Novel Insights into the Mechanism Underlying High Polysaccharide Yield in Submerged Culture of Ganoderma lucidum Revealed by Transcriptome and Proteome Analyses
Abstract
1. Introduction
2. Materials and Methods
2.1. G. lucidum Strain and Culture Conditions
2.2. Sampling and Analysis of Biomass and Polysaccharides
2.3. Transcriptome and Proteome Analyses
2.4. Gene Expression Analysis by RT-qPCR
2.5. Protein Sequence Analysis of GHs
2.6. Statistical Analysis
3. Results
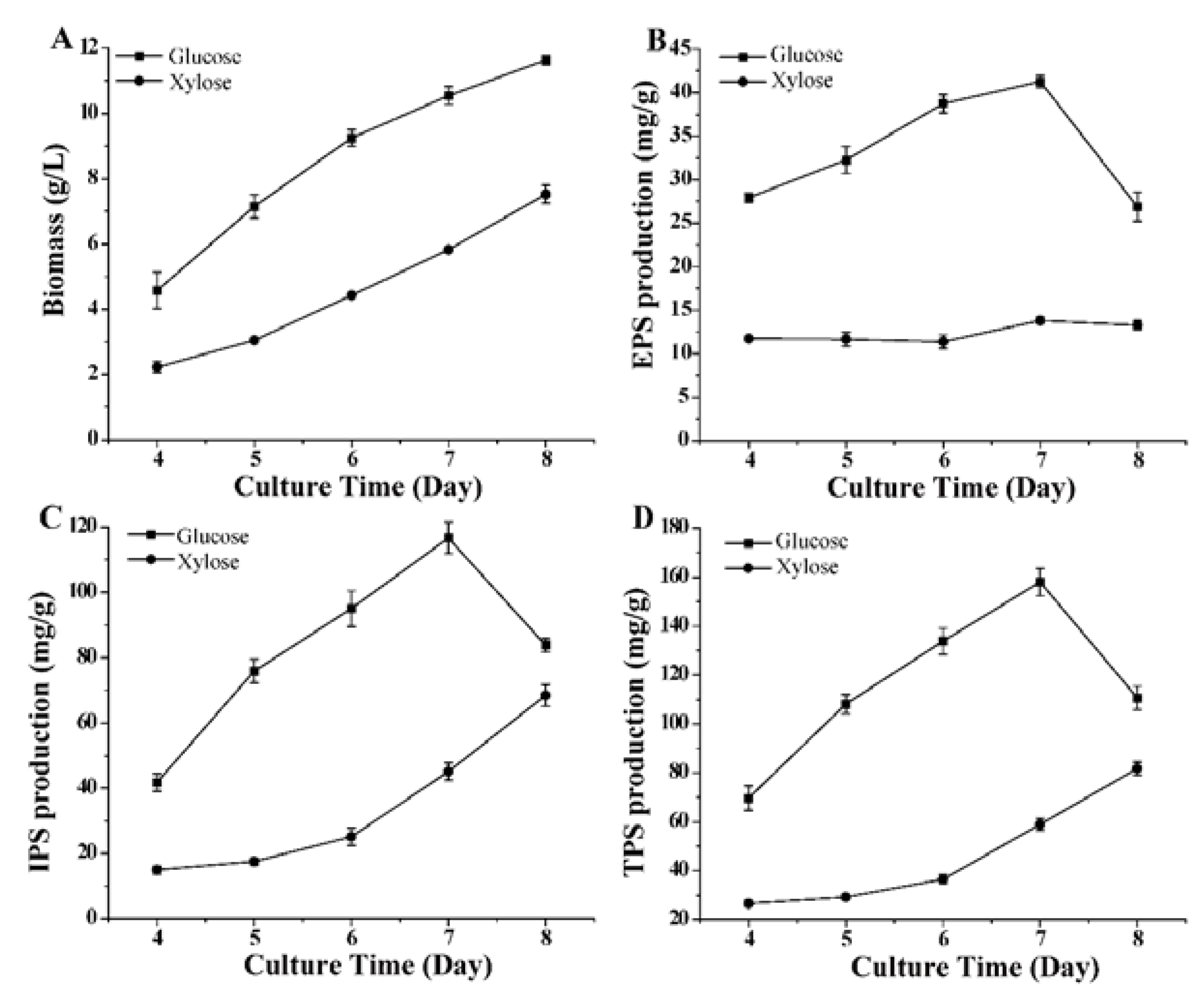
3.1. Polysaccharide Production in G. lucidum under Different Culture Conditions
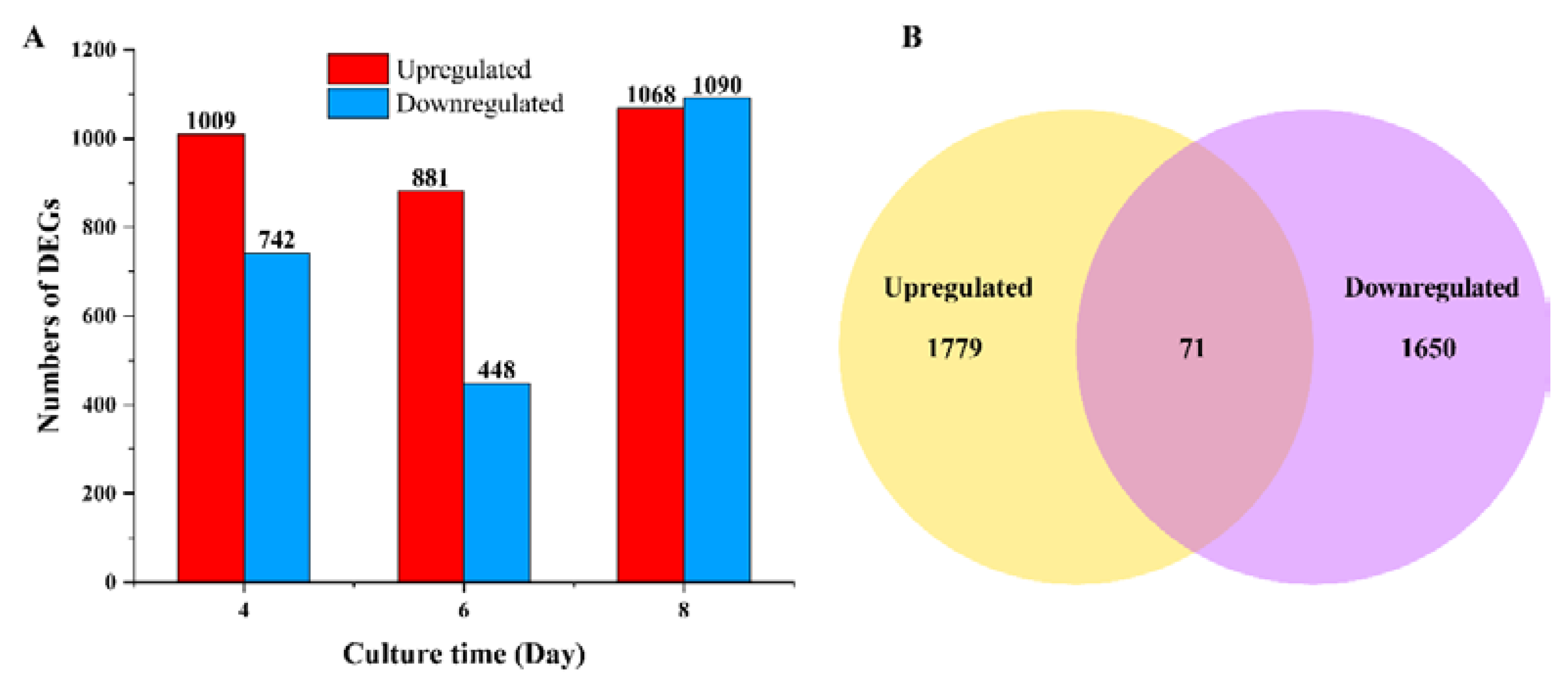
3.2. DEGs and KEGG Enrichment Analysis
3.3. Gene Expression Dynamics in Nucleotide Sugar Biosynthetic Pathway
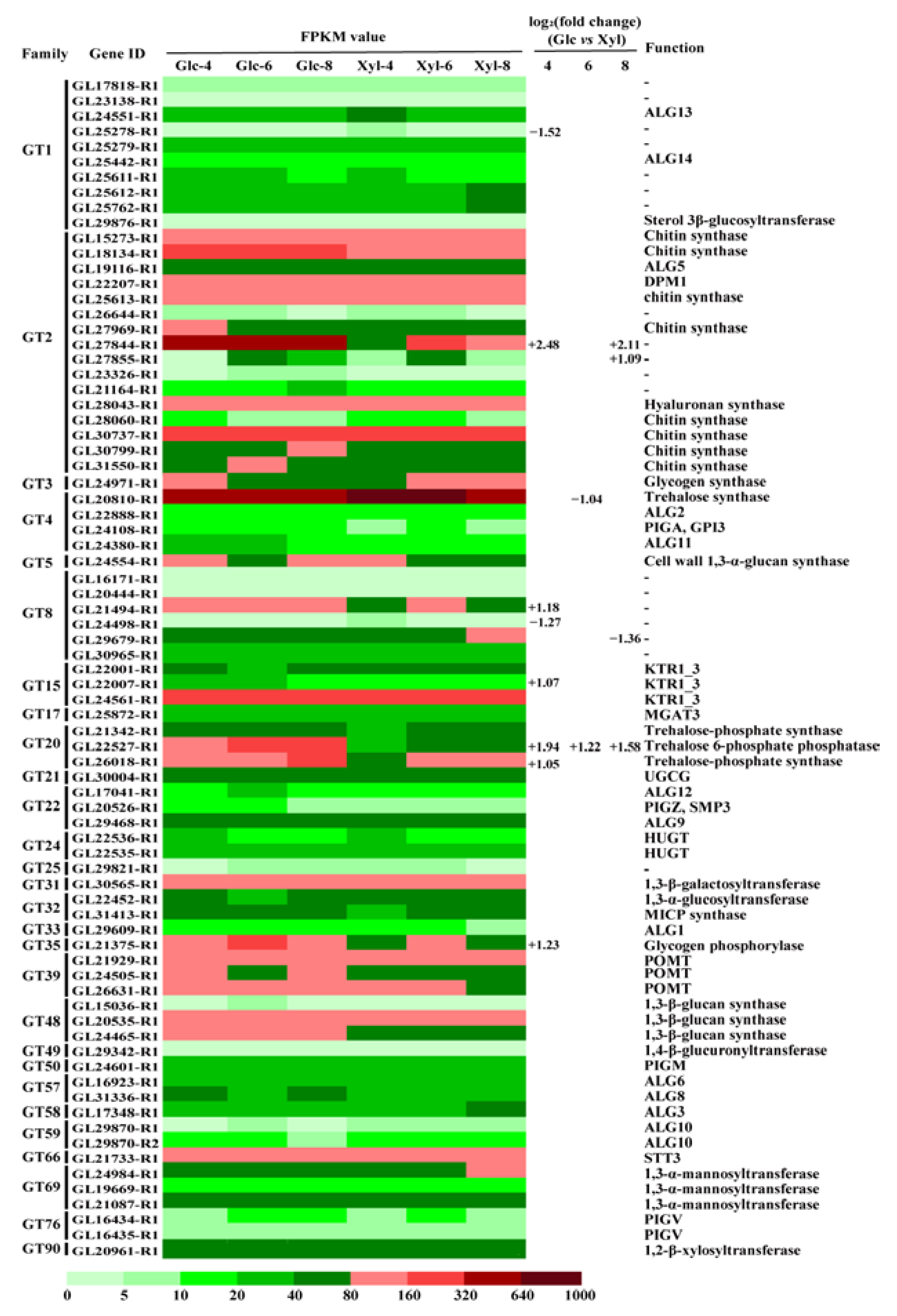
3.4. Expression Dynamics of GT Genes
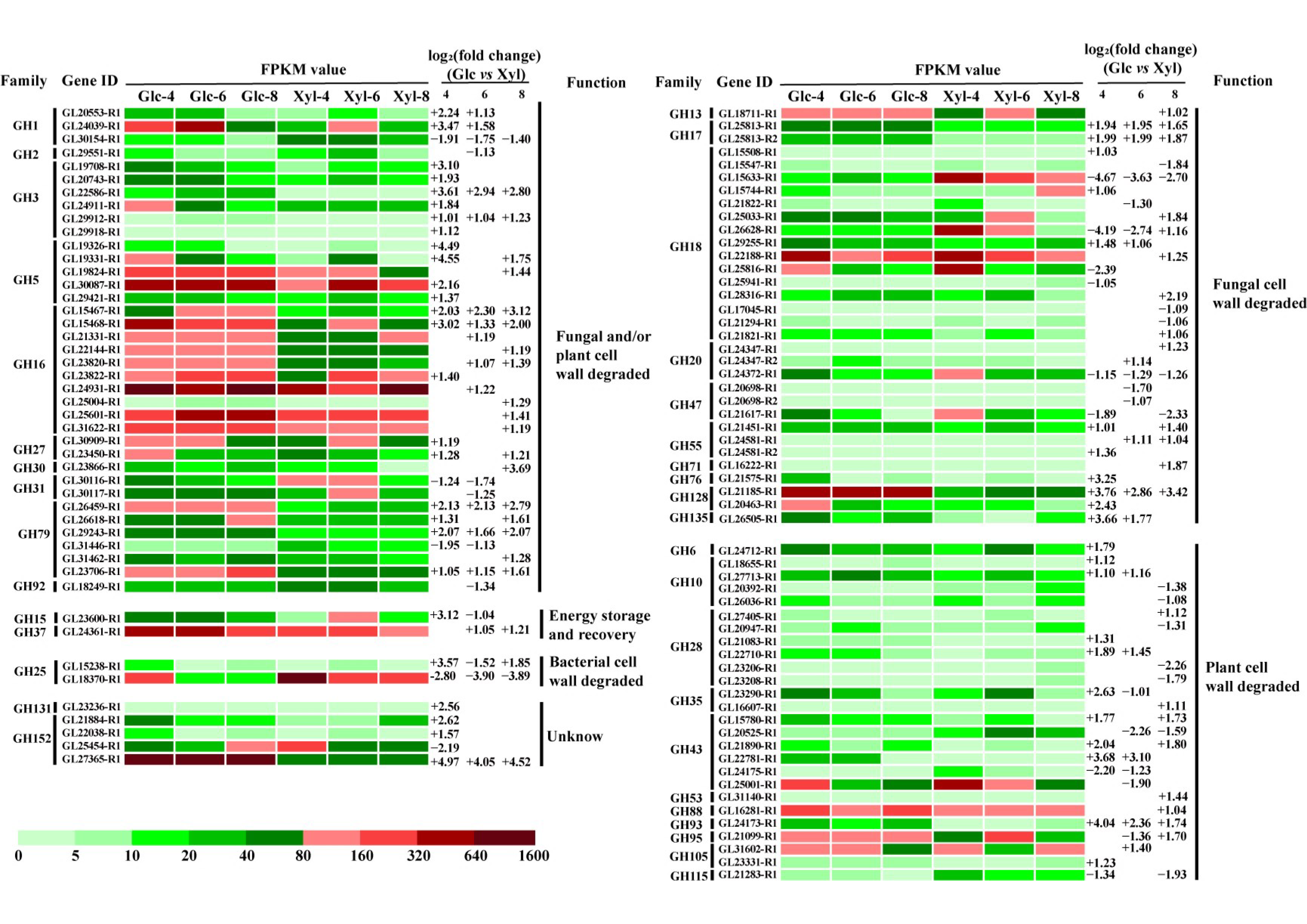
3.5. Expression Dynamics of GH DEGs
3.6. Protein Identification and Analysis
3.7. Protein Sequence Analysis of the Key GHs
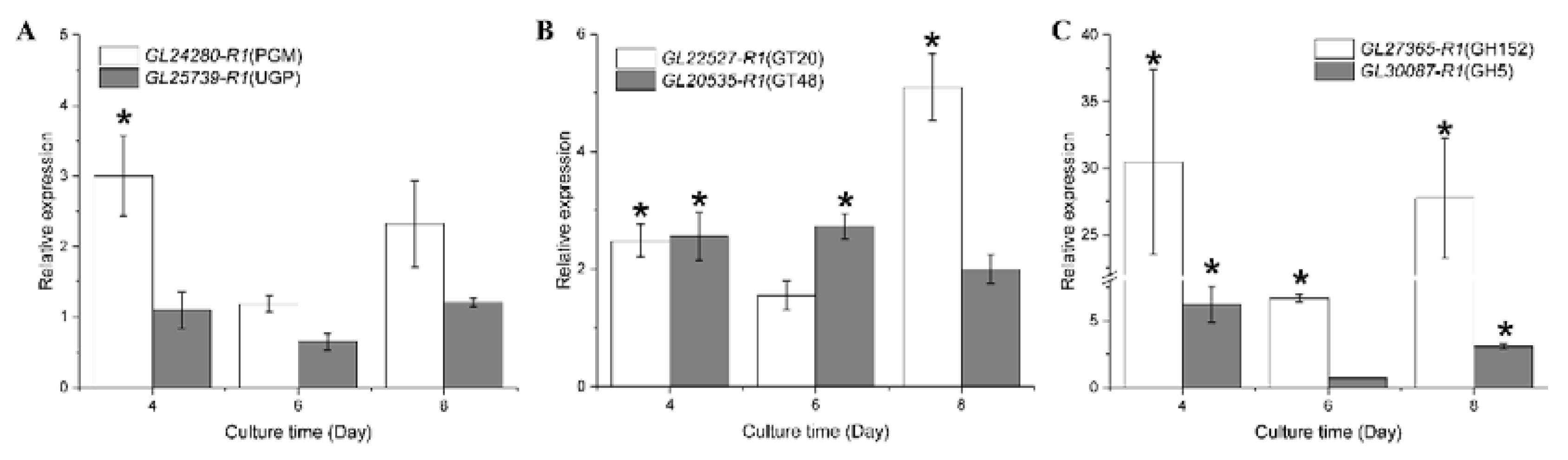
3.8. RT-qPCR Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, Z. Ganoderma (Lingzhi) in Traditional Chinese Medicine and Chinese Culture. Adv. Exp. Med. Biol. 2019, 1181, 1–13. [Google Scholar] [PubMed]
- Baby, S.; Johnson, A.J.; Govindan, B. Secondary metabolites from Ganoderma. Phytochemistry 2015, 114, 66–101. [Google Scholar] [CrossRef] [PubMed]
- Wasser, S.P. Current findings, future trends, and unsolved problems in studies of medicinal mushrooms. Appl. Microbiol. Biot. 2011, 89, 1323–1332. [Google Scholar] [CrossRef]
- Cor, D.; Knez, Z.; Knez Hrncic, M. Antitumour, antimicrobial, antioxidant and antiacetylcholinesterase effect of Ganoderma lucidum terpenoids and polysaccharides: A Review. Molecules 2018, 23, 649. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.C.; Heleno, S.A.; Reis, F.S.; Stojkovic, D.; Queiroz, M.J.; Vasconcelos, M.H.; Sokovic, M. Chemical features of Ganoderma polysaccharides with antioxidant, antitumor and antimicrobial activities. Phytochemistry 2015, 114, 38–55. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; Meng, G.; Wu, Y.; Ma, H.F.; Cui, B.K.; Dai, Y.C. Medium composition optimization, structural characterization, and antioxidant activity of exopolysaccharides from the medicinal mushroom Ganoderma lingzhi. Int. J. Biol. Macromol. 2019, 124, 1186–1196. [Google Scholar] [CrossRef]
- Xu, J.W.; Ji, S.L.; Li, H.J.; Zhou, J.S.; Duan, Y.Q.; Dang, L.Z.; Mo, M.H. Increased polysaccharide production and biosynthetic gene expressions in a submerged culture of Ganoderma lucidum by the overexpression of the homologous alpha-phosphoglucomutase gene. Bioprocess Biosyst. Eng. 2015, 38, 399–405. [Google Scholar] [CrossRef]
- Zhou, J.; Bai, Y.; Dai, R.; Guo, X.; Liu, Z.H.; Yuan, S. Improved polysaccharide production by homologous co-overexpression of phosphoglucomutase and UDP glucose pyrophosphorylase genes in the mushroom Coprinopsis cinerea. J. Agric. Food Chem. 2018, 66, 4702–4709. [Google Scholar] [CrossRef]
- Ma, Z.B.; Ye, C.; Deng, W.W.; Xu, M.M.; Wang, Q.; Liu, G.Q.; Wang, F.; Liu, L.M.; Xu, Z.H.; Shi, G.Y.; et al. Reconstruction and analysis of a genome-scale metabolic model of Ganoderma lucidum for improved extracellular polysaccharide production. Front. Microbiol. 2018, 9, 3076. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, F.; Xu, Z.H.; Ding, Z.Y. Bioactive mushroom polysaccharides: A review on monosaccharide composition, biosynthesis and regulation. Molecules 2017, 22, 955. [Google Scholar] [CrossRef]
- Peng, L.; Qiao, S.K.; Xu, Z.H.; Guan, F.; Ding, Z.Y.; Gu, Z.H.; Zhang, L.; Shi, G.Y. Effects of culture conditions on monosaccharide composition of Ganoderma lucidum exopolysaccharide and on activities of related enzymes. Carbohyd. Polym. 2015, 133, 104–109. [Google Scholar] [CrossRef]
- Peng, L.; Li, J.; Liu, Y.; Xu, Z.H.; Wu, J.Y.; Ding, Z.Y.; Gu, Z.H.; Zhang, L.; Shi, G.Y. Effects of mixed carbon sources on galactose and mannose content of exopolysaccharides and related enzyme activities in Ganoderma lucidum. Rsc. Adv. 2016, 6, 39284–39291. [Google Scholar] [CrossRef]
- Tang, Y.J.; Zhong, J.J. Exopolysaccharide biosynthesis and related enzyme activities of the medicinal fungus, Ganoderma lucidum, grown on lactose in a bioreactor. Biotechnol. Lett. 2002, 24, 743812. [Google Scholar] [CrossRef]
- Wei, Z.H.; Liu, L.L.; Guo, X.F.; Li, Y.J.; Hou, B.C.; Fan, Q.L.; Wang, K.X.; Luo, Y.D.; Zhong, J.J. Sucrose fed-batch strategy enhanced biomass, polysaccharide, and ganoderic acids production in fermentation of Ganoderma lucidum 5.26. Bioprocess Biosyst. Eng. 2016, 39, 37–44. [Google Scholar] [CrossRef]
- Zhou, H.; Bi, P.; Wu, X.; Huang, F.; Yang, H. Improved polysaccharide production in submerged culture of Ganoderma lucidum by the addition of coixenolide. Appl. Biochem. Biotechnol. 2014, 172, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.L.; Liu, R.; Ren, M.F.; Li, H.J.; Xu, J.W. Enhanced production of polysaccharide through the overexpression of homologous uridine diphosphate glucose pyrophosphorylase gene in a submerged culture of Lingzhi or Reishi medicinal mushroom, Ganoderma lucidum (higher basidiomycetes). Int. J. Med. Mushrooms 2015, 17, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.N.; Cao, Y.B.; Luo, Q.; Xu, Y.L.; Li, N.; Wang, C.X.; Xu, J.W. Overexpression of phosphomannomutase increases the production and bioactivities of Ganoderma exopolysaccharides. Carbohydr. Polym 2022, 294, 119828. [Google Scholar] [CrossRef]
- Hu, Y.R.; Li, M.J.; Wang, S.L.; Yue, S.N.; Shi, L.; Ren, A.; Zhao, M.W. Ganoderma lucidum phosphoglucomutase is required for hyphal growth, polysaccharide production, and cell wall integrity. Appl. Microbiol. Biot. 2018, 102, 1911–1922. [Google Scholar] [CrossRef]
- Li, M.; Chen, T.; Gao, T.; Miao, Z.; Jiang, A.; Shi, L.; Ren, A.; Zhao, M. UDP-glucose pyrophosphorylase influences polysaccharide synthesis, cell wall components, and hyphal branching in Ganoderma lucidum via regulation of the balance between glucose-1-phosphate and UDP-glucose. Fungal Genet. Biol. 2015, 82, 251–263. [Google Scholar] [CrossRef]
- Schuman, B.; Alfaro, J.A.; Evans, S.V. Glycosyltransferase structure and function. Top. Curr. Chem. 2007, 272, 217–257. [Google Scholar]
- Wilson, I.B.; Breton, C.; Imberty, A.; Tvaroška, I. Molecular Basis for the Biosynthesis of Oligo-And Polysaccharides. In Glycoscience; Fraser-Reid, B.O., Tatsuta, K., Thiem., J., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 2265–2323. [Google Scholar]
- Li, H.J.; Zhang, D.H.; Yue, T.H.; Jiang, L.X.; Yu, X.; Zhao, P.; Li, T.; Xu, J.W. Improved polysaccharide production in a submerged culture of Ganoderma lucidum by the heterologous expression of Vitreoscilla hemoglobin gene. J. Biotechnol. 2016, 217, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.J.; Wu, X.H.; Tao, T.L.; Zan, X.Y.; Sun, W.J.; Mu, D.S.; Yang, Y.; Wu, D. Functions of a glucan synthase gene GFGLS in mycelial growth and polysaccharide production of Grifola frondosa. J. Agric. Food Chem. 2019, 67, 8875–8883. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.H.; Li, X.F.; Zan, X.Y.; Fu, X.; Cui, F.J.; Zhu, H.A.; Sun, W.J.; Tao, T.L. The beta-1,3-glucan synthase gene GFGLS2 plays major roles in mycelial growth and polysaccharide synthesis in Grifola frondosa. Appl. Microbiol. Biotechnol. 2022, 106, 563–578. [Google Scholar] [CrossRef]
- Oka, T. Biosynthesis of galactomannans found in filamentous fungi belonging to Pezizomycotina. Biosci. Biotech. Bioch. 2018, 82, 183–191. [Google Scholar] [CrossRef]
- Sheppard, D.C.; Howell, P.L. Biofilm exopolysaccharides of pathogenic fungi: Lessons from bacteria. J. Biol. Chem. 2016, 291, 12529–12537. [Google Scholar] [CrossRef]
- Geoghegan, I.; Steinberg, G.; Gurr, S. The role of the fungal cell wall in the infection of plants. Trends Microbiol. 2017, 25, 957–967. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 5301. [Google Scholar] [CrossRef] [PubMed]
- Latge, J.P.; Beauvais, A.; Chamilos, G. The cell wall of the human fungal pathogen aspergillus fumigatus: Biosynthesis, organization, immune response, and virulence. Annu. Rev. Microbiol. 2017, 71, 99–116. [Google Scholar] [CrossRef]
- Lin, N.X.; Xu, Y.; Yu, X.W. Overview of yeast environmental stress response pathways and the development of tolerant yeasts. Synth. Syst. Biotechnol. 2022, 2, 232–245. [Google Scholar] [CrossRef]
- Jain, K.K.; Kumar, A.; Shankar, A.; Pandey, D.; Chaudhary, B.; Sharma, K.K. De novo transcriptome assembly and protein profiling of copper-induced lignocellulolytic fungus Ganoderma lucidum MDU-7 reveals genes involved in lignocellulose degradation and terpenoid biosynthetic pathways. Genomics 2020, 112, 184–198. [Google Scholar] [CrossRef]
- Cai, M.J.; Tan, Z.D.; Wu, X.X.; Liang, X.W.; Liu, Y.C.; Xie, Y.Z.; Li, X.M.; Xiao, C.; Gao, X.; Chen, S.D.; et al. Comparative transcriptome analysis of genes and metabolic pathways involved in sporulation in Ganoderma lingzhi. G3-Genes Genom. Genet. 2022, 12, jkab448. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Zhu, F.L.; Zhu, Y.X.; Li, Y.J.; Zhou, H.; Chen, S.L.; Ruan, J.S. Transcriptome profiling of transcription factors in Ganoderma lucidum in response to methyl jasmonate. Front. Microbiol. 2022, 13, 1052377. [Google Scholar] [CrossRef] [PubMed]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, M.; Zhao, L.; Wang, F.; Li, Y.; Shi, G.; Ding, Z. Transcriptome dynamics and metabolite analysis revealed the candidate genes and regulatory mechanism of ganoderic acid biosynthesis during liquid superficial-static culture of Ganoderma lucidum. Microb. Biotechnol. 2021, 14, 600–613. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Morin, E.; Kohler, A.; Baker, A.R.; Foulongne-Oriol, M.; Lombard, V.; Nagy, L.G.; Ohm, R.A.; Patyshakuliyeva, A.; Brun, A.; Aerts, A.L.; et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. USA 2012, 109, 17501–17506. [Google Scholar] [CrossRef]
- Veneault-Fourrey, C.; Commun, C.; Kohler, A.; Morin, E.; Balestrini, R.; Plett, J.; Danchin, E.; Coutinho, P.; Wiebenga, A.; de Vries, R.P.; et al. Genomic and transcriptomic analysis of Laccaria bicolor CAZome reveals insights into polysaccharides remodelling during symbiosis establishment. Fungal Genet. Biol. 2014, 72, 168–181. [Google Scholar] [CrossRef]
- Zhang, N.; Tang, Z.; Zhang, J.; Li, X.; Yang, Z.; Yang, C.; Zhang, Z.; Huang, Z. Comparative transcriptome analysis reveals the genetic basis underlying the biosynthesis of polysaccharides in Hericium erinaceus. Bot. Stud. 2019, 60, 15. [Google Scholar] [CrossRef]
- Duan, Y.; Han, H.; Qi, J.; Gao, J.M.; Xu, Z.; Wang, P.; Zhang, J.; Liu, C. Genome sequencing of Inonotus obliquus reveals insights into candidate genes involved in secondary metabolite biosynthesis. BMC Genom. 2022, 23, 314. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tang, J.; Chen, J.J.; Peng, A.Y.; Wang, Q.M.; Rao, L.Q.; Yang, H.; Zhang, X.W.; Yang, H.Z.; Zhang, C.; et al. Transcriptome analysis of three cultivars of Poria cocos reveals genes related to the biosynthesis of polysaccharides. J. Asian Nat. Prod. Res. 2019, 21, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, J.X.; Zhou, M.; Si, J.; Cui, B.K. Current advances and potential trends of the polysaccharides derived from medicinal mushrooms sanghuang. Front. Microbiol. 2022, 13, 965934. [Google Scholar] [CrossRef]
- Gao, H.; Gong, J.S.; Su, C.; Li, H.; Xu, Z.H.; Shi, J.S. Characterization, heterologous expression and engineering of trehalase for biotechnological applications. Synth. Syst. Biotechnol 2022, 2, 445–460. [Google Scholar] [CrossRef]
- Chen, J.; Fan, J.; Liu, W.; Wang, Z.; Ren, A.; Shi, L. Trehalose-6-phosphate synthase influences polysaccharide synthesis and cell wall components in Ganoderma lucidum. J. Basic Microbiol. 2022, 62, 1337–1345. [Google Scholar] [CrossRef]
- Oehme, D.P.; Shafee, T.; Downton, M.T.; Bacic, A.; Doblin, M.S. Differences in protein structural regions that impact functional specificity in GT2 family β-glucan synthases. PLoS ONE 2019, 14, e0224442. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Zhang, J.; Wang, Q.; Huang, J.; Juan, J.; Kuai, B.; Feng, Z.; Chen, H. Transcriptome and differentially expressed gene profiles in mycelium, primordium and fruiting body development in Stropharia rugosoannulata. Genes 2022, 13, 1080. [Google Scholar] [CrossRef]
- Briard, B.; Bomme, P.; Lechner, B.E.; Mislin, G.L.A.; Lair, V.; Prevost, M.C.; Latge, J.P.; Haas, H.; Beauvais, A. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci. Rep. 2015, 5, 8200. [Google Scholar] [CrossRef]
- Wu, T.; Cai, M.; Hu, H.; Jiao, C.; Zhang, Z.; Liu, Y.; Chen, J.; Xiao, C.; Li, X.; Gao, X.; et al. Whole-genome sequencing and transcriptome analysis of Ganoderma lucidum strain Yw-1-5 provides new insights into the enhanced effect of tween80 on exopolysaccharide production. J. Fungi 2022, 8, 1081. [Google Scholar] [CrossRef]
- Han, Q.; Wang, N.; Yao, G.; Mu, C.; Wang, Y.; Sang, J. Blocking beta-1,6-glucan synthesis by deleting KRE6 and SKN1 attenuates the virulence of Candida albicans. Mol. Microbiol. 2019, 111, 604–620. [Google Scholar] [CrossRef]
- Taff, H.T.; Nett, J.E.; Zarnowski, R.; Ross, K.M.; Sanchez, H.; Cain, M.T.; Hamaker, J.; Mitchell, A.P.; Andes, D.R. A Candida biofilm-induced pathway for matrix glucan delivery: Implications for drug resistance. PLoS Pathog. 2012, 8, e1002848. [Google Scholar] [CrossRef] [PubMed]
- Hartl, L.; Gastebois, A.; Aimanianda, V.; Latge, J.P. Characterization of the GPI-anchored endo beta-1,3-glucanase Eng2 of Aspergillus fumigatus. Fungal Genet. Biol. 2011, 48, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y.; Nakade, K.; Sato, S.; Yoshida, K.; Miyazaki, K.; Natsume, S.; Konno, N. Lentinula edodes genome survey and postharvest transcriptome analysis. Appl. Environ. Microbiol. 2017, 83, e02992-16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, W.; Liu, Z.; Wang, J.; Yuan, S. Purification, characterization and synergism in autolysis of a group of 1,3-beta-glucan hydrolases from the pilei of Coprinopsis cinerea fruiting bodies. Microbiology 2015, 161, 1978–1989. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.Q.; Zhang, X.W.; Liu, X.; Wang, R.; Liu, C.C.; Zhou, J.S.; Liu, Z.H.; Yuan, S. Comparative study of beta-glucan-degrading enzymes from Coprinopsis cinerea for their capacities to induce stipe cell wall extension. Int. J. Biol. Macromol. 2020, 152, 516–524. [Google Scholar] [CrossRef]
- Liu, X.; Wang, R.; Bi, J.; Kang, L.; Zhou, J.; Duan, B.; Liu, Z.; Yuan, S. A novel endo-beta-1,6-glucanase from the mushroom Coprinopsis cinerea and its application in studying of cross-linking of beta-1,6-glucan and the wall extensibility in stipe cell walls. Int. J. Biol. Macromol. 2020, 160, 612–622. [Google Scholar] [CrossRef]
- Liu, Z.H.; Xiong, Y.J.; Yi, L.; Dai, R.J.; Wang, Y.X.; Sun, M.L.; Shao, X.Y.; Zhang, Z.Q.; Yuan, S. Endo-beta-1,3-glucanase digestion combined with the HPAEC-PAD-MS/MS analysis reveals the structural differences between two laminarins with different bioactivities. Carbohyd. Polym. 2018, 194, 339–349. [Google Scholar] [CrossRef]
- Wang, J.; Kang, L.Q.; Liu, Z.H.; Yuan, S. Gene cloning, heterologous expression and characterization of a Coprinopsis cinerea endo- beta-1,3(4)-glucanase. Fungal Biol. 2017, 121, 61–68. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Watanabe, H.; Nagai, M.; Nakade, K.; Takahashi, M.; Sato, T. Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 2006, 141, 793–801. [Google Scholar] [CrossRef]

| Gene ID | Gene Name | FPKM Value | |||||
|---|---|---|---|---|---|---|---|
| Glc-4 | Glc-6 | Glc-8 | Xyl-4 | Xyl-6 | Xyl-8 | ||
| GL25253-R1 | XR | 161.66 | 914.25 * | 833.84 | 145.17 | 43.87 | 448.15 |
| GL29728-R1 | XR | 284.23 | 252.43 | 149.41 * | 301.84 | 321.09 | 561.03 |
| GL31661-R1 | XULR | 27.35 | 46.85 | 24.05 * | 30.43 | 77.50 | 79.18 |
| GL18694-R1 | XYLB | 94.59 | 250.12 | 166.04 | 174.30 | 136.04 | 297.62 |
| GL21997-R1 | XYLB | 101.59 | 102.22 | 97.52 | 129.53 | 128.80 | 114.71 |
| GL26014-R1 | TKTA | 62.44 * | 52.35 * | 54.24 | 17.52 | 25.08 | 32.27 |
| GL26783-R1 | GK | 50.78 | 51.30 | 60.06 | 65.72 | 61.88 | 59.12 |
| GL20491-R1 | HK | 26.99 | 27.72 | 26.47 | 29.47 | 28.72 | 35.44 |
| GL24280-R1 | PGM | 279.26 | 265.65 | 264.92 | 278.27 | 308.83 | 274.51 |
| GL25739-R1 | UGP | 506.58 | 501.29 | 416.90 | 279.62 | 542.14 | 348.39 |
| GL29575-R1 | GALE | 75.59 | 82.80 * | 120.30 | 73.47 | 23.00 | 107.93 |
| GL30389-R1 | GALE | 135.67 | 165.89 * | 267.17 | 180.65 | 53.07 | 275.96 |
| GL18437-R1 | UGDH | 257.30 | 222.58 | 202.82 | 272.26 | 176.89 | 281.65 |
| GL26652-R1 | UXS | 177.33 | 174.14 | 133.19 | 139.76 | 144.57 | 188.09 |
| GL22245-R1 | GPI | 329.20 | 297.68 | 245.43 | 281.02 | 290.33 | 386.39 |
| GL22193-R1 | PMI | 139.69 | 126.56 | 112.25 | 93.81 | 136.62 | 112.32 |
| GL17878-R1 | PMI | 19.70 | 13.89 | 21.62 | 8.50 | 19.43 | 24.38 |
| GL20742-R1 | PMM | 106.29 | 93.07 | 77.94 | 86.58 | 89.79 | 93.87 |
| GL21817-R1 | PMM | 98.82 | 92.67 | 83.43 | 88.77 | 96.24 | 89.87 |
| GL25424-R1 | GMPP | 133.36 | 101.89 | 112.82 | 114.84 | 105.41 | 92.70 |
| GL20928-R1 | GMD | 269.46 | 251.06 | 210.68 | 178.67 | 214.09 | 131.50 |
| GL21002-R1 | FS | 91.93 | 78.47 | 72.66 | 80.75 | 91.68 | 90.04 |
| GH Family | Gene ID | Putative Enzymes | Signal Peptide | GPI | Likely Destination |
|---|---|---|---|---|---|
| GH1 | GL20553-R1 | β-glucosidase | No | No | Cytoplasm |
| GL24039-R1 | β-glucosidase | No | No | Extracellular space | |
| GH3 | GL20743-R1 | β-glucosidase | No | No | Extracellular space |
| GL22586-R1 | β-xylosidase | Yes | No | Extracellular space | |
| GL24911-R1 | β-glucosidase | No | No | Extracellular space | |
| GL29912-R1 | β-glucosidase | Yes | No | Extracellular space | |
| GH5 | GL19331-R1 | - | No | No | Cytoplasm |
| GL30087-R1 | Exo-1,3-β-glucanase | Yes | No | Extracellular space | |
| GH13 | GL18711-R1 | Glycogen debranching enzyme | No | No | Cytoplasm |
| GL31461-R1 | 1,4-α-glucan-branching enzyme | No | No | Cytoplasm | |
| GH16 | GL15467-R1 | Fungal 1,3(4)-β-D-glucanases | Yes | Highly probable | Anchored component of plasma membrane |
| GL15468-R1 | Fungal 1,3(4)-β-D-glucanases | Yes | Highly probable | Anchored component of plasma membrane | |
| GL23820-R1 | Fungal 1,3(4)-β-D-glucanases | Yes | No | Extracellular space | |
| GH17 | GL25813-R1 | Exo-1,3-β-glucanase | No | No | Plasma membrane |
| GL25813-R2 | Exo-1,3-β-glucanase | No | No | Plasma membrane | |
| GL25603-R1 | 1,6-β-glucan synthase | Yes | No | Extracellular space | |
| GH18 | GL25033-R1 | Chitinase | No | No | Extracellular space |
| GL29255-R1 | Chitinase | Yes | No | Extracellular space | |
| GL22188-R1 | Chitinase | Yes | No | Extracellular space | |
| GH55 | GL21451-R1 | Exo-1,3-β-glucanase | Yes | No | Extracellular space |
| GL24581-R1 | Exo-1,3-β-glucanase | Yes | No | Extracellular space | |
| GH79 | GL26459-R1 | - | Yes | Weakly probable | Extracellular space |
| GL26618-R1 | - | Yes | Highly probable | Anchored component of plasma membrane | |
| GL29243-R1 | - | Yes | Weakly probable | Plasma membrane | |
| GL23706-R1 | - | No | No | Extracellular space | |
| GH128 | GL21185-R1 | - | Yes | No | Extracellular space |
| GH152 | GL27365-R1 | 1,3-β-glucanase | Yes | No | Extracellular space |
| GH154 | GL25647-R1 | - | No | No | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Xu, M.; Zhao, L.; Chen, L.; Ding, Z. Novel Insights into the Mechanism Underlying High Polysaccharide Yield in Submerged Culture of Ganoderma lucidum Revealed by Transcriptome and Proteome Analyses. Microorganisms 2023, 11, 772. https://doi.org/10.3390/microorganisms11030772
Wang Q, Xu M, Zhao L, Chen L, Ding Z. Novel Insights into the Mechanism Underlying High Polysaccharide Yield in Submerged Culture of Ganoderma lucidum Revealed by Transcriptome and Proteome Analyses. Microorganisms. 2023; 11(3):772. https://doi.org/10.3390/microorganisms11030772
Chicago/Turabian StyleWang, Qiong, Mengmeng Xu, Liting Zhao, Lei Chen, and Zhongyang Ding. 2023. "Novel Insights into the Mechanism Underlying High Polysaccharide Yield in Submerged Culture of Ganoderma lucidum Revealed by Transcriptome and Proteome Analyses" Microorganisms 11, no. 3: 772. https://doi.org/10.3390/microorganisms11030772
APA StyleWang, Q., Xu, M., Zhao, L., Chen, L., & Ding, Z. (2023). Novel Insights into the Mechanism Underlying High Polysaccharide Yield in Submerged Culture of Ganoderma lucidum Revealed by Transcriptome and Proteome Analyses. Microorganisms, 11(3), 772. https://doi.org/10.3390/microorganisms11030772





