Dendritic Cells and Cryptosporidium: From Recognition to Restriction
Abstract
1. Introduction
2. Dendritic Cells
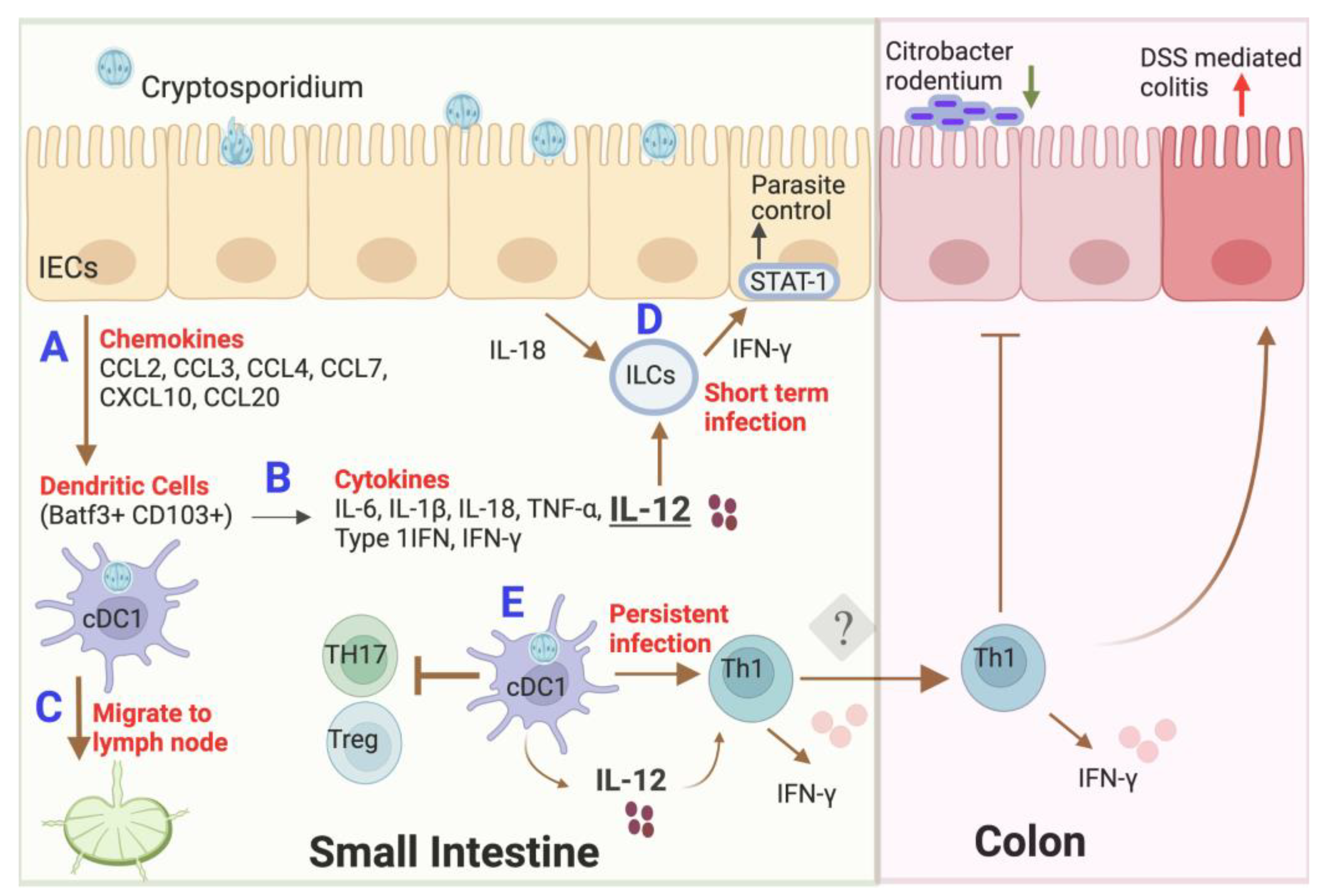
3. Interaction of Dendritic Cells with Cryptosporidium
4. Outcome of Dendritic Cell Interactions with Cryptosporidium
5. Effect of Microbial Dysbiosis on Dendritic Cells during Infection
6. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Chen, X.M.; Keithly, J.S.; Paya, C.V.; LaRusso, N.F. Cryptosporidiosis. N. Engl. J. Med. 2002, 346, 1723–1731. [Google Scholar] [CrossRef] [PubMed]
- Khalil, I.A.; Troeger, C.; Rao, P.C.; Blacker, B.F.; Brown, A.; Brewer, T.G.; Colombara, D.V.; De Hostos, E.L.; Engmann, C.; Guerrant, R.L.; et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: A meta-analyses study. Lancet Glob Health 2018, 6, e758–e768. [Google Scholar] [CrossRef]
- Levine, M.M.; Nasrin, D.; Acácio, S.; Bassat, Q.; Powell, H.; Tennant, S.M.; Sow, S.O.; Sur, D.; Zaidi, A.K.M.; Faruque, A.S.G.; et al. Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: Analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Glob Health 2020, 8, e204–e214. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Nasrin, D.; Blackwelder, W.C.; Wu, Y.; Farag, T.; Panchalingham, S.; Sow, S.O.; Sur, D.; Zaidi, A.K.M.; Faruque, A.S.G.; et al. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: A 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS). Lancet Glob Health 2019, 7, e568–e584. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Feng, Y.; Xiao, L. An Update on Zoonotic Cryptosporidium Species and Genotypes in Humans. Animals 2021, 11, 3307. [Google Scholar] [CrossRef]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 2010, 124, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Tzipori, S.; Griffiths, J.K. Natural history and biology of Cryptosporidium parvum. Adv. Parasitol. 1998, 40, 5–36. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Banerjee, S.; Steffen, M.; O’Connor, R.M.; Ward, H.D.; Robbins, P.W.; Samuelson, J. Evidence for mucin-like glycoproteins that tether sporozoites of Cryptosporidium parvum to the inner surface of the oocyst wall. Eukaryot. Cell 2010, 9, 84–96. [Google Scholar] [CrossRef]
- Martínez-Ocaña, J.; Maravilla, P.; Olivo-Díaz, A. Interaction between human mucins and parasite glycoproteins: The role of lectins and glycosidases in colonization by intestinal protozoa. Rev. Inst. Med. Trop. Sao Paulo 2020, 62, e64. [Google Scholar] [CrossRef] [PubMed]
- Bouzid, M.; Hunter, P.R.; Chalmers, R.M.; Tyler, K.M. Cryptosporidium pathogenicity and virulence. Clin. Microbiol. Rev. 2013, 26, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.R.; Petry, F. Cryptosporidium parvum: Structural components of the oocyst wall. J. Parasitol. 1999, 85, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Guérin, A.; Striepen, B. The Biology of the Intestinal Intracellular Parasite Cryptosporidium. Cell Host Microbe 2020, 28, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Laurent, F.; McCole, D.; Eckmann, L.; Kagnoff, M.F. Pathogenesis of Cryptosporidium parvum infection. Microbes Infect. 1999, 1, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; LaRusso, N.F. Cryptosporidiosis and the pathogenesis of AIDS-cholangiopathy. Semin. Liver Dis. 2002, 22, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Argenzio, R.A.; Liacos, J.A.; Levy, M.L.; Meuten, D.J.; Lecce, J.G.; Powell, D.W. Villous atrophy, crypt hyperplasia, cellular infiltration, and impaired glucose-Na absorption in enteric cryptosporidiosis of pigs. Gastroenterology 1990, 98, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Farthing, M.J. Clinical aspects of human cryptosporidiosis. Contrib. Microbiol. 2000, 6, 50–74. [Google Scholar] [CrossRef]
- Borad, A.; Ward, H. Human immune responses in cryptosporidiosis. Future Microbiol. 2010, 5, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Laurent, F.; Lacroix-Lamandé, S. Innate immune responses play a key role in controlling infection of the intestinal epithelium by Cryptosporidium. Int. J. Parasitol. 2017, 47, 711–721. [Google Scholar] [CrossRef]
- McDonald, V.; Korbel, D.S.; Barakat, F.M.; Choudhry, N.; Petry, F. Innate immune responses against Cryptosporidium parvum infection. Parasite Immunol. 2013, 35, 55–64. [Google Scholar] [CrossRef]
- Crawford, C.K.; Kol, A. The Mucosal Innate Immune Response to Cryptosporidium parvum, a Global One Health Issue. Front. Cell Infect. Microbiol. 2021, 11, 689401. [Google Scholar] [CrossRef] [PubMed]
- Gibson, A.R.; Sateriale, A.; Dumaine, J.E.; Engiles, J.B.; Pardy, R.D.; Gullicksrud, J.A.; O’Dea, K.M.; Doench, J.G.; Beiting, D.P.; Hunter, C.A.; et al. A genetic screen identifies a protective type III interferon response to Cryptosporidium that requires TLR3 dependent recognition. PLoS Pathog. 2022, 18, e1010003. [Google Scholar] [CrossRef] [PubMed]
- Auray, G.; Lacroix-Lamandé, S.; Mancassola, R.; Dimier-Poisson, I.; Laurent, F. Involvement of intestinal epithelial cells in dendritic cell recruitment during C. parvum infection. Microbes Infect. 2007, 9, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, B.D.; Noel, F.; Rouzier, P.D.; Powell, J.L.; Pape, J.W.; Bois, G.; Alston, W.K.; Larsson, C.J.; Tenney, K.; Ventrone, C.; et al. Childhood cryptosporidiosis is associated with a persistent systemic inflammatory response. Clin. Infect. Dis. 2006, 43, 604–608. [Google Scholar] [CrossRef]
- Sasahara, T.; Maruyama, H.; Aoki, M.; Kikuno, R.; Sekiguchi, T.; Takahashi, A.; Satoh, Y.; Kitasato, H.; Takayama, Y.; Inoue, M. Apoptosis of intestinal crypt epithelium after Cryptosporidium parvum infection. J. Infect. Chemother 2003, 9, 278–281. [Google Scholar] [CrossRef]
- McCole, D.F.; Eckmann, L.; Laurent, F.; Kagnoff, M.F. Intestinal epithelial cell apoptosis following Cryptosporidium parvum infection. Infect. Immun. 2000, 68, 1710–1713. [Google Scholar] [CrossRef]
- Laurent, F.; Kagnoff, M.F.; Savidge, T.C.; Naciri, M.; Eckmann, L. Human intestinal epithelial cells respond to Cryptosporidium parvum infection with increased prostaglandin H synthase 2 expression and prostaglandin E2 and F2alpha production. Infect. Immun. 1998, 66, 1787–1790. [Google Scholar] [CrossRef]
- Gookin, J.L.; Duckett, L.L.; Armstrong, M.U.; Stauffer, S.H.; Finnegan, C.P.; Murtaugh, M.P.; Argenzio, R.A. Nitric oxide synthase stimulates prostaglandin synthesis and barrier function in C. parvum-infected porcine ileum. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G571–G581. [Google Scholar] [CrossRef]
- Zaalouk, T.K.; Bajaj-Elliott, M.; George, J.T.; McDonald, V. Differential regulation of beta-defensin gene expression during Cryptosporidium parvum infection. Infect. Immun. 2004, 72, 2772–2779. [Google Scholar] [CrossRef]
- Tarver, A.P.; Clark, D.P.; Diamond, G.; Russell, J.P.; Erdjument-Bromage, H.; Tempst, P.; Cohen, K.S.; Jones, D.E.; Sweeney, R.W.; Wines, M.; et al. Enteric beta-defensin: Molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosporidium parvum infection. Infect. Immun. 1998, 66, 1045–1056. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; O’Hara, S.P.; Nelson, J.B.; Splinter, P.L.; Small, A.J.; Tietz, P.S.; Limper, A.H.; LaRusso, N.F. Multiple TLRs are expressed in human cholangiocytes and mediate host epithelial defense responses to Cryptosporidium parvum via activation of NF-kappaB. J. Immunol. 2005, 175, 7447–7456. [Google Scholar] [CrossRef]
- Castellanos-Gonzalez, A.; Yancey, L.S.; Wang, H.C.; Pantenburg, B.; Liscum, K.R.; Lewis, D.E.; White, A.C., Jr. Cryptosporidium infection of human intestinal epithelial cells increases expression of osteoprotegerin: A novel mechanism for evasion of host defenses. J. Infect. Dis. 2008, 197, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Mead, J.R.; You, X. Susceptibility differences to Cryptosporidium parvum infection in two strains of gamma interferon knockout mice. J. Parasitol. 1998, 84, 1045–1048. [Google Scholar] [CrossRef] [PubMed]
- Lacroix-Lamandé, S.; Mancassola, R.; Naciri, M.; Laurent, F. Role of gamma interferon in chemokine expression in the ileum of mice and in a murine intestinal epithelial cell line after Cryptosporidium parvum infection. Infect. Immun. 2002, 70, 2090–2099. [Google Scholar] [CrossRef]
- Barakat, F.M.; McDonald, V.; Foster, G.R.; Tovey, M.G.; Korbel, D.S. Cryptosporidium parvum infection rapidly induces a protective innate immune response involving type I interferon. J. Infect. Dis. 2009, 200, 1548–1555. [Google Scholar] [CrossRef]
- Pollok, R.C.; Farthing, M.J.; Bajaj-Elliott, M.; Sanderson, I.R.; McDonald, V. Interferon gamma induces enterocyte resistance against infection by the intracellular pathogen Cryptosporidium parvum. Gastroenterology 2001, 120, 99–107. [Google Scholar] [CrossRef]
- Gullicksrud, J.A.; Sateriale, A.; Engiles, J.B.; Gibson, A.R.; Shaw, S.; Hutchins, Z.A.; Martin, L.; Christian, D.A.; Taylor, G.A.; Yamamoto, M.; et al. Enterocyte-innate lymphoid cell crosstalk drives early IFN-γ-mediated control of Cryptosporidium. Mucosal. Immunol. 2022, 15, 362–372. [Google Scholar] [CrossRef]
- Gomez Morales, M.A.; La Rosa, G.; Ludovisi, A.; Onori, A.M.; Pozio, E. Cytokine profile induced by Cryptosporidium antigen in peripheral blood mononuclear cells from immunocompetent and immunosuppressed persons with cryptosporidiosis. J. Infect. Dis. 1999, 179, 967–973. [Google Scholar] [CrossRef] [PubMed][Green Version]
- White, A.C.; Robinson, P.; Okhuysen, P.C.; Lewis, D.E.; Shahab, I.; Lahoti, S.; DuPont, H.L.; Chappell, C.L. Interferon-gamma expression in jejunal biopsies in experimental human cryptosporidiosis correlates with prior sensitization and control of oocyst excretion. J. Infect. Dis. 2000, 181, 701–709. [Google Scholar] [CrossRef]
- Bonilla, F.A.; Oettgen, H.C. Adaptive immunity. J. Allergy Clin. Immunol. 2010, 125 (Suppl. S2), S33–S40. [Google Scholar] [CrossRef]
- O’Connor, R.M.; Shaffie, R.; Kang, G.; Ward, H.D. Cryptosporidiosis in patients with HIV/AIDS. Aids 2011, 25, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Harp, J.A.; Harmsen, A.G. Requirements for CD4+ cells and gamma interferon in resolution of established Cryptosporidium parvum infection in mice. Infect. Immun. 1993, 61, 3928–3932. [Google Scholar] [CrossRef]
- Sateriale, A.; Šlapeta, J.; Baptista, R.; Engiles, J.B.; Gullicksrud, J.A.; Herbert, G.T.; Brooks, C.F.; Kugler, E.M.; Kissinger, J.C.; Hunter, C.A.; et al. A Genetically Tractable, Natural Mouse Model of Cryptosporidiosis Offers Insights into Host Protective Immunity. Cell Host Microbe 2019, 26, 135–146.e135. [Google Scholar] [CrossRef] [PubMed]
- Pantenburg, B.; Castellanos-Gonzalez, A.; Dann, S.M.; Connelly, R.L.; Lewis, D.E.; Ward, H.D.; White, A.C., Jr. Human CD8(+) T cells clear Cryptosporidium parvum from infected intestinal epithelial cells. Am. J. Trop. Med. Hyg. 2010, 82, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Cohn, I.S.; Henrickson, S.E.; Striepen, B.; Hunter, C.A. Immunity to Cryptosporidium: Lessons from Acquired and Primary Immunodeficiencies. J. Immunol. 2022, 209, 2261–2268.e2547. [Google Scholar] [CrossRef]
- Russler-Germain, E.V.; Jung, J.; Miller, A.T.; Young, S.; Yi, J.; Wehmeier, A.; Fox, L.E.; Monte, K.J.; Chai, J.N.; Kulkarni, D.H.; et al. Commensal Cryptosporidium colonization elicits a cDC1-dependent Th1 response that promotes intestinal homeostasis and limits other infections. Immunity 2021, 54, 2547–2564.e2547. [Google Scholar] [CrossRef]
- History of Dendritic Cells. Dendritic Cells in Clinics; Springer: Tokyo, Japan, 2008; pp. 1–4. [Google Scholar] [CrossRef]
- Steinman, R.M.; Cohn, Z.A. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J. Exp. Med. 1973, 137, 1142–1162. [Google Scholar] [CrossRef]
- Ni, K.; O’Neill, H.C. The role of dendritic cells in T cell activation. Immunol. Cell Biol. 1997, 75, 223–230. [Google Scholar] [CrossRef]
- Steinman, R.M.; Idoyaga, J. Features of the dendritic cell lineage. Immunol. Rev. 2010, 234, 5–17. [Google Scholar] [CrossRef]
- Heath, W.R.; Carbone, F.R. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat. Immunol. 2009, 10, 1237–1244. [Google Scholar] [CrossRef]
- Mildner, A.; Jung, S. Development and function of dendritic cell subsets. Immunity 2014, 40, 642–656. [Google Scholar] [CrossRef]
- Domínguez, P.M.; Ardavín, C. Differentiation and function of mouse monocyte-derived dendritic cells in steady state and inflammation. Immunol. Rev. 2010, 234, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Villani, A.C.; Satija, R.; Reynolds, G.; Sarkizova, S.; Shekhar, K.; Fletcher, J.; Griesbeck, M.; Butler, A.; Zheng, S.; Lazo, S.; et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017, 356, eaah4573. [Google Scholar] [CrossRef]
- Liu, K.; Victora, G.D.; Schwickert, T.A.; Guermonprez, P.; Meredith, M.M.; Yao, K.; Chu, F.F.; Randolph, G.J.; Rudensky, A.Y.; Nussenzweig, M. In vivo analysis of dendritic cell development and homeostasis. Science 2009, 324, 392–397. [Google Scholar] [CrossRef]
- Hildner, K.; Edelson, B.T.; Purtha, W.E.; Diamond, M.; Matsushita, H.; Kohyama, M.; Calderon, B.; Schraml, B.U.; Unanue, E.R.; Diamond, M.S.; et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 2008, 322, 1097–1100. [Google Scholar] [CrossRef]
- Cisse, B.; Caton, M.L.; Lehner, M.; Maeda, T.; Scheu, S.; Locksley, R.; Holmberg, D.; Zweier, C.; den Hollander, N.S.; Kant, S.G.; et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 2008, 135, 37–48. [Google Scholar] [CrossRef]
- Reis e Sousa, C. Dendritic cells in a mature age. Nat. Rev. Immunol. 2006, 6, 476–483. [Google Scholar] [CrossRef]
- Steinman, R.M.; Hemmi, H. Dendritic cells: Translating innate to adaptive immunity. Curr. Top. Microbiol. Immunol. 2006, 311, 17–58. [Google Scholar] [CrossRef]
- Campbell, L.D.; Stewart, J.N.; Mead, J.R. Susceptibility to Cryptosporidium parvum infections in cytokine- and chemokine-receptor knockout mice. J. Parasitol. 2002, 88, 1014–1016. [Google Scholar] [CrossRef]
- Lantier, L.; Lacroix-Lamandé, S.; Potiron, L.; Metton, C.; Drouet, F.; Guesdon, W.; Gnahoui-David, A.; Le Vern, Y.; Deriaud, E.; Fenis, A.; et al. Intestinal CD103+ dendritic cells are key players in the innate immune control of Cryptosporidium parvum infection in neonatal mice. PLoS Pathog. 2013, 9, e1003801. [Google Scholar] [CrossRef]
- Wang, H.C.; Dann, S.M.; Okhuysen, P.C.; Lewis, D.E.; Chappell, C.L.; Adler, D.G.; White, A.C., Jr. High levels of CXCL10 are produced by intestinal epithelial cells in AIDS patients with active cryptosporidiosis but not after reconstitution of immunity. Infect. Immun. 2007, 75, 481–487. [Google Scholar] [CrossRef]
- Guesdon, W.; Auray, G.; Pezier, T.; Bussière, F.I.; Drouet, F.; Le Vern, Y.; Marquis, M.; Potiron, L.; Rabot, S.; Bruneau, A.; et al. CCL20 Displays Antimicrobial Activity Against Cryptosporidium parvum, but Its Expression Is Reduced During Infection in the Intestine of Neonatal Mice. J. Infect. Dis. 2015, 212, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Perez-Cordon, G.; Yang, G.; Zhou, B.; Nie, W.; Li, S.; Shi, L.; Tzipori, S.; Feng, H. Interaction of Cryptosporidium parvum with mouse dendritic cells leads to their activation and parasite transportation to mesenteric lymph nodes. Pathog. Dis. 2014, 70, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.A.; Rogers, A.B.; Leav, B.A.; Sanchez, A.; Vannier, E.; Uematsu, S.; Akira, S.; Golenbock, D.; Ward, H.D. MyD88-dependent pathways mediate resistance to Cryptosporidium parvum infection in mice. Infect. Immun. 2006, 74, 549–556. [Google Scholar] [CrossRef]
- Xu, Q.M.; Fang, F.; Wu, S.H.; Shi, Z.Q.; Liu, Z.; Zhoa, Y.J.; Zheng, H.W.; Lu, G.X.; Kong, H.R.; Wang, G.J.; et al. Dendritic cell TLR4 induces Th1-type immune response against Cryptosporidium parvum infection. Trop. Biomed 2021, 38, 172–179. [Google Scholar] [CrossRef]
- O’Hara, S.P.; Bogert, P.S.; Trussoni, C.E.; Chen, X.; LaRusso, N.F. TLR4 promotes Cryptosporidium parvum clearance in a mouse model of biliary cryptosporidiosis. J. Parasitol. 2011, 97, 813–821. [Google Scholar] [CrossRef]
- Park, B.S.; Lee, J.O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 2013, 45, e66. [Google Scholar] [CrossRef]
- Debierre-Grockiego, F.; Campos, M.A.; Azzouz, N.; Schmidt, J.; Bieker, U.; Resende, M.G.; Mansur, D.S.; Weingart, R.; Schmidt, R.R.; Golenbock, D.T.; et al. Activation of TLR2 and TLR4 by glycosylphosphatidylinositols derived from Toxoplasma gondii. J. Immunol. 2007, 179, 1129–1137. [Google Scholar] [CrossRef]
- Dunst, J.; Azzouz, N.; Liu, X.; Tsukita, S.; Seeberger, P.H.; Kamena, F. Interaction between Plasmodium Glycosylphosphatidylinositol and the Host Protein Moesin Has No Implication in Malaria Pathology. Front. Cell. Infect. Microbiol. 2017, 7, 183. [Google Scholar] [CrossRef] [PubMed]
- de Sablet, T.; Potiron, L.; Marquis, M.; Bussière, F.I.; Lacroix-Lamandé, S.; Laurent, F. Cryptosporidium parvum increases intestinal permeability through interaction with epithelial cells and IL-1β and TNFα released by inflammatory monocytes. Cell. Microbiol. 2016, 18, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Bedi, B.; McNair, N.N.; Mead, J.R. Dendritic cells play a role in host susceptibility to Cryptosporidium parvum infection. Immunol. Lett. 2014, 158, 42–51. [Google Scholar] [CrossRef]
- Watowich, S.S.; Liu, Y.J. Mechanisms regulating dendritic cell specification and development. Immunol. Rev. 2010, 238, 76–92. [Google Scholar] [CrossRef]
- Potiron, L.; Lacroix-Lamandé, S.; Marquis, M.; Levern, Y.; Fort, G.; Franceschini, I.; Laurent, F. Batf3-Dependent Intestinal Dendritic Cells Play a Critical Role in the Control of Cryptosporidium parvum Infection. J. Infect. Dis. 2019, 219, 925–935. [Google Scholar] [CrossRef]
- Worbs, T.; Hammerschmidt, S.I.; Förster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2017, 17, 30–48. [Google Scholar] [CrossRef] [PubMed]
- David, R. Dendritic cells: The true face of migratory DCs. Nat. Rev. Immunol. 2014, 14, 649. [Google Scholar] [CrossRef] [PubMed]
- Marcial, M.A.; Madara, J.L. Cryptosporidium: Cellular localization, structural analysis of absorptive cell-parasite membrane-membrane interactions in guinea pigs, and suggestion of protozoan transport by M cells. Gastroenterology 1986, 90, 583–594. [Google Scholar] [CrossRef]
- Niess, J.H.; Brand, S.; Gu, X.; Landsman, L.; Jung, S.; McCormick, B.A.; Vyas, J.M.; Boes, M.; Ploegh, H.L.; Fox, J.G.; et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 2005, 307, 254–258. [Google Scholar] [CrossRef]
- Bedi, B.; Mead, J.R. Cryptosporidium parvum antigens induce mouse and human dendritic cells to generate Th1-enhancing cytokines. Parasite Immunol. 2012, 34, 473–485. [Google Scholar] [CrossRef]
- Lantier, L.; Drouet, F.; Guesdon, W.; Mancassola, R.; Metton, C.; Lo-Man, R.; Werts, C.; Laurent, F.; Lacroix-Lamandé, S. Poly(I:C)-induced protection of neonatal mice against intestinal Cryptosporidium parvum infection requires an additional TLR5 signal provided by the gut flora. J. Infect. Dis. 2014, 209, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Tessema, T.S.; Schwamb, B.; Lochner, M.; Förster, I.; Jakobi, V.; Petry, F. Dynamics of gut mucosal and systemic Th1/Th2 cytokine responses in interferon-gamma and interleukin-12p40 knock out mice during primary and challenge Cryptosporidium parvum infection. Immunobiology 2009, 214, 454–466. [Google Scholar] [CrossRef]
- Petry, F.; Jakobi, V.; Tessema, T.S. Host immune response to Cryptosporidium parvum infection. Exp. Parasitol. 2010, 126, 304–309. [Google Scholar] [CrossRef]
- Sateriale, A.; Gullicksrud, J.A.; Engiles, J.B.; McLeod, B.I.; Kugler, E.M.; Henao-Mejia, J.; Zhou, T.; Ring, A.M.; Brodsky, I.E.; Hunter, C.A.; et al. The intestinal parasite Cryptosporidium is controlled by an enterocyte intrinsic inflammasome that depends on NLRP6. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Saraav, I.; Cervantes-Barragan, L.; Olias, P.; Fu, Y.; Wang, Q.; Wang, L.; Wang, Y.; Mack, M.; Baldridge, M.T.; Stappenbeck, T.; et al. Chronic Toxoplasma gondii infection enhances susceptibility to colitis. Proc. Natl. Acad. Sci. USA 2021, 118, e2106730118. [Google Scholar] [CrossRef] [PubMed]
- Palm, N.W.; de Zoete, M.R.; Flavell, R.A. Immune-microbiota interactions in health and disease. Clin. Immunol. 2015, 159, 122–127. [Google Scholar] [CrossRef]
- Sherwood, D.; Angus, K.W.; Snodgrass, D.R.; Tzipori, S. Experimental cryptosporidiosis in laboratory mice. Infect. Immun. 1982, 38, 471–475. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Funkhouser-Jones, L.J.; Akey, M.E.; Schaefer, D.A.; Ackman, K.; Riggs, M.W.; Stappenbeck, T.S.; Sibley, L.D. Neonatal Mouse Gut Metabolites Influence Cryptosporidium parvum Infection in Intestinal Epithelial Cells. mBio 2020, 11, e02582-20. [Google Scholar] [CrossRef]
- Schulfer, A.; Blaser, M.J. Risks of Antibiotic Exposures Early in Life on the Developing Microbiome. PLoS Pathog. 2015, 11, e1004903. [Google Scholar] [CrossRef]
- Al Nabhani, Z.; Eberl, G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. 2020, 13, 183–189. [Google Scholar] [CrossRef]
- Widmer, G.; Carmena, D.; Kváč, M.; Chalmers, R.M.; Kissinger, J.C.; Xiao, L.; Sateriale, A.; Striepen, B.; Laurent, F.; Lacroix-Lamandé, S.; et al. Update on Cryptosporidium spp.: Highlights from the Seventh International Giardia and Cryptosporidium Conference. Parasite 2020, 27, 14. [Google Scholar] [CrossRef]
- Mammeri, M.; Chevillot, A.; Thomas, M.; Julien, C.; Auclair, E.; Pollet, T.; Polack, B.; Vallée, I.; Adjou, K.T. Cryptosporidium parvum-Infected Neonatal Mice Show Gut Microbiota Remodelling Using High-Throughput Sequencing Analysis: Preliminary Results. Acta Parasitol. 2019, 64, 268–275. [Google Scholar] [CrossRef]
- Carey, M.A.; Medlock, G.L.; Alam, M.; Kabir, M.; Uddin, M.J.; Nayak, U.; Papin, J.; Faruque, A.S.G.; Haque, R.; Petri, W.A.; et al. Megasphaera in the Stool Microbiota Is Negatively Associated With Diarrheal Cryptosporidiosis. Clin. Infect. Dis. 2021, 73, e1242–e1251. [Google Scholar] [CrossRef]
- Ichikawa-Seki, M.; Motooka, D.; Kinami, A.; Murakoshi, F.; Takahashi, Y.; Aita, J.; Hayashi, K.; Tashibu, A.; Nakamura, S.; Iida, T.; et al. Specific increase of Fusobacterium in the faecal microbiota of neonatal calves infected with Cryptosporidium parvum. Sci. Rep. 2019, 9, 12517. [Google Scholar] [CrossRef]
- Shetty, S.A.; Marathe, N.P.; Lanjekar, V.; Ranade, D.; Shouche, Y.S. Comparative genome analysis of Megasphaera sp. reveals niche specialization and its potential role in the human gut. PLoS ONE 2013, 8, e79353. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Keelaghan, A.P.; Charania, R.; Mead, J.R. The Effect of Short-Chain Fatty Acids on Growth of Cryptosporidium parvum In Vitro. Microorganisms 2022, 10, 1822. [Google Scholar] [CrossRef]
- Yokanovich, L.T.; Newberry, R.D.; Knoop, K.A. Regulation of oral antigen delivery early in life: Implications for oral tolerance and food allergy. Clin. Exp. Allergy 2021, 51, 518–526. [Google Scholar] [CrossRef]
- Zhang, X.; Borbet, T.C.; Fallegger, A.; Wipperman, M.F.; Blaser, M.J.; Müller, A. An Antibiotic-Impacted Microbiota Compromises the Development of Colonic Regulatory T Cells and Predisposes to Dysregulated Immune Responses. mBio 2021, 12, e03335-20. [Google Scholar] [CrossRef]
- Britton, G.J.; Contijoch, E.J.; Mogno, I.; Vennaro, O.H.; Llewellyn, S.R.; Ng, R.; Li, Z.; Mortha, A.; Merad, M.; Das, A.; et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 2019, 50, 212–224.e214. [Google Scholar] [CrossRef]
- Zhang, X.T.; Gong, A.Y.; Wang, Y.; Chen, X.; Lim, S.S.; Dolata, C.E.; Chen, X.M. Cryptosporidium parvum infection attenuates the ex vivo propagation of murine intestinal enteroids. Physiol. Rep. 2016, 4, e13060. [Google Scholar] [CrossRef] [PubMed]
- DeCicco RePass, M.A.; Chen, Y.; Lin, Y.; Zhou, W.; Kaplan, D.L.; Ward, H.D. Novel Bioengineered Three-Dimensional Human Intestinal Model for Long-Term Infection of Cryptosporidium parvum. Infect. Immun. 2017, 85, e00731-16. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Dutta, D.; Schaefer, D.A.; Iakobachvili, N.; Artegiani, B.; Sachs, N.; Boonekamp, K.E.; Bowden, G.; Hendrickx, A.P.A.; Willems, R.J.L.; et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018, 3, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Fajardo-Moser, M.; Berzel, S.; Moll, H. Mechanisms of dendritic cell-based vaccination against infection. Int. J. Med. Microbiol. 2008, 298, 11–20. [Google Scholar] [CrossRef]
- Zitvogel, L.; Angevin, E.; Tursz, T. Dendritic cell-based immunotherapy of cancer. Ann. Oncol. 2000, 11 (Suppl. S3), 199–205. [Google Scholar] [CrossRef]
- Yu, J.; Sun, H.; Cao, W.; Song, Y.; Jiang, Z. Research progress on dendritic cell vaccines in cancer immunotherapy. Exp. Hematol. Oncol. 2022, 11, 3. [Google Scholar] [CrossRef]
| Dendritic Cell Subset | Conventional Type-I (cDC1). | Conventional Type-II (cDC2) | Plasmacytoid DCs (pDCs) | Monocyte Derived DC (Mo-DCs) |
|---|---|---|---|---|
| Transcription factor | BATF3, ID2, IRF8, ZFBTB46 | IRF2, IRF4, RelB, ZFBTB46 | E2-2, IRF4, IRF7 | KLF4, IRF4, |
| Key markers for identification | CD11c+, MHC-II+, FLT3+, CD11b–, CD8a+, CD103+, XCR1+ | CD11c+, MHC-II+, CD24+ CD135+ | CD11clow, MHC-IIint B220+ PDCA1+ Siglec H+, Lyc6C+ | CD11b+ Lyc6Chi/lo, CD64+, CD14+ |
| Functions | Act against intracellular pathogens IL-12 production; cross-presentation to CD8 T cells (MHC Class I) | Act against extracellular pathogens IL-12, IL-23, and TNF-α production Presentation to CD4 T cells (MHC Class II) | Antiviral response-type-I and type-III IFNs against viral infection. TLR7 and TLR9 induced CD4 and CD8 T-cell response | Differentiates from monocytes during inflammation with dendritic cell morphology. TNF-α, iNOS, and ROI production; Presentation to CD4 T cells (MHC Class II) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saraav, I.; Sibley, L.D. Dendritic Cells and Cryptosporidium: From Recognition to Restriction. Microorganisms 2023, 11, 1056. https://doi.org/10.3390/microorganisms11041056
Saraav I, Sibley LD. Dendritic Cells and Cryptosporidium: From Recognition to Restriction. Microorganisms. 2023; 11(4):1056. https://doi.org/10.3390/microorganisms11041056
Chicago/Turabian StyleSaraav, Iti, and L. David Sibley. 2023. "Dendritic Cells and Cryptosporidium: From Recognition to Restriction" Microorganisms 11, no. 4: 1056. https://doi.org/10.3390/microorganisms11041056
APA StyleSaraav, I., & Sibley, L. D. (2023). Dendritic Cells and Cryptosporidium: From Recognition to Restriction. Microorganisms, 11(4), 1056. https://doi.org/10.3390/microorganisms11041056





