Investigating Catheter-Related Infections in Southern Benin Hospitals: Identification, Susceptibility, and Resistance Genes of Involved Bacterial Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Study Design
2.3. Samples Collection
2.4. Isolation and Identification of Bacterial Strains
2.5. Antibiotic Susceptibility Testing of Identified Species
2.6. Molecular Identification of Resistance and Virulence Genes
2.7. Data Analysis
3. Results
3.1. Venous Catheter Infection
3.1.1. Catheterization in the Departmental Hospital of Lokossa and the Comè Zone Hospital
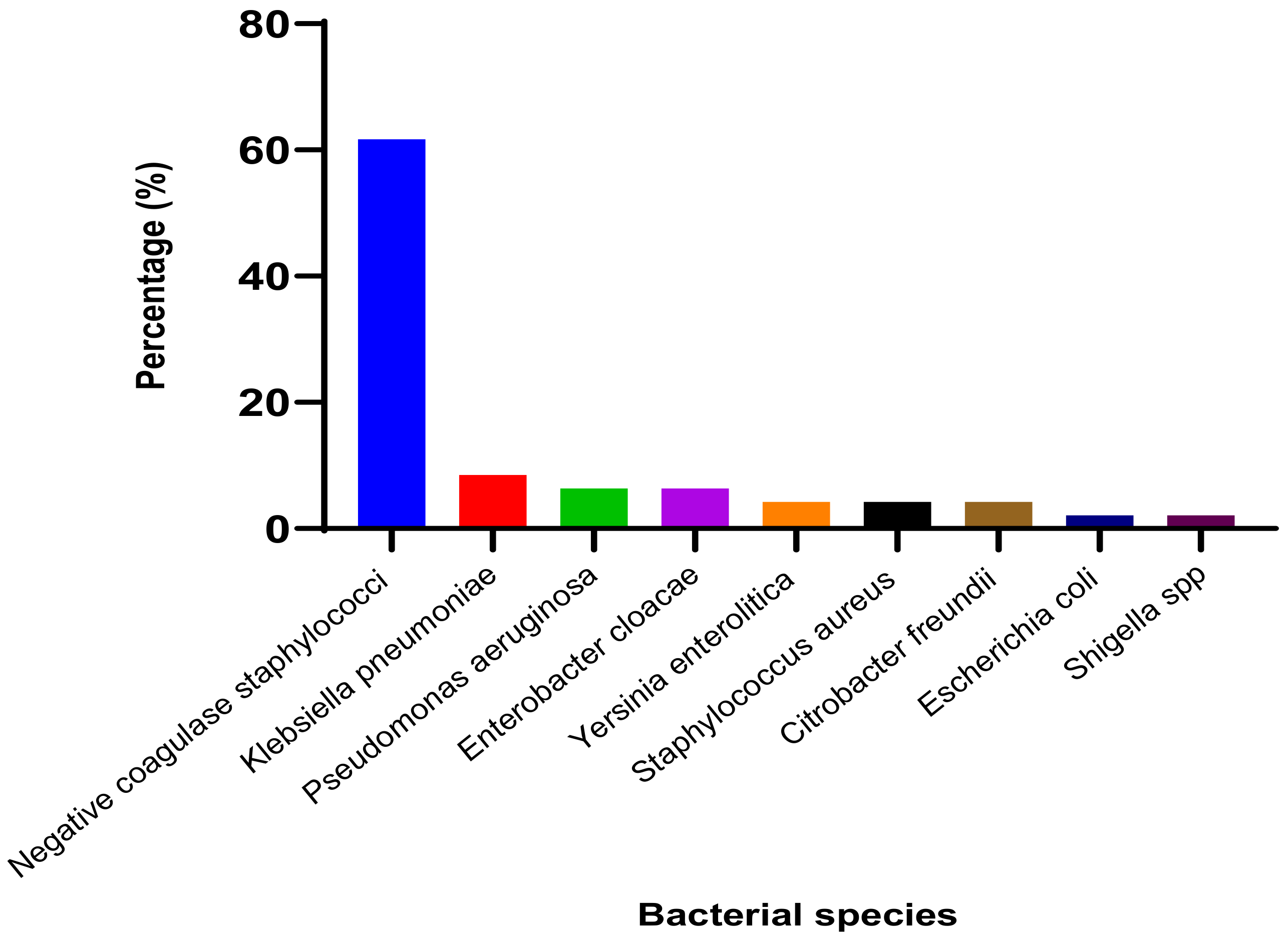
3.1.2. Identified Bacterial Species and Their Resistance Profile
3.1.3. Resistance and Virulence Genes Detected
3.2. Bladder Catheter Infections
3.2.1. Sociodemographic Data and Clinical Factors Related to the Occurrence of Urinary Tract Infections in Cotonou Hospitals in Benin
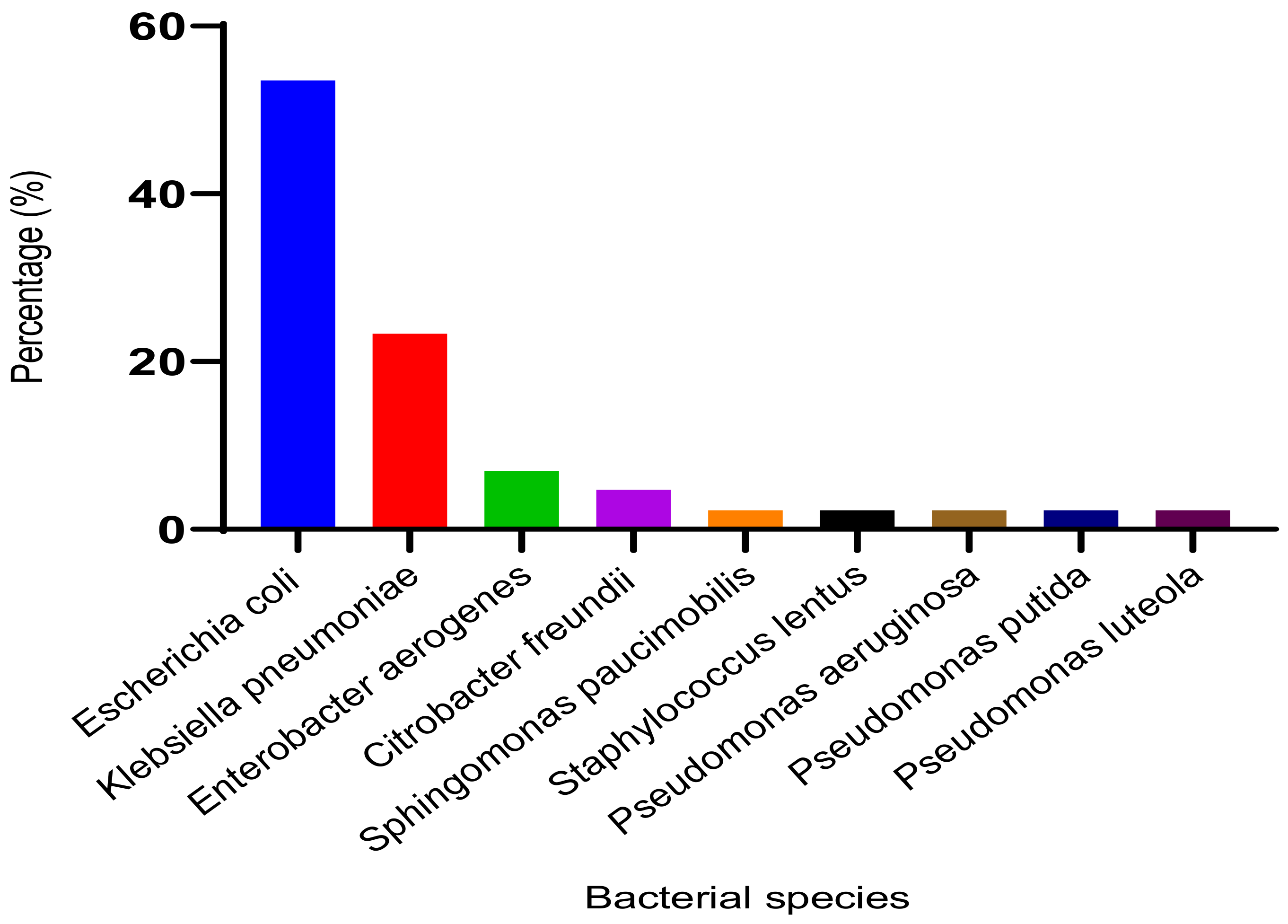
3.2.2. Identified Bacterial Species and Their Resistance Profile
3.2.3. Resistance and Virulence Genes Detected
4. Discussion
4.1. Epidemiological Characteristics of the Study Population and Aspects Related to In-Hospital Care
4.2. Bacterial Species Identified and Sensitivity to Antibiotics
4.3. Resistance and Virulence Genes Detected
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forrester, J.; Maggio, P.; Tennakoon, L. Cost of Health Care-Associated Infections in the United States. J. Patient Saf. 2022, 18, e477–e479. [Google Scholar] [CrossRef] [PubMed]
- Bagley, K.; Severud, L. Preventing Catheter-Associated Urinary Tract Infections with Incontinence Management Alternatives: PureWick and Condom Catheter. Nurs. Clin. N. Am. 2021, 56, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Chenoweth, C.E. Urinary Tract Infections: Update. Infect. Dis. Clin. 2021, 35, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Zikria, S.; Brian, G.; Mohamed, A.H.; Furqan, K.H.; Faiza, A.; Inayat, R. Point prevalence surveys of health-care associated infections: A systematic review. Pathog. Glob. Health 2019, 113, 191–205. [Google Scholar]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Aissaoui, Y.; Chouaib, N.; Chouikh, C.; Rafai, M.; Azendour, H.; Balkhi, H.; Haimeur, C.; Drissi Kamili, N. Central venous catheter-related bacteraemia: Prospective study in a Moroccan medical intensive care unit. Ann. Fr. d’Anesthésie Réanim. 2010, 29, 897–901. [Google Scholar] [CrossRef]
- Jiang, J.; DU, L.; Wang, X.; Huang, S.; Hu, W.; Zhou, L.; Liu, X. Specific nursing care improves postoperative urinary control function and self-efficacy in patients undergoing radical prostatectomy. Am. J. Transl. Res. 2022, 14, 1695. [Google Scholar]
- Merrer, J. Epidemiology of catheter–related infections in intensive care unit. Ann. Fr. d’Anesthésie Réanim. 2005, 24, 278–281. [Google Scholar] [CrossRef]
- Meddings, J.; Rogers, M.A.; Macy, M.; Saint, S. Systematic review and meta-analysis: Reminder systems to reduce catheter-associated urinary tract infections and urinary catheter use in hospitalized patients. Clin. Infect. Dis. 2010, 51, 550–560. [Google Scholar] [CrossRef]
- Elahi, R.; Siddiqui, M.H.; Rana, M.A.; Qayyum, M.A.; Iqbal, W.; Khalid, M.S.; Pervaiz, R.; Hafeez, M.M. The efficacy of taurolidine citrate solution versus heparin blocking solution instilled into the catheter lumens of end-stage renal disease. Pak. J. Med. Health Sci. 2022, 16, 152. [Google Scholar]
- Sikora, A.; Zahra, F. Nosocomial infections. StatPearls 2022, 5, 14–17. [Google Scholar]
- Gad, H.M.; Abdel-Aziz, H.H. Catheter-Associated Urinary Tract Infections in the Adult Patient Group: A Qualitative Systematic Review on the Adopted Preventative and Interventional Protocols from the Literature. Cureus 2021, 13, e16284. [Google Scholar] [CrossRef] [PubMed]
- Degbey, C.; Aguemon, B.; Ouendo, E.M.; Makoutode, M.; Simon, A. Study of the quality of medical-technical equipment used in operating theaters for the prevention of infections associated with care and services at the National Hospital and University Center of Cotonou in Benin. J. Soc. Clin. Biol. 2013, 18, 29–35. [Google Scholar]
- Dougnon, T.V.; Koudokpon, H.; Yossounon, C.; Fabiyi, K.; Legba, B.; Dougnon, J.; Baba-Moussa, L. Infection Risks and Antimicrobial Resistance in Tertiary Hospitals in Benin: Study Cases of Sakété-Ifangni and Menontin Hospitals. Int. J. Infect. 2020, 7, e99649. [Google Scholar] [CrossRef]
- Afle, F.C.D.; Agbankpe, A.J.; Johnson, R.C.; Houngbégnon, O.; Houssou, S.C.; Bankole, H.S. Healthcare-associated infections: Bacteriological characterization of the hospital surfaces in the University Hospital of Abomey-Calavi/So-ava in South Benin (West Africa). BMC Infect. Dis. 2019, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Legba, B.B.; Dougnon, V.; Koudokpon, H.; Mero, S.; Elovainio, R.; Parry, M.; Bankole, H.; Haukka, K. Assessment of blood cultures and antibiotic susceptibility testing for bacterial sepsis diagnosis and utilization of results by clinicians in Benin: A qualitative study. Front. Public Health 2023, 10, 1088590. [Google Scholar] [CrossRef]
- Dougnon, V.; Koudokpon, H.; Hounmanou, Y.M.; Azonbakin, S.; Fabiyi, K.; Oussou, A.; Codjia, I.; Ahoyo, A.; Baba-Moussa, L.; Bankolé, H. High Prevalence of Multidrug-Resistant Bacteria in the Centre Hospitalier et Universitaire de la Mère et de l’Enfant Lagune (CHU-MEL) Reveals Implications of Poor Hygiene Practices in Healthcare. SN Compr. Clin. Med. 2019, 1, 1029–1037. [Google Scholar] [CrossRef]
- Schwartz, D. La méthode statistique en médecine: Les enquêtes éthiologiques. Rev. Stat. Appl. 1960, 8, 5–27. [Google Scholar]
- CASFM “Comité de l’Antibiogramme de la Société Française de Microbilogie: Recommandations. 2021. Available online: https://www.sfmmicrobiologie.org/2021/05/06/casfm-eucast-2021-v2/ (accessed on 25 September 2022).
- Memariani, M.; Peerayeh, S.N.; Salehi, T.Z.; Mostafavi, S.K.S. Occurrence of SHV, TEM and CTX-M β-lactamase genes among Enteropathogenic Escherichia coli strains isolated from children with diarrhea. Jundishapur J. Microbiol. 2015, 8, e15620. [Google Scholar] [CrossRef]
- Cerezales, M.; Biniossek, L.; Gerson, S.; Xanthopoulou, K.; Wille, J.; Wohlfarth, E.; Kaase, M.; Seifert, H.; Higgins, P.G. Novel multiplex PCRs for detection of the most prevalent carbapenemase genes in gram-negative bacteria within Germany. J. Med. Microbiol. 2021, 70, 001310. [Google Scholar] [CrossRef]
- Hassan, D.; Omolo, C.A.; Gannimani, R.; Waddad, A.Y.; Mocktar, C.; Rambharose, S.; Agrawal, N.; Govender, T. Delivery of novel Vancomycin nanoplexes for combating methicillin resistant Staphylococcus aureus (MRSA) infections. Int. J. Pharm. 2019, 558, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ji, S.; Chen, Y.; Zhou, W.; Wei, Z.; Li, L.; Ma, Y. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J. Infect. 2007, 54, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Hassoun-Kheir, N.; Stabholz, Y.; Kreft, J.U.; De La Cruz, R.; Romalde, J.L.; Nesme, J.; Sørensen, S.J.; Smets, B.F.; Graham, D.; Paul, M. Comparison of antibiotic-resistant bacteria and antibiotic resistance genes abundance in hospital and community wastewater: A systematic review. Sci. Total Environ. 2020, 743, 140804. [Google Scholar] [CrossRef] [PubMed]
- Harbarth, S.; Balkhy, H.H.; Goossens, H.; Jarlier, V.; Kluytmans, J.; Laxminarayan, R.; Saam, M.; Van Belkum, A.; Pittet, D. For the World Healthcare-Associated Infections Resistance Forum participants. Antimicrobial resistance: One world, one fight! Antimicrob. Resist. Infect. Control 2015, 18, 49. [Google Scholar] [CrossRef]
- Kim, B.; Pai, H.; Choi, W.S.; Kim, Y.; Kweon, K.T.; Kim, H.A.; Ryu, S.Y.; Wie, S.H.; Kim, J. Current status of indwelling urinary catheter utilization and catheter-associated urinary tract infection throughout hospital wards in Korea: A multicenter prospective observational study. PLoS ONE 2017, 12, e0185369. [Google Scholar] [CrossRef] [PubMed]
- Kakaria, B.A.; Ashish, K.; Tushar, R. Study of incidence and risk factors for urinary tract infection in catheterized patients admitted at tertiary care. Int. J. Res. Med. Sci. 2015, 6, 1730–1733. [Google Scholar] [CrossRef]
- Li, F.; Song, M.; Xu, L.; Deng, B.; Zhu, S.; Li, X. Risk factors for catheter-associated urinary tract infection among hospitalized patients: A systematic review and meta-analysis of observational studies. J. Adv. Nurs. 2019, 75, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Faiq, A.; Najem, J.N. Some of clinical characteristics of neonatal sepsis in Children’s hospital at Kirkuk city, Iraq. Eurasian Med. Res. Period. 2022, 9, 133–138. [Google Scholar]
- Douard, M.C. Infections liées aux cathéters (ILC): Moyens diagnostiques. Nutr. Clin. Métab. 2002, 16, 59–61. [Google Scholar] [CrossRef]
- Vincitorio, D.; Barbadoro, P.; Pennacchietti, L.; Pellegrini, I.; David, S.; Ponzio, E.; Prospero, E. Risk factors for catheter-associated urinary tract infection in Italian elderly. Am. J. Infect. Control 2014, 42, 898–901. [Google Scholar] [CrossRef]
- Lachassinne, E.; Letamendia-Richard, E.; Gaudelus, J. Epidemiology of nosocomial infections in neonates. Arch. Pediatr. Organe Off. Soc. Fr. Pediatr. 2004, 11, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Faye, F.A.; Bingen, E. Place des infections nosocomiales en pédiatrie. Méd. Thér. Pédiatr. 2012, 15, 3–11. [Google Scholar]
- Kakupa, D.K.; Muenze, P.K.; Byl, B.; Wilmet, M.D. Etude de la prévalence des infections nosocomiales et des facteurs associés dans les deux hôpitaux universitaires de Lubumbashi, République Démocratique du Congo: Cas des Cliniques Universitaires de Lubumbashi et l’Hôpital Janson Sendwe. Pan Afr. Med. J. 2016, 24, 275. [Google Scholar] [CrossRef] [PubMed]
- Zemmour, H.; Derbale, F.Z. Incidence des Infections Liées aux Cathéters Veineux Centraux et Périphériques et Facteurs de Risque Attribuables au Niveau du Service de Chirurgie Générale «A» au CHU de Tlemcen. Ph.D. Thesis, Département de Pharmacie, Université Abou Bekr Belkaid, Tlemcen, Algeria, 2016; 150p. [Google Scholar]
- Floret, N.; Bertrand, X.; Thouverez, M.; Talon, D. Infections nosocomiales à Pseudomonas aeruginosa: Origine exogène ou endogène de la bactérie responsable? Pathol. Biol. 2009, 57, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Lukuke, H.M.; Kasamba, E.; Mahuridi, A.; Ngatu, N.R.; Narufumi, S.; Mukengeshayi, A.N.; Malou, V.; Makoutode, M.; Kaj, F.M. L’incidence des infections nosocomiales urinaires et des sites opératoires dans la maternité de l’Hôpital Général de Référence de Katuba à Lubumbashi en République Démocratique du Congo. Pan Afr. Med. J. 2017, 28. [Google Scholar] [CrossRef]
- Koçak, M.; Büyükkaragöz, B.; Çelebi Tayfur, A.; Çaltik, A.; Köksoy, A.Y.; Çizmeci, Z. Causative pathogens and antibiotic resistance in children hospitalized for urinary tract infection. Pediatr. Int. 2016, 58, 467–471. [Google Scholar] [CrossRef]
- Moutachakkir, M.; Chinbo, M.; Elkhoudri, N.; Soraa, N. Antibiotic resistance in uropathogenic Enterobacteriaceae in a pediatric environment at the University Hospital of Marrakech. J. Pediatr. Child. 2015, 28, 16–22. [Google Scholar]
- Raji, M.A.; Jamal, W.; Ojemhen, O.; Rotimi, V.O. Point-surveillance of antibiotic resistance in Enterobacteriaceae isolates from patients in a Lagos Teaching Hospital, Nigeria. J. Infect. Public Health 2013, 6, 431–437. [Google Scholar] [CrossRef]
- Rangaiahagari, A.; Uwizeyimana, J.P.; Nyirabanzi, J.; Ngoga, E.; Wane, J. Antibiotic sensitivity patterns of Enterobacteriaceae isolated at King Faisal Hospital, Kigali: A three years study. Rwanda Med. J. 2013, 70, 11–14. [Google Scholar]
- Sekhsokh, Y.; Chadli, M.; El Hamzaoui, S.A. Frequency and antibiotic susceptibility of bacteria identified in urine. Med. Infect. Dis. 2008, 38, 324–327. [Google Scholar]
- Sushitha, T.S.; Suseela, K.V. Catheter Associated Urinary Tract Infections: A Cross-sectional Study from a Tertiary Care Center in Kerala, India. Natl. J. Lab. Med. 2022, 11, 9–12. [Google Scholar] [CrossRef]
- Al-azawi, I.H.; Al-hamadan, A.H.; Hasson, S.O. Association between biofilm formation and antibiotic sensitivity in strains of Staphylococccus lentus isolated from urinary catheterized patients. Nano. Biomed. Eng. 2018, 10, 97–103. [Google Scholar] [CrossRef]
- Al-Salamy, M.H.; Darweesh, M.F.; Almousawi, A. Study of Staphylococcus lentus isolated from end stage renal failure patients and their antibiotic susceptibility patterns. World J. Pharm. Res. 2017, 6, 186–196. [Google Scholar]
- Zahir, H.; Draiss, G.; Rada, N.; Abourrahouat, A.; Sbihi, M.; Bouskraoui, M.; Soraa, N. Microbial ecology and sensitivity to antibiotics of bacteria isolated from urinary tract infections in children in Morocco. Francoph. Rev. Lab. 2019, 211, 65–70. [Google Scholar]
- Chaussade, H.; Sunder, S.; Bernard, L.; Coloby, P.; Guy, L.; Karsenty, G. Antibiotic drugs in urology. Progrès Urol. 2013, 23, 1327–1341. [Google Scholar] [CrossRef]
- Ouedraogo, A.S.; Pierre, H.J.; Banuls, A.-L.; Ouédraogo, R.; Godreuil, S. Emergence and spread of antibiotic resistance in West Africa: Contributing factors and threat assessment. Trop. Med. Health 2017, 27, 147–154. [Google Scholar] [CrossRef]
- Bernabe, K.J.; Langendorf, C.; Ford, N.; Ronat, J.B.; Murphy, R.A. Antimicrobial resistance in West Africa: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2017, 50, 629–639. [Google Scholar] [CrossRef]
- Konaré, S. Sensitivity to Antibiotics of Enterobacteriaceae Strains Isolated in 2016 at the Laboratory of Medical Biology and Hospital Hygiene of the CHU du Point G. Thesis, University of Sciences, Techniques and Technologies of Bamako, Bamako, Mali, 2016. [Google Scholar]
- Kbirou, A.; Alafifi, M.; Nedjim, S.A.; Abdi, E.M.; Moataz, A.; Dakir, M.; Debbagh, A.; Aboutaieb, R. Catheter-Associated Urinary Tract Infection, approximately 321 Cases. IJCMCR 2021, 17, 0045. [Google Scholar]
- Bitsori, M.; Maraki, S.; Koukouraki, S.; Galanakis, E. Pseudomonas aeruginosa urinary tract infection in children: Risk factors and outcomes. J. Urol. 2012, 187, 260–264. [Google Scholar] [CrossRef]
- Ajayi, A.O.; Osanyinlusi, S.A.; Ojerinde, A.; Ogeneh, B. Detection of Extended Spectrum Beta-lactamase Gene (CTX-M) among Representative Multidrug-Resistant Gram-Negative Bacterial Isolates from Patients with Urinary Tract Infection in Ekiti State, Nigeria. Int. J. Trop. Dis. Health 2022, 42, 59–64. [Google Scholar] [CrossRef]
- Rajivgandhi, G.; Maruthupandy, M.; Ramachandran, G.; Priyanga, M.; Manoharan, N. Detection of ESBL genes from ciprofloxacin resistant gram-negative bacteria isolated from urinary tract infections (UTIs). Front. Lab. Med. 2018, 2, 5–13. [Google Scholar] [CrossRef]
- Singh, S.B.; Barrett, J.F. Empirical antibacterial drug discovery—Foundation in natural products. Biochem. Pharmacol. 2006, 71, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Davies, J. Microbes have the last word: A drastic re-evaluation of antimicrobial treatment is needed to overcome the threat of antibiotic-resistant bacteria. EMBO Rep. 2007, 8, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Conen, A.; Frei, R.; Adler, H.; Dange, I.M.; Fux, C.A.; Widmer, A.F. Microbiological screening is necessary to distinguish carriers of plasmid-mediated AmpC beta-lactamase-producing enterobacteriaceae and extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae because of clinical similarity. PLoS ONE 2015, 10, e0120688. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, I.; Yushau, M.; Sharif, A.A.; Getso, M.I.; Yahaya, H.; Bala, J.A.; Aliyu, A.I.; Haruna, M. Detection of metallo-betalactamases among gram-negative bacterial isolates from Murtala Muhammad Specialist Hospital, Kano and Almadina Hospital Kaduna, Nigeria. Bayero J. Pure Appl. Sci. 2012, 5, 84–88. [Google Scholar] [CrossRef]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef]
- Martens, E.; Demain, A.L. The antibiotic resistance crisis, with a focus on the United States. J. Antibiot. 2017, 70, 520–526. [Google Scholar] [CrossRef]
- Al-Mahfoodh, W.J.M.; Pekacar, F.S.; Abbas, A.H. The molecular study for evaluation the antibiotic resistance of Escherichia coli and Klebsiella pneumoniae bacteria isolated from urinary tract infection patients. Gene Rep. 2021, 25, 101423. [Google Scholar] [CrossRef]
- El-Houssaini, H.H.; Elnabawy, O.M.; Nasser, H.A.; Elkhatib, W.F. Correlation between antifungal resistance and virulence factors in Candida albicans recovered from vaginal specimens. Microb. Pathog. 2019, 128, 13–19. [Google Scholar] [CrossRef]
| Genes | Primers | Sequence 5′-3′ | References |
|---|---|---|---|
| blaTEM | TEM F | ATGAGTATTCAACATTTCCGC | [20] |
| TEM R | CAATGCTTAATCAGTGAGG | ||
| blaSHV | SHV F | AAGATCCACTATCGCCAGCAG | |
| SHV R | ATTCAGTTCCGTTTCCCAGCGG | ||
| blaCTX-M-1 | CTX-M-1 F | GGTTAAAAAATCACTGCGTC | |
| CTX-M-1 R | TTGGTGACGATTTTAGCCGC | ||
| blaCTX-M-15 | CTX-M-15 F | CACACGTGGAATTTAGGGACT | |
| CTX-M-15 R | GCCGTCTAAGGCGATAAACA | ||
| blaKPC | KPC F | CGCCAATTTGTTGCTGAAGG | [21] |
| KPC R | CAGGTTCCGGTTTTGTCTCC | ||
| blaNDM | NDM F | GTTTGATCGTCAGGGATGGC | |
| NDM R | CTCATCACGATCATGCTGGC | ||
| blaVIM | VIM F | GATGGTGTTTGGTCGCATATC | |
| VIM R | CGTCATGAAAGTGCGTGGAG | ||
| blaIMP | IMP F | GAAGGCGTTTATGTTCATAC | |
| IMP R | GTACGTTTCAAGAGTGATGC | ||
| blaOXA-48 | OXA-48 F | GGTAGCAAAGGAATGGCAAGAA | |
| OXA-48 R | CGACCCACCAGCCAATCTTA | ||
| vanA | vanA F | GGGCTGTGAGGTCGGTTG | [22] |
| vanA R | TTCAGTACAATGCGGCCGTTA | ||
| vanB | vanB F | TTGTCGGCGAAGTGGATCA | |
| vanB R | AGCCTTTTTCCGGCTCGTT | ||
| mecA | mecA F | GTTAGATTGGGATCATAGCGTCATT | [23] |
| mecA R | TGCCTAATCTCATATGTGTTCCTGTAT | ||
| fimH | fimH F | TACTGCTGATGGGGCTGGTC | [20,24] |
| fimH R | TACTGCTGATGGGGCTGGTC | ||
| ISS | ISS F | GGCAATGCTTATTACAGGATGTGC | |
| ISS R | GAGCAATATACCCGGGGCTTCC |
| Positive | Negative | Total | Chi2 | Odds Ratio | Z | |
|---|---|---|---|---|---|---|
| Sex | ||||||
| Male | 9 (9.5%) | 27 (28.4%) | 36 (37.9%) | 0.005 | 0.068 | −3.53 |
| Female | 32 (33.7%) | 27 (28.4%) | 59 (62.1%) | |||
| Age | ||||||
| (0–10) | 13 (13.7%) | 12 (12.6) | 25 (26.3%) | 0.330 | 0.91 | −0.53 |
| (10–20) | 4 (4.2%) | 6 (6.3%) | 10 (10.5%) | |||
| (20–30) | 11 (11.6%) | 13 (13.7%) | 24 (25.3%) | |||
| (30–40) | 2 (2.1%) | 11 (11.6%) | 13 (13.7%) | |||
| (40–50) | 7 (7.4%) | 4 (4.2%) | 11 (11.6%) | |||
| (50–60) | 2 (2.1%) | 2 (2.1%) | 4 (4.2%) | |||
| (60–70) | 1 (1.1%) | 3 (3.2%) | 4 (4.2%) | |||
| (70–80) | 1 (1.1%) | 3 (3.2%) | 4 (4.2%) | |||
| Professional category | ||||||
| Nurse | 33 (34.7%) | 42 (44.2%) | 75 (79%) | 0.552 | 0.818 | −0.45 |
| Health care aide | 1 (1.1%) | 4 (4.2%) | 5 (5.3%) | |||
| Midwife | 7 (7.4%) | 8 (8.4%) | 15 (15.8 | |||
| Catheter size | ||||||
| G18 | 5 (5.3%) | 11 (11.6%) | 16 (16.8%) | 0.173 | 0.348 | −239 |
| G20 | 16 (16.8%) | 27 (28.4%) | 45 (45.3%) | |||
| G22 | 7 (7.37%) | 3 (3.16%) | 10 (10.5%) | |||
| G24 | 13 (13.7%) | 13 (13.7%) | 26 (27.4%) | |||
| Type of veins | ||||||
| Peripheral | 40 (42.1%) | 53 (55.8%) | 93 (97.9%) | 0.843 | 0.15 | −1.04 |
| Central | 1 (1.1%) | 1 (1.1%) | 2 (2.1%) | |||
| Placement site | ||||||
| Upper extremity | 39 (41.05%) | 50 (52.6%) | 89 (93.7%) | 0.744 | 13.83 | 1.44 |
| Lower limb | 1 (1.1%) | 3 (3.2%) | 4 (4.2%) | |||
| Jugular | 1 (1.01%) | 1 (1.1%) | 2 (2.1%) | |||
| Use of antiseptics before | ||||||
| Yes | 10 (10.5%) | 10 (10.5%) | 20 (21.1%) | 0.487 | 3.56 | 1.56 |
| No | 31 (32.6%) | 44 (46.3%) | 75 (79%) | |||
| Used solution | ||||||
| Alcohol 70 | 39 (41.1%) | 48 (50.5%) | 87 (91.6%) | 0.396 | 1.5 | 0.63 |
| Beta-block alcohol | 1 (1.1%) | 1 (1.1%) | 2 (2.1%) | |||
| Others | 1 (1.1%) | 5 (5.3%) | 6 (6.3%) | |||
| Disinfection quality | ||||||
| Good | 41 (43.2%) | 54 (56.8%) | 95 (100%) | |||
| Duration of catheterization | ||||||
| 1 | 1 (1.1%) | 0 (0%) | 1 (1.1%) | 0.492 | 1.2 | 0.90 |
| 2 | 7 (7.4%) | 9 (9.5%) | 16 (16.8%) | |||
| 3 | 15 (15.8%) | 13 (13.7%) | 28 (29.45%) | |||
| 4 | 10 (10.5%) | 14 (14.7%) | 24 (25.3%) | |||
| 5 | 6 (6.3%) | 12 (122%) | 18 (19%) | |||
| 6 | 1 (1.1%) | 3 (3.2%) | 4 (4.2%) | |||
| 7 | 0 (0%) | 2 (2.1%) | 2 (2.1%) | |||
| 8 | 0 (0%) | 1 (1.1%) | 1 (1.1%) | |||
| >8 | 1 (1.1%) | 0 (0%) | 1 (1.1%) | |||
| Presence of varnish after catheter placement | ||||||
| Present | 4 (4.2%) | 8 (8.4%) | 12 (12.6%) | 0.462 | 0.66 | −0.51 |
| Absent | 37 (39%) | 46 (48.4%) | 83 (83.4%) | |||
| Hand washing before catheter insertion | ||||||
| Yes | 32 (33.7%) | 39 (41.1%) | 71 (74.7%) | 0.517 | 1.89 | 0.90 |
| No | 9 (9.5%) | 15 (15.8%) | 24 (25.3%) | |||
| Appearance of the bandages upon removal | ||||||
| Dirty | 22 (23.2%) | 23 (24.2%) | 45 (47.4%) | 0.0013 | 1.185 | 0.73 |
| Unstuck | 3 (3.2%) | 0 (0%) | 3 (3.2%) | |||
| Unstuck and dirty | 2 (2.1%) | 14 (14.7%) | 16 (16.8%) | |||
| Hermeneutical and clean | 14 (14.7%) | 17 (17.9%) | 31 (32.6%) | |||
| Nature of the injected product | ||||||
| Antibiotic | 34 (35.8%) | 52 (54.7%) | 86 (90.5%) | 0.028 | 0.087 | −2.21 |
| Not antibiotic | 7 (7.4%) | 2 (2.1%) | 9 (9.3%) | |||
| AMP | OX | TOB | C | E | NA | FOX | VAN | |
|---|---|---|---|---|---|---|---|---|
| S. aureus | - | 1 (50%) | - | 2 (100%) | 1 (50%) | 1 (50%) | - | 1 (50%) |
| NCS | 5 (17%) | 9 (31%) | 11 (38%) | 15 (52%) | 11 (38%) | 20 (72%) | 4 (14%) | 4 (14%) |
| Total | 5 (16.1%) | 10 (32.4%) | 11 (35.5%) | 17 (54.8%) | 12 (38.7%) | 21 (67.7%) | 4 (12.9%) | 5 (16.1%) |
| AMP | AMC | CRO | IMP | KAN | GEN | NA | FOX | |
|---|---|---|---|---|---|---|---|---|
| Yersinia enterolitica | 1 (50%) | 1 (50%) | 1 (50%) | - | 1 (50%) | 1 (50%) | - | - |
| Shigella spp. | - | 1 (100%) | - | - | - | - | 1 (100%) | - |
| Klebsiella pneumoniae | 4 (100%) | 4 (100%) | 2 (50%) | - | 2 (50%) | 2 (50%) | 2 (50%) | - |
| Enterobacter cloaceae | 3 (100%) | 2 (75%) | 3 (100%) | - | 3 (100%) | 3 (100%) | - | 1 (25%) |
| Pseudomonas aeruginosa | 3 (100%) | 1 (33%) | 3 (100%) | - | 1 (33%) | 1 (33%) | 1 (33%) | 3 (100%) |
| Escherichia coli | 1 (100%) | 1 (100%) | 1 (100%) | - | 1 (100%) | 1 (100%) | - | - |
| Citrobacter freundii | 1 (50%) | - | 1 (50%) | - | - | 2 (100%) | 1 (50%) | - |
| Total | 13 (81.3%) | 10 (62.5%) | 11 (68.8%) | - | 8 (50%) | 10 (62.5%) | 5 (31.3%) | 4 (25%) |
| Resistance Genes | Virulence Genes | ||||
|---|---|---|---|---|---|
| blaTEM | blaSHV | blaCTX-M-15 | fimH | ISS | |
| Yersinia enterolitica | 1 (50%) | 1 (50%) | 2 (100%) | - | - |
| Shigella spp. | 1 (100%) | 1 (100%) | - | - | - |
| Klebsiella pneumoniae | - | 4 (100%) | 1 (25%) | 4 (100%) | - |
| Enterobacter cloaceae | 3 (100%) | - | 3 (100%) | - | - |
| Pseudomonas aeruginosa | - | - | - | - | - |
| Escherichia coli | 1 (100%) | - | 1 (100%) | - | - |
| Citrobacter freundii | 1 (50%) | - | 2 (100%) | - | - |
| Total | 7 (54%) | 6 (37.5%) | 8 (50%) | 4 (25%) | - |
| Variable | Negative | Positive | Total | Chi2 | OR |
|---|---|---|---|---|---|
| Sex | |||||
| Female | 176 (50%) | 46 (14.7%) | 202 (64.7%) | 0.070 | 1.72 |
| Male | 70 (22.4%) | 40 (12.8%) | 110 (35.3%) | ||
| Total | 226 (72.4%) | 86 (27.6%) | 312 (100%) | ||
| Age | |||||
| (20, 30) | 52 (16.7%) | 18 (5.7%) | 70 (22.4%) | 0.103 | 0.96 |
| (30, 40) | 62 (19.87%) | 14 (4.5%) | 76 (24.4%) | ||
| (40, 50) | 38 (12.2%) | 16 (5.1%) | 54 (17.3%) | ||
| (50, 60) | 24 (7.7%) | 22 (7.1%) | 46 (14.7%) | ||
| (60, 70) | 30 (9.62%) | 14 (4.50%) | 44 (14.1%) | ||
| (70, 80) | 18 (5.8%) | 0 (0.00%) | 18 (5.6%) | ||
| (80, 90) | 2 (0.6%) | 2 (0.6%) | 4 (1.3%) | ||
| Variable | Negative | Positive | Total | Pr Chi2 | OR |
|---|---|---|---|---|---|
| Routine antibiotic therapy | |||||
| Yes | 110 (35.3%) | 56 (18%) | 166 (53.2%) | 0.066 | 0.36 |
| No | 116 (37.2%) | 30 (9.6%) | 146 (46.8%) | ||
| Total | 226 (72.4%) | 86 (27.6%) | 312 (100%) | ||
| Type of antibiotic therapy | |||||
| C3G + Quinolones | 6 (1.9%) | 6 (1.9%) | 12 (3.9%) | 0.145 | 0.96 |
| 3G + SXT | 22 (7.1%) | 2 (0.6%) | 24 (7.7%) | ||
| C3G + Aminosides | 4 (1.3%) | 2 (0.6%) | 6 (1.9%) | ||
| C3G | 58 (18.7%) | 38 (12.3%) | 96 (31%) | ||
| No ATB | 122 (38.7%) | 32 (10.3%) | 154 (49%) | ||
| Aminosides | 6 (1.9%) | 4 (1.3%) | 10 (3.2%) | ||
| Quinolones | 2 (0.6%) | 2 (0.6%) | 4 (1.3%) | ||
| Glycopeptides | 6 (1.9%) | 0 (0.0%) | 6 (1.9%) | ||
| Total | 226 (72.4%) | 86 (27.6%) | 312 (100%) | ||
| Reason for hospitalization | |||||
| Uncomplicated delivery | 8 (2.6%) | 0 (0.00%) | 8 (2.6%) | 0.517 | 0.97 |
| Diabetic ketoacidosis | 8 (2.6%) | 8 (2.6%) | 16 (5.1%) | ||
| Change in general condition | 24 (7.70%) | 6 (1.9%) | 30 (9.6%) | ||
| Stroke | 24 (7.7%) | 6 (1.9%) | 30 (9.6%) | ||
| Cesarean section | 62 (19.9%) | 14 (4.5%) | 76 (24.4%) | ||
| Coma | 2 (0.6%) | 2 (0.6%) | 4 (1.3%) | ||
| Gastroenteritis | 6 (1.9%) | 4 (1.3%) | 10 (3.2%) | ||
| Head trauma | 26 (8.3%) | 8 (2.6%) | 34 (10.9%) | ||
| Infectious syndrome | 8 (2.6%) | 8 (2.6%) | 16 (5.1%) | ||
| Postoperative monitoring | 16 (5.1%) | 10 (3.2%) | 26 (8.3%) | ||
| Acute retention of urine | 12 (3.9%) | 4 (1.3%) | 16 (5.1%) | ||
| Renal failure | 8 (2.6%) | 2 (0.6%) | 10 (3.2%) | ||
| Other conditions | 22 (7.1%) | 14 (4.5%) | 36 (11.5%) | ||
| Total | 226 (72.4%) | 86 (27.6%) | 312 (100%) | ||
| Antibiotics | AMX | CMA | TIC | TZP | CF | FOX | CTX | CAZ | ERT | IMI | AN | GEN | TOB | N/A | CIP | OFX | TIN | SXT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | 100 | 21.74 | 100 | 17.39 | 32.56 | 30.43 | 43.48 | 43.48 | - | - | - | 56.52 | 30.43 | 86.96 | 73.91 | 69.56 | 21.74 | 95.65 |
| Klebsiella pneumoniae | 100 | 40 | 100 | 30 | 80 | 30 | 80 | 80 | - | 30 | 10 | 80 | 20 | 70 | 50 | 50 | 30 | 70 |
| Enterobacter aerogenes | 25 | 75 | 100 | - | 75 | 75 | 50 | - | - | - | - | 50 | 50 | 75 | 50 | 50 | 25 | 100 |
| Citrobacter freundii | - | 100 | 50 | - | 100 | 100 | - | - | - | - | - | 50 | 50 | 100 | 50 | 50 | - | 50 |
| Pseudomonas lutola | - | - | - | 100 | - | - | - | 100 | - | - | 100 | - | 100 | - | 100 | 100 | - | 100 |
| Pseudomonas aeruginosa | - | - | - | - | - | - | 100 | - | - | - | - | - | - | - | - | - | - | - |
| Pseudomonas putida | - | - | 100 | 100 | - | - | 100 | - | - | 100 | - | 100 | - | - | 100 | 100 | - | 100 |
| Sphingomonas paucimobilis | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 0 | 100 | 100 | 100 | 100 | 0 | 100 | 100 | 0 | 100 |
| Isolated Species | Genes | ||||
|---|---|---|---|---|---|
| blaTEM | blaSHV | blaCTXM15 | blaNDM | blaCTXM1 | |
| Escherichia coli | 20 (50%) | 10 (25%) | 6 (15%) | 4 (10%) | 12 (30%) |
| Klebsiella pneumoniae | 10 (55.6%) | 8 (44.4%) | 4 (22.2%) | 2 (11.1%) | 6 (33.3%) |
| Citrobacter freundii | 2 (100%) | - | 2 (100%) | - | 2 (100%) |
| Pseudomonas putida | 2 (100%) | 2 (100%) | 2 (100%) | - | 2 (100%) |
| Sphingomonas paucimobilis | - | - | - | - | - |
| Enterobacter aerogenes | 2 (100%) | 2 (100%) | - | - | - |
| Total | 36 (42.9%) | 22 (26.2%) | 14 (16.7%) | 6 (7.2%) | 24 (28.6%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dougnon, V.T.; Sintondji, K.; Koudokpon, C.H.; Houéto, M.; Agbankpé, A.J.; Assogba, P.; Oussou, A.; Gnamy, A.; Legba, B.; Idrissou, A.; et al. Investigating Catheter-Related Infections in Southern Benin Hospitals: Identification, Susceptibility, and Resistance Genes of Involved Bacterial Strains. Microorganisms 2023, 11, 617. https://doi.org/10.3390/microorganisms11030617
Dougnon VT, Sintondji K, Koudokpon CH, Houéto M, Agbankpé AJ, Assogba P, Oussou A, Gnamy A, Legba B, Idrissou A, et al. Investigating Catheter-Related Infections in Southern Benin Hospitals: Identification, Susceptibility, and Resistance Genes of Involved Bacterial Strains. Microorganisms. 2023; 11(3):617. https://doi.org/10.3390/microorganisms11030617
Chicago/Turabian StyleDougnon, Victorien Tamègnon, Kevin Sintondji, Charles Hornel Koudokpon, Morènikè Houéto, Alidehou Jerrold Agbankpé, Phénix Assogba, Alida Oussou, Anderson Gnamy, Boris Legba, Abdoulaye Idrissou, and et al. 2023. "Investigating Catheter-Related Infections in Southern Benin Hospitals: Identification, Susceptibility, and Resistance Genes of Involved Bacterial Strains" Microorganisms 11, no. 3: 617. https://doi.org/10.3390/microorganisms11030617
APA StyleDougnon, V. T., Sintondji, K., Koudokpon, C. H., Houéto, M., Agbankpé, A. J., Assogba, P., Oussou, A., Gnamy, A., Legba, B., Idrissou, A., & Bankole, H. S. (2023). Investigating Catheter-Related Infections in Southern Benin Hospitals: Identification, Susceptibility, and Resistance Genes of Involved Bacterial Strains. Microorganisms, 11(3), 617. https://doi.org/10.3390/microorganisms11030617





