Abstract
Gardnerella vaginalis is the main pathogen that causes bacterial vaginosis. In the healthy vaginal microecological environment of a woman, the lactobacilli produce lactate and hydrogen peroxide to inhibit the growth of pathogens such as G. vaginalis. The lack of lactobacilli results in a high pH and low hydrogen peroxide in the vagina which facilitate G. vaginalis growth, leading to the imbalance of the vaginal microecology. In this study, lactate and hydrogen peroxide were added to a G. vaginalis culture medium to simulate the co-culture of the lactobacilli and G. vaginalis, and then the genes related to the stress response of G. vaginalis were identified using transcriptomics and proteomics. It was indicated that, among all the upregulated genes, most of them encoded transporters associated with the efflux of harmful substances, and the majority of the downregulated genes were related to the biofilm formation and epithelial cell adhesion. This study may help find new drug targets for G. vaginalis for the development of novel therapies for bacterial vaginosis.
1. Introduction
The reproductive tract is an open cavity that houses a large number of microorganisms and is closely related to female reproductive health. It is a dynamic environment in which drug stimulation, host hormone levels and immunity affect the microbiome composition, resulting in a microecological imbalance and bacterial vaginosis (BV) [1].
A loss of vaginal lactobacilli and an overgrowth of diverse anaerobes are two characteristics of a vaginal microecological imbalance. To cause BV, the anaerobic microbiota produce enzymes (such as sialidase) that degrade the mucosa, produce biofilm and secrete short-chain fatty acids [2]. BV is associated with pelvic inflammatory disease [3] and infection following gynecological surgery and cervical cancer. Patients can develop complications whether or not they have clinical symptoms. BV in pregnancy increases the risks of spontaneous abortion, amniotic chorionitis, premature rupture of the membranes and premature delivery, all of which have serious consequences for a woman’s health [4,5]. The incidence rate of BV is approximately 23–29%, with significant differences between ethnic groups [6,7]. The standard treatment for BV is metronidazole and clindamycin, and the recurrence rate is as high as 30% within 3 months [8]. Probiotics therapy has recently achieved some success. The vaginal flora of healthy women is dominated by the lactobacilli while patients with BV have a diverse microbiota containing anaerobic and uncultivable species [9]. In clinical and laboratory features, vaginal microbiome transplantation from healthy donors resulted in four patients obtaining full long-term remission and one patient achieving incomplete remission [10]. A study of Lactobacillus crispatus CTV-05 after vaginal metronidazole treatment found a significantly lower incidence of BV recurrence [11]. Even Saccharomyces cerevisiae-based probiotics obtained beneficial effects for preventing and/or treating vulvovaginal candidiasis (VVC) and BV [12]. However, effective prevention and treatment strategies must still be developed.
Gardnerella vaginalis plays an important role in the establishment of the BV bacterial community, for which it can adhere to the host epithelium, produce biofilm and secrete vaginolysin [13]. In a healthy vagina, the lactobacilli directly inhibit the growth of G. vaginalis by secreting lactate, hydrogen peroxide and bacteriocin, and promote the integrity of the epithelial cells by stimulating the mucus secretion and regulating the immune response [14].
To date, how lactate and hydrogen peroxide stress affect G. vaginalis and how G. vaginalis responds to stress are unknown. To simulate the co-culture of the lactobacilli and G. vaginalis, we cultivated G. vaginalis in the presence of lactate and hydrogen peroxide. As a deprived species in cell signal transduction, there were no c-di-A/GMP and only six two-component system (TCS) genes in the genome of G. vaginalis [15]. Therefore, the shk (sensor histidine kinase), which encodes a membrane protein belonging to the TCS and is related to the regulation of the biofilm formation [16], and the trx (thioredoxin disulfide reductase), which encodes an intracellular protein responding to lactic acid and hydrogen peroxide [17], were selected as the markers to indicate the exact moment when the cells exhibited the strongest response. Transcriptomics and proteomics were then used to identify the genes involved in the G. vaginalis response to the acidic pH and hydrogen peroxide stress.
2. Materials and Methods
2.1. Strains and Growth Conditions
G. vaginalis ATCC 14,019 was purchased from the American Type Culture Collection. The planktonic cells were grown in an sBHI [10.0 g of peptone, 12.5 g of dehydrated calf brain extract, 5.0 g of dehydrated beef heart extract, 5.0 g of NaCl, 2.0 g of glucose, 2.5 g of disodium hydrogen phosphate in 1 L ddH2O, pH 7.4] for 12 h at 37 °C with 5% CO2.
2.2. Growth Curve Test
A two-percent seed solution was inoculated into the sBHI without sheep’s blood and cultured to the early exponential phase when the cell density reached an OD600 of approximately 1.0. At this point, the lactate (Sigma Aldrich L6661, Shanghai, China) and hydrogen peroxide (Sigma Aldrich H1009, Shanghai, China) were added to the pH 5.5 and 0.5 mM, respectively, to obtain the sub-lethal cells [18,19]. Then, 5 min, 10 min, 30 min and 1 h after the start of stress, the culture samples were collected to measure the cell density and transcription levels of the response marker genes, including the shk and trx. The transcriptome and proteome analyses were performed on the samples with the highest level of the response marker gene transcription. Three biologic replicates were carried out for each condition.
2.3. cDNA Library Preparation and Sequencing
A total amount of 3 μg of RNA per sample was prepared as the input material for the RNA sample preparations. A Vazyme Ribo-off rRNA depletion kit (bacteria) (Vazyme biotech, Piscataway, NJ, USA) was used to remove rRNA. The sequencing libraries were generated using an NEB Next UltraTM RNA library Prep Kit (NEB, Ipswich, MA, USA).
After purification, the RNA samples were fragmented into small pieces using divalent cations under an elevated temperature. The cleaved RNA fragments were reversely transcribed into first strand cDNA using random primers. The strand specificity was achieved by replacing dTTP with dUTP in the Second Strand Marking Mix (SMM), followed by a second strand cDNA synthesis using DNA Polymerase I and RNase H. The incorporation of dUTP in the second strand synthesis quenched the second strand during the amplification since the DNA polymerase used in the assay did incorporate nucleotide after dUMP. The addition of Actinomycin D to the First Stand Synthesis Act D mix (FSA) prevented spurious DNA-dependent synthesis, while allowing for RNA-dependent synthesis and improving the strand specificity. These cDNA fragments were then added to a single ‘A’ base at the 3′ end for subsequent ligation to the adapter. The products were then purified and enriched with PCR to create the final cDNA library. The library quality was assessed using the Agilent Bioanalyzer 2100 system. The library preparations were sequenced on an Illumina Hiseq 4000 platform by the Beijing Allwegene Technology Company Limited (Beijing, China) and the paired-end 150 bp reads were generated.
2.4. RNA-Sequencing Data Analysis
The raw data (raw reads) of the fastq format were firstly processed using in-house perl scripts. In this step, the clean data (clean reads) were obtained by removing the reads containing adapter, the reads containing ploy-N (N > 10%) and the low-quality reads (Q < 5) greater than 50% from the raw data. At the same time, the Q20, Q30 and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with a high quality. The adaptor sequences and low-quality sequence reads were removed from the data sets. The raw sequences were transformed into clean reads after data processing. These clean reads were then mapped to the reference genome sequence by Bowtie2 v2.2.6. Only the reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. The Bowtie2 mapping results were assembled using the Rockhopper software v2.0.3 and compared to the annotated gene models to find new transcript regions. The Blastx program was compared to the NR library (the e-value is set to le-5). The newly predicted transcript region was annotated and the annotated transcript region was regarded as a new transcript region with coding potential.
2.5. Quantitative PCR
The total RNA was extracted using the MiniBEST kit (TaKaRa, Dalian, China) and was reverse transcribed using the TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix kit (Transgen Biotech, Beijing, China). In order to confirm the stress response time of the strain and validate the RNA-seq data, a qPCR was performed to quantify the transcription level of the signal transduction genes and virulence genes, respectively. The gyr gene was used as the reference gene. The oligonucleotide primers were designed using SnapGene (Table S1). The qPCR reaction was prepared by mixing together 10 μL of SYBR green supermix (Bio-Rad, Hercules, CA, USA), 1 μL of 1:800 diluted cDNA, 0.5 μL of 10 μM forward and reverse primers and water up to 20 μL. The run was performed using a CFX96TM thermal cycler (Jena, Germany) with the following cycling parameters: 3 min at 95 °C, followed by 40 cycles of 10 s at 95 °C, 10 s at 60 °C and 15 s at 72 °C. The normalized gene expression was determined using the delta Ct method (EΔCt), a variation of the Livak method, where ΔCt = Ct (reference gene)−Ct (target gene) and E stands for the experimentally determined reaction efficiency. Three biologic replicates of each condition were analyzed.
2.6. Relative Analysis of the TMT-Marked Proteins
For each sample, a 100 μg peptide mixture was labeled using the TMT reagent according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). Each fraction was injected for a nano LC-MS/MS analysis. The peptide mixture was loaded onto a reverse phase trap column (Thermo Scientific Acclaim PepMap100, 100 μm * 2 cm, nanoViper C18) connected to the C18-reversed phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% formic acid) at a flow rate of 300 nL/min controlled by the IntelliFlow technology. The LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Fisher Scientific) that was coupled with the Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) for 60/90 min (determined by the project proposal). The MS/MS spectra were searched using the MASCOT engine (Matrix Science, London, UK; version 2.2) embedded into Proteome Discoverer 1.4.
3. Results
3.1. Acidic pH and Hydrogen Peroxide Significantly Inhibit G. vaginalis Growth
To determine the effect of the acidic pH and hydrogen peroxide stress on G. vaginalis development, the cell growth curves for G. vaginalis were obtained in the sBHI medium, Sbhi + acidic pH and sBHI + acidic pH + hydrogen peroxide. The three cultures had nearly identical growth curves during the lag and initial log phases. After adding lactate to one group of samples at 5 h to adjust the pH to 5.5, the cells grew slower for about 2 h, then declined for 3 h before beginning to recover. In the transcriptomic and proteomic analysis, these samples were named as the Lac group. When both lactate and H2O2 were added to another group of samples to achieve a pH of 5.5 and a final concentration of 0.5 mM of H2O2, the cells grew identically to the Lac group for 1 h before entering the stable phase with a cell density lower than the stress-free control group. In the following study, the samples subjected to the acidic and oxidative stress were labeled as the LacHyd group (Figure 1A). The expression levels of the response marker genes shk and trx peaked 10 min after the stress began, according to the RT-PCR data. After one generation time (1 h), the expression level of these genes began to decline and reached its lowest resting level (Figure S1). Therefore, the Lac and LacHyd group 10-min cultures were subjected to the transcriptomics and proteomics analysis.
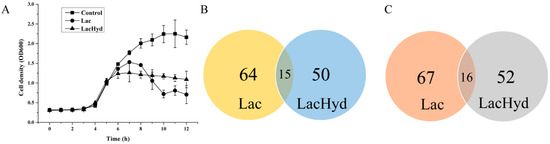
Figure 1.
General comparison of the control, Lac group and LacHyd group. (A) Growth curves of the three groups. (B) Venn analysis of the shared genes that were upregulated compared to the control in the two groups. (C) Venn analysis of the shared genes that were downregulated compared to the control in the two groups. The pH values of the three groups were 7.0., 5.5 and 5.5, respectively; the H2O2 concentration of the LacHyd group was 0.5 mM.
3.2. Genes Related to Biofilm Formation and Epithelial Adhesion Were Downregulated during Stress
The transcriptomics data revealed that only eight, three and one gene were uniquely expressed in the control, Lac and LacHyd groups, respectively (Figure 1B). Comparing the Lac group to the control group revealed 79 upregulated and 83 downregulated genes. Comparing the LacHyd group to the control group revealed 65 upregulated and 68 downregulated genes. This was unsurprising given that G. vaginalis did not have a complex signaling system. Surprisingly, the comparison of the Lac group to the LacHyd group revealed only one upregulated and six downregulated genes. The similar stress response under the two conditions suggests that the lactate stimulation may represent the lactobacilli stimulation in G. vaginalis (Table S2). The genes with significant differences are listed in Table 1 and Table S3. The RT-PCR was used to confirm the expression levels of the eight virulence genes, revealing that, in addition to the stress response genes being upregulated, the genes related to the epithelial cell adhesion and biofilm formation were actually downregulated (Figure 2). This could be the mechanism by which the lactobacilli probiotics inhibit GV cell adhesion to the epithelial cells via biofilm.

Table 1.
List of the 10 genes with the highest fold change values among the differentially expressed genes in G. vaginalis cultured under lactate and hydrogen peroxide. All p values were less than 0.02.
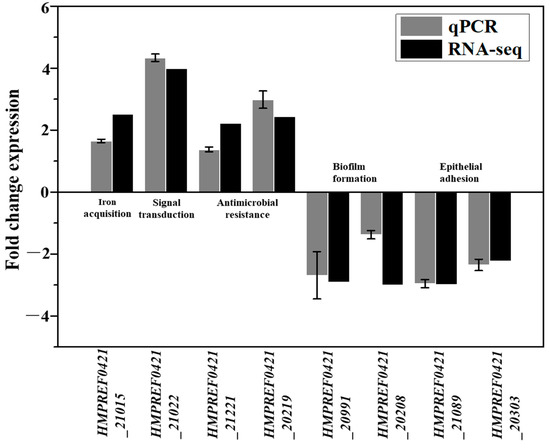
Figure 2.
Quantification of the transcription of the virulence genes that were expressed differentially in G. vaginalis cultured under lactate and hydrogen peroxide. Bars represent the mean, and the error bars the standard error of the mean (mean ± SEM).
3.3. The Proteomics Data Are Consistent with the Transcriptomic Data
The TMT quantitative proteomics were performed on the LacHyd group to determine whether the protein regulation was consistent with the transcription level after stimulation. The results revealed that 2740 proteins were identified at the species level, which was more than twice the number of the genome genes, indicating the species’ high genetic heterogeneity. According to the numbers of the differentially expressed proteins, the standard screening of the proteins with differential expression multiples was adjusted to more than 1.2 times (upregulated by more than 1.2 times or downregulated by less than 0.83 times) and a p value of <0.05. There were 86 upregulated differentially expressed proteins and 55 downregulated differential expression proteins in the LacHyd group compared to the control group. The analysis of the GO and KEGG pathways revealed that the function of these differentially expressed proteins was primarily involved in the processes of metabolism, cell division, localization, response to stimulus and transcriptional regulation. Since the main pathogenic factors found in the upregulated proteins were still efflux transporters, it was hypothesized that ATCC 14,019 actively effluxed harmful ions and small molecules by upregulating transporters (HMPREF0421_20219 and HMPREF0421_21025). Furthermore, while some virulence factors were upregulated, the biofilm formation-related proteins were downregulated (Table 2), implying that GV was intended to gather weapons and energy to combat the lactobacilli whether or not it worked.

Table 2.
List of the 10 genes with the highest fold change values among the differentially expressed proteins in G. vaginalis cultured under lactate and hydrogen peroxide. All p values were less than 0.05.
4. Discussion
Many upregulated genes of the strain ATCC 14,019 in the LacHyd and Lac groups belonged to bacterial efflux pumps, including the ABC transporter, MFS transporter, sulfonate ABC transporter osmotic enzyme and cobalt ABC transporter (HMPREF0421_21024, HMPREF0421_20219, HMPREF0421_21221 and HMPREF0421_20844). The ABC transporters used ATP hydrolysis to provide energy for the cells, and their upregulation helped in the removal of harmful substances via the ABC export system (HMPREF0421_21024) [20,21]. The MFS transporter was most commonly described as an efflux pump that removes antibiotics (e.g., TetB and TetK) from the cells [22,23,24]. Various families were responsible for the efflux of various types of small molecules. This gene was only upregulated in the LacHyd group, which was thought to be responsible for the hydrogen peroxide efflux (HMPREF0421_20219). A small molecular compound and intracellular ion efflux was mediated by the cobalt transporters (HMPREF0421_20844) [25]. It is worth noting that histidine kinase, as one of the strain’s six pairs of two-component systems, was found to be highly upregulated in the Lac and LacHyd groups, implying that it could be responsible for the lactate signal transduction on the cell membrane (HMPREF0421_21022). Furthermore, 3-Oxoacetyl-ACP reductase was involved in the synthesis of siderophores, and its upregulation indicated that the strain can obtain more iron from the environment to inhibit the lactobacilli iron uptake (HMPREF0421_21015) [15,26]. The ATCC 14,019 downregulated genes were mostly involved in the adhesion and biofilm syntheses (HMPRE0421_21089, HMPRE0421_20208, HMPRE0421_20303 and HMPRE0421_20991; Figure 2). G. vaginalis can promote cell aggregation via the biofilm formation and adhere to the host cells via pili and surface-anchored proteins. The gene expression level of the Lac group was nearly identical to that of the LacHyd group, with only five different genes, and the fold change was less than 2 (Table S4). In other words, hydrogen peroxide had little antimicrobial effect in mixing with lactate, which was consistent with the conclusion of a previous study [27], implying that the strains took response measures to the lactobacilli signals and dealt with more risks based on lactate.
Apart from the efflux transporters, the expressional level of vaginolysin, a cholesterol-dependent hemolysin (HMPREF0421_20066), had been upregulated by 1.43 times according to the proteomics data [28]. CHAP is a protein domain found in many extracellular proteins and receptors. It was initially identified in enzymes that degraded bacterial cell walls, and it recognizes the polysaccharides involved in N-acetylglucosamine synthesis, including peptidoglycan (HMPREF0421_20542) [29]. When stimulated, the virulence factors increased their competitive capacity against the lactobacilli. The pathogenic factors found in the downregulated proteins included the pili, biofilm syntheses and adhesion proteins, which were consistent with the transcriptomics results. The PTS EII transmembrane protein, which was mostly downregulated, was primarily responsible for the glucose transport and was critical for maintaining normal cell growth and biofilm synthesis (HMPREF0421_20893) [30]. RelE, a toxin–antitoxin pair encoded by the RelBE operon, was a very effective translation inhibitor in vivo and in vitro. RelB can reverse the RelE inhibitory effects on the protein synthesis in vivo, and RelE plays a regulatory role on bacteria in the process of adapting to poor growth conditions and forming persistent cells by controlling the cell dormancy (HMPREF0421_21052) [31]. The maltose-binding protein was responsible for the maltodextrin absorption (HMPREF0421_20232), which provided raw materials for the biofilm synthesis. The sugar ABC transporters transported monosaccharides or oligosaccharides, providing oligosaccharides units for cell growth or biofilm formation (HMPREF0421_20098) [32]. Therefore, we can speculate that, while the formed biofilm increased the G. vaginalis tolerance to lactate and hydrogen peroxide [33], the lactobacilli were able to inhibit G. vaginalis cell growth and biofilm formation in vivo.
In conclusion, in a healthy vaginal microecological environment, the lactobacilli, as the dominant flora, secrete a high concentration of lactate and hydrogen peroxide to the extracellular environment, forcing G. vaginalis to expend considerable energy to excrete these two small molecules. There is no excess energy for its own growth and proliferation (Figure 3A). When the vaginal microecology is out of balance, the number of lactobacilli and the concentrations of lactate and hydrogen peroxide are significantly reduced. Anaerobic microorganisms, primarily G. vaginalis, begin to upregulate the expression levels of the pili, adhesion proteins and biofilm formation, and gradually form biofilm adhered to the vaginal epithelial cells, resulting in BV (Figure 3B). Therefore, inhibiting the expression of the adhesion proteins and biofilm components using G. vaginalis at the onset of a vaginal microecological imbalance could be the key to preventing bacterial vaginitis.
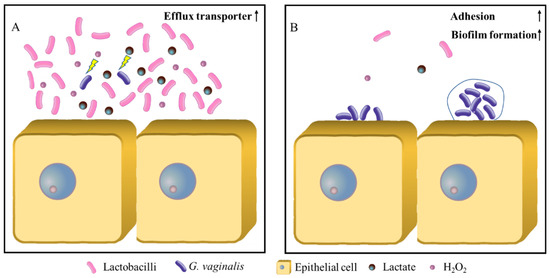
Figure 3.
The effect model of the lactobacilli on the biofilm formation of G. vaginalis. (A) Healthy vaginal microecosystem; (B) BV vaginal microecosystem.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11030695/s1. Figure S1: The expressional level of response markers after adding lactate and H2O2. Bars represent the mean and the error bars the standard error of the mean (mean ± SEM); Table S1: Primers for QPCR; Table S2: Unique genes in the Venn analysis; Table S3: List of the genes with fold changes above two among the differentially expressed genes in G. vaginalis cultured under lactate. All P values were less than 0.02; Table S4: List of the differentially expressed genes in the Lac group compared to the LacHyd group. All p values were less than 0.02.
Author Contributions
Methodology, K.Z. and X.Z.; validation, M.L. and Y.Q.; formal analysis, H.W. and Y.H.; investigation, H.D.; resources, X.Z.; writing—original draft preparation, K.Z.; writing—review and rditing, L.G.; visualization, K.Z.; supervision, L.G.; project administration, L.G.; funding acquisition, K.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the China Postdoctoral Science Foundation (No. 2018M642648), the State Key Laboratory of the Microbial Technology Open Projects Fund (No. M2021-17), the Natural Science Foundation of Shandong Province (No. ZR2021MC038) and the Science and Technology Project of the Jinan Municipal Health Commission (No. 2019-1-25).
Data Availability Statement
The transcriptomics data have been submitted to NCBI with the Accession No. PRJNA865867.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, C.; Song, X.; Wei, W.; Zhong, H.; Dai, J.; Lan, Z.; Li, F.; Yu, X.; Feng, Q.; Wang, Z.; et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 2017, 8, 875. [Google Scholar] [CrossRef]
- Anahtar, M.N.; Gootenberg, D.B.; Mitchell, C.M.; Kwon, D.S. Cervicovaginal Microbiota and Reproductive Health: The Virtue of Simplicity. Cell Host Microbe 2018, 23, 159–168. [Google Scholar] [CrossRef]
- Taylor, B.D.; Darville, T.; Haggerty, C.L. Does Bacterial Vaginosis Cause Pelvic Inflammatory Disease? Sex. Transm. Dis. 2013, 40, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Turovskiy, Y.; Sutyak Noll, K.; Chikindas, M.L. The aetiology of bacterial vaginosis. J. Appl. Microbiol. 2011, 110, 1105–1128. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Jorgensen, J.S.; Lamont, R.F. The contribution of bacteriophages to the aetiology and treatment of the bacterial vaginosis syndrome. Fac. Rev. 2022, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Peebles, K.; Velloza, J.; Balkus, J.E.; McClelland, R.S.; Barnabas, R.V. High Global Burden and Costs of Bacterial Vaginosis: A Systematic Review and Meta-Analysis. Sex. Transm. Dis. 2019, 46, 304–311. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef]
- Workowski, K.A.; Bolan, G.A.; Centers for Disease Control & Prevention. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm. Rep. 2015, 64, 1–137. [Google Scholar]
- Abou Chacra, L.; Fenollar, F.; Diop, K. Bacterial Vaginosis: What Do We Currently Know? Front. Cell. Infect. Microbiol. 2021, 11, 672429. [Google Scholar] [CrossRef]
- Lev-Sagie, A.; Goldman-Wohl, D.; Cohen, Y.; Dori-Bachash, M.; Leshem, A.; Mor, U.; Strahilevitz, J.; Moses, A.E.; Shapiro, H.; Yagel, S.; et al. Vaginal microbiome transplantation in women with intractable bacterial vaginosis. Nat. Med. 2019, 25, 1500–1504. [Google Scholar] [CrossRef]
- Cohen, C.R.; Wierzbicki, M.R.; French, A.L.; Morris, S.; Newmann, S.; Reno, H.; Green, L.; Miller, S.; Powell, J.; Parks, T.; et al. Randomized Trial of Lactin-V to Prevent Recurrence of Bacterial Vaginosis. N. Engl. J. Med. 2020, 382, 1906–1915. [Google Scholar] [CrossRef] [PubMed]
- Gaziano, R.; Sabbatini, S.; Roselletti, E.; Perito, S.; Monari, C. Saccharomyces cerevisiae-Based Probiotics as Novel Antimicrobial Agents to Prevent and Treat Vaginal Infections. Front. Microbiol. 2020, 11, 718. [Google Scholar] [CrossRef] [PubMed]
- Schwebke, J.R.; Muzny, C.A.; Josey, W.E. Role of Gardnerella vaginalis in the pathogenesis of bacterial vaginosis: A conceptual model. J. Infect. Dis. 2014, 210, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Younes, J.A.; Lievens, E.; Hummelen, R.; van der Westen, R.; Reid, G.; Petrova, M.I. Women and Their Microbes: The Unexpected Friendship. Trends Microbiol. 2018, 26, 16–32. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Yildirim, S.; Thomas, S.M.; Durkin, A.S.; Torralba, M.; Sutton, G.; Buhay, C.J.; Ding, Y.; Dugan-Rocha, S.P.; Muzny, D.M.; et al. Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential. PLoS ONE 2010, 5, e12411. [Google Scholar] [CrossRef]
- Liu, C.; Sun, D.; Zhu, J.; Liu, W. Two-Component Signal Transduction Systems: A Major Strategy for Connecting Input Stimuli to Biofilm Formation. Front. Microbiol. 2018, 9, 3279. [Google Scholar] [CrossRef]
- Kim, J.; Cho, Y.; Jang, I.A.; Park, W. Molecular mechanism involved in the response to hydrogen peroxide stress in Acinetobacter oleivorans DR1. Appl. Microbiol. Biotechnol. 2015, 99, 10611–10626. [Google Scholar] [CrossRef]
- Vaneechoutte, M. The human vaginal microbial community. Res. Microbiol. 2017, 168, 811–825. [Google Scholar] [CrossRef]
- Strus, M.; Brzychczy-Wloch, M.; Gosiewski, T.; Kochan, P.; Heczko, P.B. The in vitro effect of hydrogen peroxide on vaginal microbial communities. FEMS Immunol. Med. Microbiol. 2006, 48, 56–63. [Google Scholar] [CrossRef]
- Rees, D.C.; Johnson, E.; Lewinson, O. ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Perez, A.M.; Gomez, M.M.; Kalvapalle, P.; O’Brien-Gilbert, E.; Bennett, M.R.; Shamoo, Y. Using cellular fitness to map the structure and function of a major facilitator superfamily effluxer. Mol. Syst. Biol. 2017, 13, 964. [Google Scholar] [CrossRef]
- Mahey, N.; Tambat, R.; Verma, D.K.; Chandal, N.; Thakur, K.G.; Nandanwar, H. Antifungal Azoles as Tetracycline Resistance Modifiers in Staphylococcus aureus. Appl. Environ. Microbiol. 2021, 87, e00155-21. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Xie, G.; Daligault, H.; Davenport, K.; Gleasner, C.; Jacobs, L.; Kubicek-Sutherland, J.; LeCuyer, T.; Otieno, V.; Raballah, E.; et al. Genome Sequence of a Staphylococcus xylosus Clinical Isolate, Strain SMA0341-04 (UGA5), from Siaya County Referral Hospital in Siaya, Kenya. Microbiol. Resour. Announc. 2019, 8, e01625-18. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Qi, X.; Hong, S.; Xu, K.; He, F.; Zhang, M.; Chen, J.; Chao, D.; Zhao, W.; Li, D.; et al. Structure and mechanism of a group-I cobalt energy coupling factor transporter. Cell Res. 2017, 27, 675–687. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Sitnik, M.; Wojcieszynska, D. Protocatechuate 3,4-dioxygenase: A wide substrate specificity enzyme isolated from Stenotrophomonas maltophilia KB2 as a useful tool in aromatic acid biodegradation. J. Mol. Microbiol. Biotechnol. 2014, 24, 150–160. [Google Scholar] [CrossRef]
- Tachedjian, G.; O’Hanlon, D.E.; Ravel, J. The implausible “in vivo” role of hydrogen peroxide as an antimicrobial factor produced by vaginal microbiota. Microbiome 2018, 6, 29. [Google Scholar] [CrossRef]
- Zvirbliene, A.; Pleckaityte, M.; Lasickiene, R.; Kucinskaite-Kodze, I.; Zvirblis, G. Production and characterization of monoclonal antibodies against vaginolysin: Mapping of a region critical for its cytolytic activity. Toxicon 2010, 56, 19–28. [Google Scholar] [CrossRef]
- Joris, B.; Englebert, S.; Chu, C.P.; Kariyama, R.; Daneo-Moore, L.; Shockman, G.D.; Ghuysen, J.M. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 1992, 70, 257–264. [Google Scholar] [CrossRef]
- Mitchell, W.J.; Tangney, M. Carbohydrate Uptake by the Phosphotransferase System and Other Mechanisums; Peter, D., Ed.; CRC Press: Boca Raton, FL, USA, 2005; pp. 155–175. [Google Scholar]
- Pedersen, K.; Zavialov, A.V.; Pavlov, M.Y.; Elf, J.; Gerdes, K.; Ehrenberg, M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 2003, 112, 131–140. [Google Scholar] [CrossRef]
- Wolber, J.M.; Urbanek, B.L.; Meints, L.M.; Piligian, B.F.; Lopez-Casillas, I.C.; Zochowski, K.M.; Woodruff, P.J.; Swarts, B.M. The trehalose-specific transporter LpqY-SugABC is required for antimicrobial and anti-biofilm activity of trehalose analogues in Mycobacterium smegmatis. Carbohydr. Res. 2017, 450, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.L.; Girerd, P.H.; Karjane, N.W.; Jefferson, K.K. Effect of biofilm phenotype on resistance of Gardnerella vaginalis to hydrogen peroxide and lactic acid. Am. J. Obstet. Gynecol. 2007, 197, 170.e1–170.e7. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).