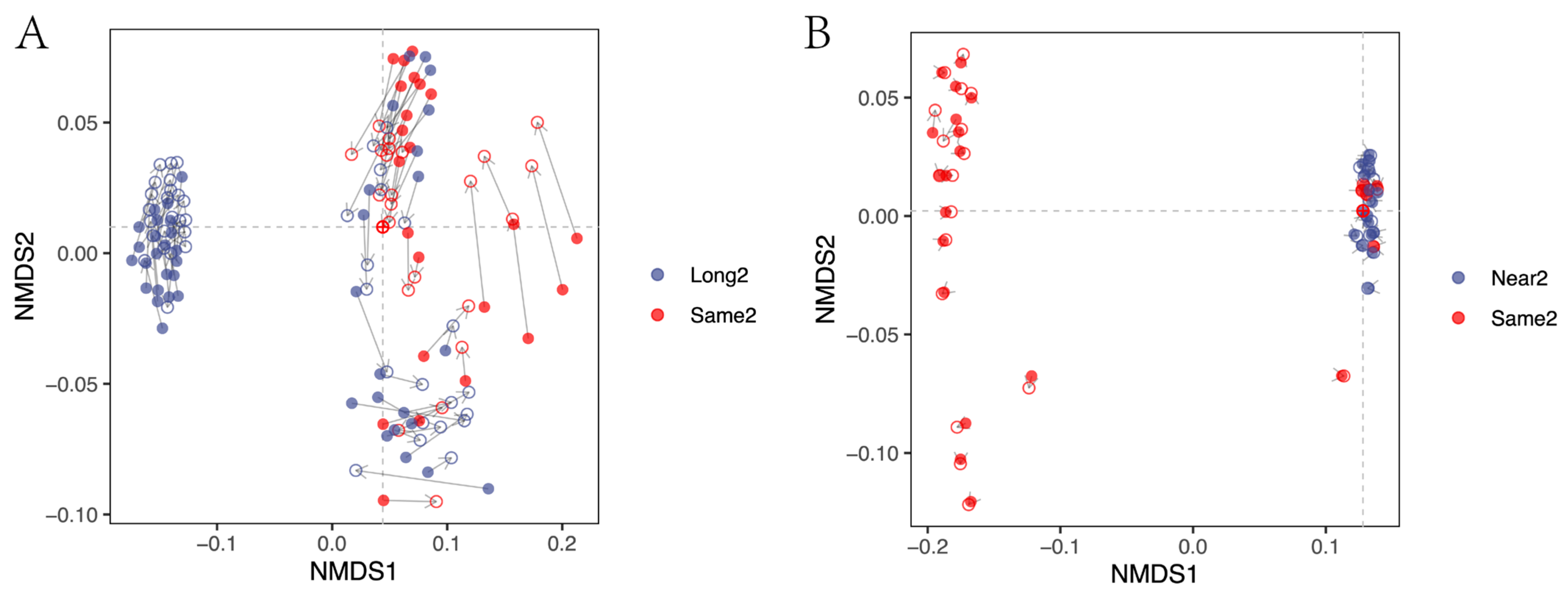
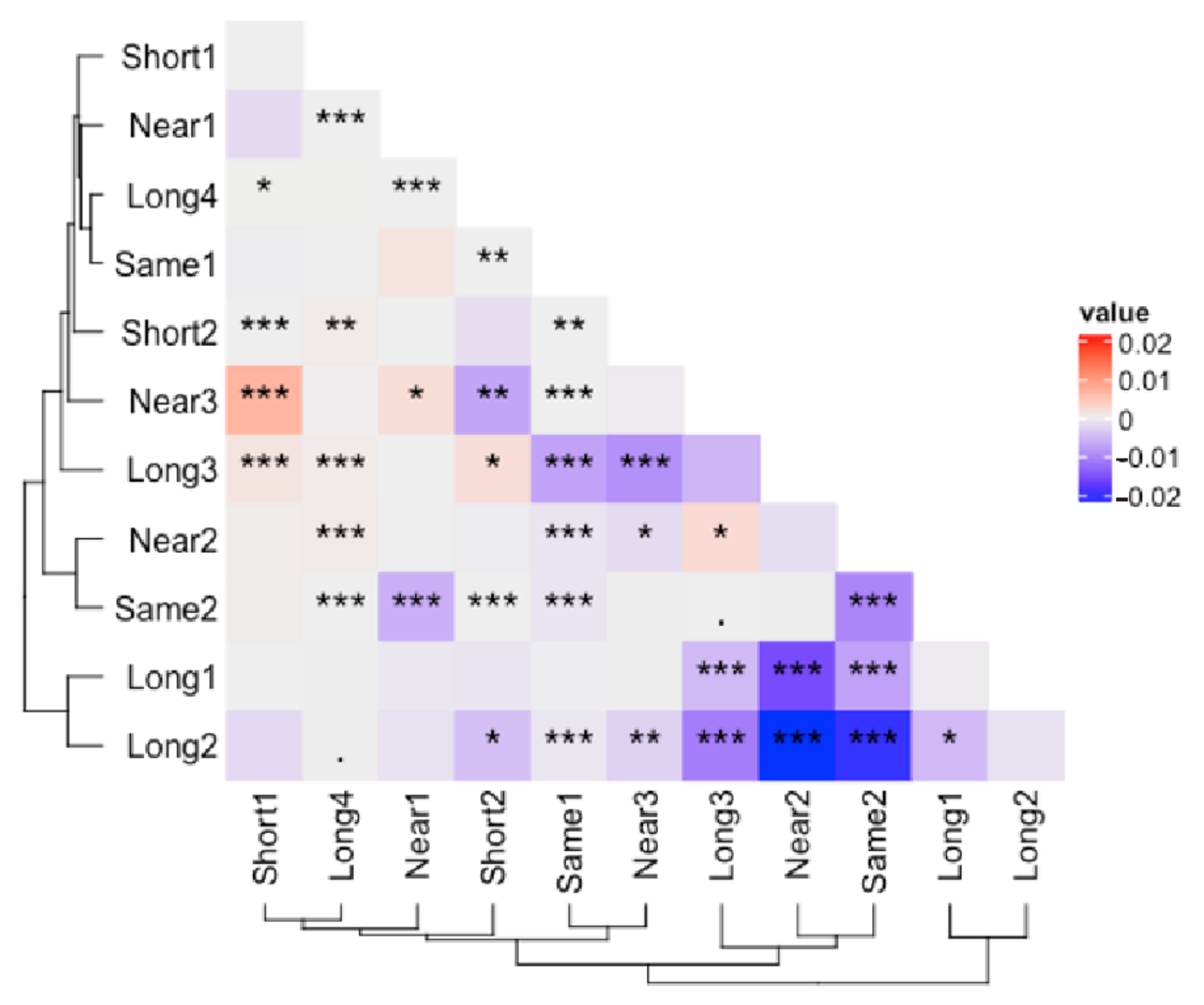
3.1. Correction Effect Comparison
3.2. Corrected and Non-Differential Effects
3.3. Correction Effect in Relation to Amplicon Region
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tara Ocean Foundation; Abreu, A.; Bourgois, E.; Gristwood, A.; Troublé, R.; Tara Oceans; Acinas, S.G.; Bork, P.; Boss, E.; Bowler, C.; et al. Priorities for Ocean Microbiome Research. Nat. Microbiol. 2022, 7, 937–947. [Google Scholar] [CrossRef]
- Cordier, T.; Angeles, I.B.; Henry, N.; Lejzerowicz, F.; Berney, C.; Morard, R.; Brandt, A.; Cambon-Bonavita, M.-A.; Guidi, L.; Lombard, F.; et al. Patterns of Eukaryotic Diversity from the Surface to the Deep-Ocean Sediment. Sci. Adv. 2022, 8, eabj9309. [Google Scholar] [CrossRef]
- Villarino, E.; Watson, J.R.; Jönsson, B.; Gasol, J.M.; Salazar, G.; Acinas, S.G.; Estrada, M.; Massana, R.; Logares, R.; Giner, C.R.; et al. Large-Scale Ocean Connectivity and Planktonic Body Size. Nat. Commun. 2018, 9, 142. [Google Scholar] [CrossRef]
- Jamy, M.; Foster, R.; Barbera, P.; Czech, L.; Kozlov, A.; Stamatakis, A.; Bending, G.; Hilton, S.; Bass, D.; Burki, F. Long-read Metabarcoding of the Eukaryotic RDNA Operon to Phylogenetically and Taxonomically Resolve Environmental Diversity. Mol. Ecol. Resour. 2020, 20, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Schlaeppi, K.; Bender, S.F.; Mascher, F.; Russo, G.; Patrignani, A.; Camenzind, T.; Hempel, S.; Rillig, M.C.; Heijden, M.G.A. High-resolution Community Profiling of Arbuscular Mycorrhizal Fungi. New Phytol. 2016, 212, 780–791. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.; et al. Greengenes2 Enables a Shared Data Universe for Microbiome Studies. bioRxiv 2022. [Google Scholar] [CrossRef]
- Franzén, O.; Hu, J.; Bao, X.; Itzkowitz, S.H.; Peter, I.; Bashir, A. Improved OTU-Picking Using Long-Read 16S RRNA Gene Amplicon Sequencing and Generic Hierarchical Clustering. Microbiome 2015, 3, 43. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.J.; Soubrier, J.; Weyrich, L.S.; Cooper, A. Environmental Metabarcodes for Insects: In Silico PCR Reveals Potential for Taxonomic Bias. Mol. Ecol. Resour. 2014, 14, 1160–1170. [Google Scholar] [CrossRef]
- Hugerth, L.W.; Pereira, M.; Zha, Y.; Seifert, M.; Kaldhusdal, V.; Boulund, F.; Krog, M.C.; Bashir, Z.; Hamsten, M.; Fransson, E.; et al. Assessment of In Vitro and In Silico Protocols for Sequence-Based Characterization of the Human Vaginal Microbiome. mSphere 2020, 5, e00448-20. [Google Scholar] [CrossRef]
- Brasseur, M.V.; Astrin, J.J.; Geiger, M.F.; Mayer, C. MitoGeneExtractor: Efficient Extraction of Mitochondrial Genes from Next-generation Sequencing Libraries. Methods Ecol. Evol. 2023, 1–8. [Google Scholar] [CrossRef]
- Allio, R.; Schomaker-Bastos, A.; Romiguier, J.; Prosdocimi, F.; Nabholz, B.; Delsuc, F. MitoFinder: Efficient Automated Large-scale Extraction of Mitogenomic Data in Target Enrichment Phylogenomics. Mol. Ecol. Resour. 2020, 20, 892–905. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved Software Detection and Extraction of ITS1 and ITS2 from Ribosomal ITS Sequences of Fungi and Other Eukaryotes for Analysis of Environmental Sequencing Data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Hartmann, M.; Eriksson, K.M.; Pal, C.; Thorell, K.; Larsson, D.G.J.; Nilsson, R.H. metaxa 2: Improved Identification and Taxonomic Classification of Small and Large Subunit RRNA in Metagenomic Data. Mol. Ecol. Resour. 2015, 15, 1403–1414. [Google Scholar] [CrossRef]
- Hartmann, M.; Howes, C.G.; Abarenkov, K.; Mohn, W.W.; Nilsson, R.H. V-Xtractor: An Open-Source, High-Throughput Software Tool to Identify and Extract Hypervariable Regions of Small Subunit (16S/18S) Ribosomal RNA Gene Sequences. J. Microbiol. Methods 2010, 83, 250–253. [Google Scholar] [CrossRef]
- Büttner, M.; Miao, Z.; Wolf, F.A.; Teichmann, S.A.; Theis, F.J. A Test Metric for Assessing Single-Cell RNA-Seq Batch Correction. Nat. Methods 2019, 16, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Karst, S.M.; Ziels, R.M.; Kirkegaard, R.H.; Sørensen, E.A.; McDonald, D.; Zhu, Q.; Knight, R.; Albertsen, M. High-Accuracy Long-Read Amplicon Sequences Using Unique Molecular Identifiers with Nanopore or PacBio Sequencing. Nat. Methods 2021, 18, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Song, X.; Zhang, C.-Y.; Chen, G.-F.; Lao, Y.-M.; Jin, H.; Cai, Z.-H. Distribution Patterns of Microbial Community Structure Along a 7000-Mile Latitudinal Transect from the Mediterranean Sea Across the Atlantic Ocean to the Brazilian Coastal Sea. Microb. Ecol. 2018, 76, 592–609. [Google Scholar] [CrossRef]
- Vaulot, D.; Geisen, S.; Mahé, F.; Bass, D. Pr2-primers: An 18S RRNA Primer Database for Protists. Mol. Ecol. Resour. 2022, 22, 168–179. [Google Scholar] [CrossRef]
- Brown, M.S.; Bowman, J.S.; Lin, Y.; Feehan, C.J.; Moreno, C.M.; Cassar, N.; Marchetti, A.; Schofield, O.M. Low Diversity of a Key Phytoplankton Group along the West Antarctic Peninsula. Limnol. Oceanogr. 2021, 66, 2470–2480. [Google Scholar] [CrossRef]
- Annenkova, N.V.; Giner, C.R.; Logares, R. Tracing the Origin of Planktonic Protists in an Ancient Lake. Microorganisms 2020, 8, 543. [Google Scholar] [CrossRef]
- Enberg, S.; Majaneva, M.; Autio, R.; Blomster, J.; Rintala, J. Phases of Microalgal Succession in Sea Ice and the Water Column in the Baltic Sea from Autumn to Spring. Mar. Ecol. Prog. Ser. 2018, 599, 19–34. [Google Scholar] [CrossRef]
- Fiore-Donno, A.M.; Rixen, C.; Rippin, M.; Glaser, K.; Samolov, E.; Karsten, U.; Becker, B.; Bonkowski, M. New Barcoded Primers for Efficient Retrieval of Cercozoan Sequences in High-Throughput Environmental Diversity Surveys, with Emphasis on Worldwide Biological Soil Crusts. Mol. Ecol. Resour. 2018, 18, 229–239. [Google Scholar] [CrossRef]
- Fadeev, E.; Salter, I.; Schourup-Kristensen, V.; Nöthig, E.-M.; Metfies, K.; Engel, A.; Piontek, J.; Boetius, A.; Bienhold, C. Microbial Communities in the East and West Fram Strait During Sea Ice Melting Season. Front. Mar. Sci. 2018, 5, 429. [Google Scholar] [CrossRef]
- Belevich, T.A.; Ilyash, L.V.; Milyutina, I.A.; Logacheva, M.D.; Goryunov, D.V.; Troitsky, A.V. Photosynthetic Picoeukaryotes in the Land-Fast Ice of the White Sea, Russia. Microb. Ecol. 2018, 75, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Boscaro, V.; Rossi, A.; Vannini, C.; Verni, F.; Fokin, S.I.; Petroni, G. Strengths and Biases of High-Throughput Sequencing Data in the Characterization of Freshwater Ciliate Microbiomes. Microb. Ecol. 2017, 73, 865–875. [Google Scholar] [CrossRef]
- Bradley, I.M.; Pinto, A.J.; Guest, J.S. Design and Evaluation of Illumina MiSeq-Compatible, 18S RRNA Gene-Specific Primers for Improved Characterization of Mixed Phototrophic Communities. Appl. Environ. Microbiol. 2016, 82, 5878–5891. [Google Scholar] [CrossRef]
- Kwong, W.K.; del Campo, J.; Mathur, V.; Vermeij, M.J.A.; Keeling, P.J. A Widespread Coral-Infecting Apicomplexan with Chlorophyll Biosynthesis Genes. Nature 2019, 568, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Geisen, S.; Snoek, L.B.; ten Hooven, F.C.; Duyts, H.; Kostenko, O.; Bloem, J.; Martens, H.; Quist, C.W.; Helder, J.A.; der Putten, W.H. Integrating Quantitative Morphological and Qualitative Molecular Methods to Analyse Soil Nematode Community Responses to Plant Range Expansion. Methods Ecol. Evol. 2018, 9, 1366–1378. [Google Scholar] [CrossRef]
- Venter, P.C.; Nitsche, F.; Domonell, A.; Heger, P.; Arndt, H. The Protistan Microbiome of Grassland Soil: Diversity in the Mesoscale. Protist 2017, 168, 546–564. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-Accuracy Alignment Ensembles Enable Unbiased Assessments of Sequence Homology and Phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. Nhmmer: DNA Homology Search with Profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [PubMed]
- Pages, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: String objects representing biological sequences, and matching algorithms. R Package Version 2016, 2, 10–18129. [Google Scholar]
- Schloss, P.D. Amplicon Sequence Variants Artificially Split Bacterial Genomes into Separate Clusters. mSphere 2021, 6, e00191-21. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved Error-Correction for Illumina 16S and ITS Amplicon Sequencing. bioRxiv 2016, 15, 081257. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Pitz, K.J.; Guo, J.; Johnson, S.B.; Campbell, T.L.; Zhang, H.; Vrijenhoek, R.C.; Chavez, F.P.; Geller, J. Zooplankton Biogeographic Boundaries in the California Current System as Determined from Metabarcoding. PLoS ONE 2020, 15, e0235159. [Google Scholar] [CrossRef] [PubMed]
- Harder, C.B.; Rønn, R.; Brejnrod, A.; Bass, D.; Al-Soud, W.A.; Ekelund, F. Local Diversity of Heathland Cercozoa Explored by In-Depth Sequencing. ISME J. 2016, 10, 2488–2497. [Google Scholar] [CrossRef]
- Balzano, S.; Abs, E.; Leterme, S. Protist Diversity along a Salinity Gradient in a Coastal Lagoon. Aquat. Microb. Ecol. 2015, 74, 263–277. [Google Scholar] [CrossRef]
- Xu, Z.; Li, Y.; Lu, Y.; Li, Y.; Yuan, Z.; Dai, M.; Liu, H. Impacts of the Zhe-Min Coastal Current on the Biogeographic Pattern of Microbial Eukaryotic Communities. Prog. Oceanogr. 2020, 183, 102309. [Google Scholar] [CrossRef]


| Group ID | Forward Primer | Start Position | Reverse Primer | End Position | Sample Size | Source (ID) | Reference |
|---|---|---|---|---|---|---|---|
| Same1 | CCAGCASCYGCGGTAATTCC | 564 | ACTTTCGTTCTTGAT | 980 | 120 | EBI-ENA (PRJNA508517) | [19] |
| Same2 | CCAGCASCYGCGGTAAT | 564 | ACTTTCGTTCTTGATYRA | 980 | 23 | NCBI-SRA (PRJEB24415) | [20] |
| Short1 | CYGCGGTAATTCCAGCTC | 571 | TCYDAGAATTYCACCTCT | 914 | 73 | NCBI-SRA (PRJEB21047) | [21] |
| Short2 | TTAAAAAGCTCGTAGTTG | 616 | AAGAAGACATCCTTGGTG | 963 | 27 | NCBI-SRA (SRR5189947) | [22] |
| Near1 | GCGGTAATTCCAGCTCCAA | 573 | ACTTTCGTTCTTGATYRR | 980 | 33 | EBI-ENA (PRJEB26288) | [23] |
| Near2 | CCAGCASCCGCGGTAATWCC | 564 | AKCCCCYAACTTTCGTTCTTGAT | 988 | 17 | NCBI-SRA (PRJNA368621) | [24] |
| Near3 | CCAGCASCCGCGGTAATWCC | 564 | TCTGRTYGTCTTTGATCCCYTA | 1002 | 12 | EBI-ENA (PRJEB12534) | [25] |
| Long1 | CGGTAAYTCCAGCTCYAV | 574 | CCGTCAATTHCTTYAART | 1149 | 62 | NCBI-SRA (SRP071862) | [26] |
| Long2 | GTGCCAGCAGCCGCG | 561 | TTTAAGTTTCAGCCTTGCG | 1138 | 44 | NCBI-SRA (PRJNA482746) | [27] |
| Long3 | GGCAAGTCTGGTGCCAG | 551 | TCCGTCAATTYCTTTAAGT | 1149 | 36 | EBI-ENA (PRJEB24755) | [28] |
| Long4 | CGGTAATTCCAGCTCCAATAGC | 574 | CACCAACTAAGAACGGCCATGC | 1293 | 150 | NCBI-SRA (SRP101780) | [29] |
| Silva | PR2 | |
|---|---|---|
| Version | 18S v123 | v4.14.0 |
| Total reads | 138,553 | 197,602 |
| Full length (>1600 bp) | 112,110 (80.91%) | 94,283 (47.71%) |
| Partical (<1000 bp) | 293 (0.21%) | 48,002 (24.29%) |
| v4 by VnFinder | 136,228 (98.32%) | 174,646 (88.38%) |
| v4 by V-Xtractor | 132,770 (95.83%) | 151,393 (76.62%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Zhao, F.; Xu, K. Information Scale Correction for Varying Length Amplicons Improves Eukaryotic Microbiome Data Integration. Microorganisms 2023, 11, 949. https://doi.org/10.3390/microorganisms11040949
Zhou T, Zhao F, Xu K. Information Scale Correction for Varying Length Amplicons Improves Eukaryotic Microbiome Data Integration. Microorganisms. 2023; 11(4):949. https://doi.org/10.3390/microorganisms11040949
Chicago/Turabian StyleZhou, Tong, Feng Zhao, and Kuidong Xu. 2023. "Information Scale Correction for Varying Length Amplicons Improves Eukaryotic Microbiome Data Integration" Microorganisms 11, no. 4: 949. https://doi.org/10.3390/microorganisms11040949
APA StyleZhou, T., Zhao, F., & Xu, K. (2023). Information Scale Correction for Varying Length Amplicons Improves Eukaryotic Microbiome Data Integration. Microorganisms, 11(4), 949. https://doi.org/10.3390/microorganisms11040949





