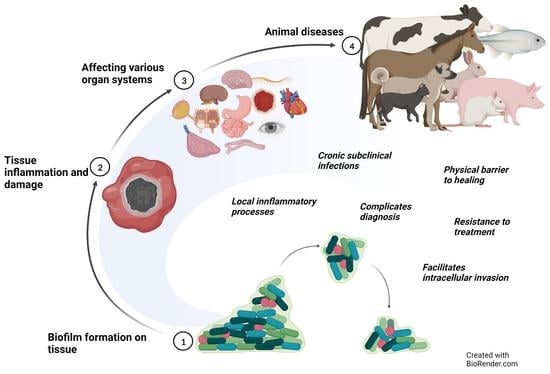
The Role of Biofilms in the Pathogenesis of Animal Bacterial Infections
Abstract
1. Introduction
2. Auditory System
Canine Otitis Externa (OE)
3. Cardiovascular System
Bovine Myocarditis
4. Central Nervous System
Chronic Streptococcal Meningoencephalitis in Fish
5. Digestive System
Salmonella Infections
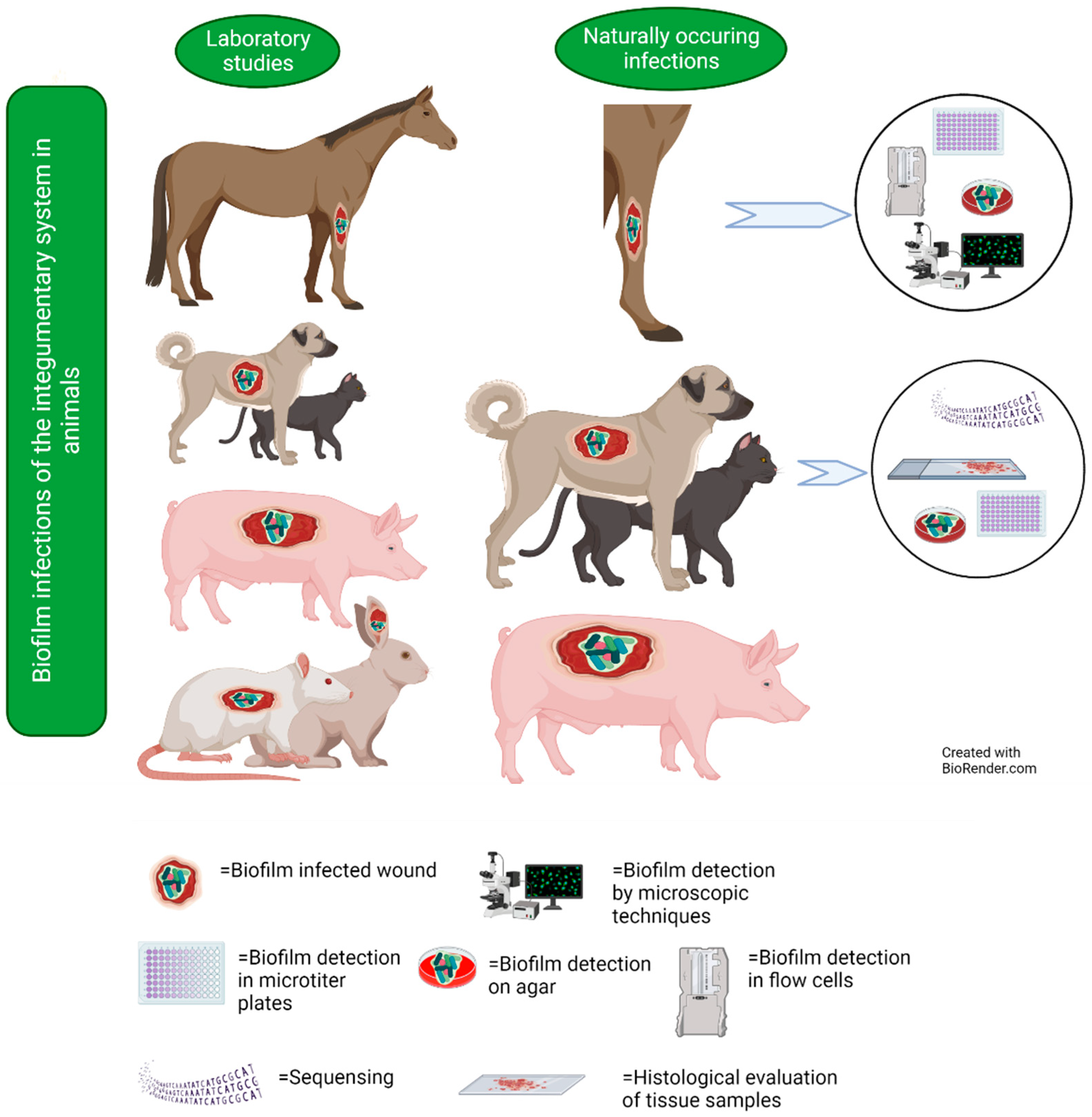
6. Integumentary System
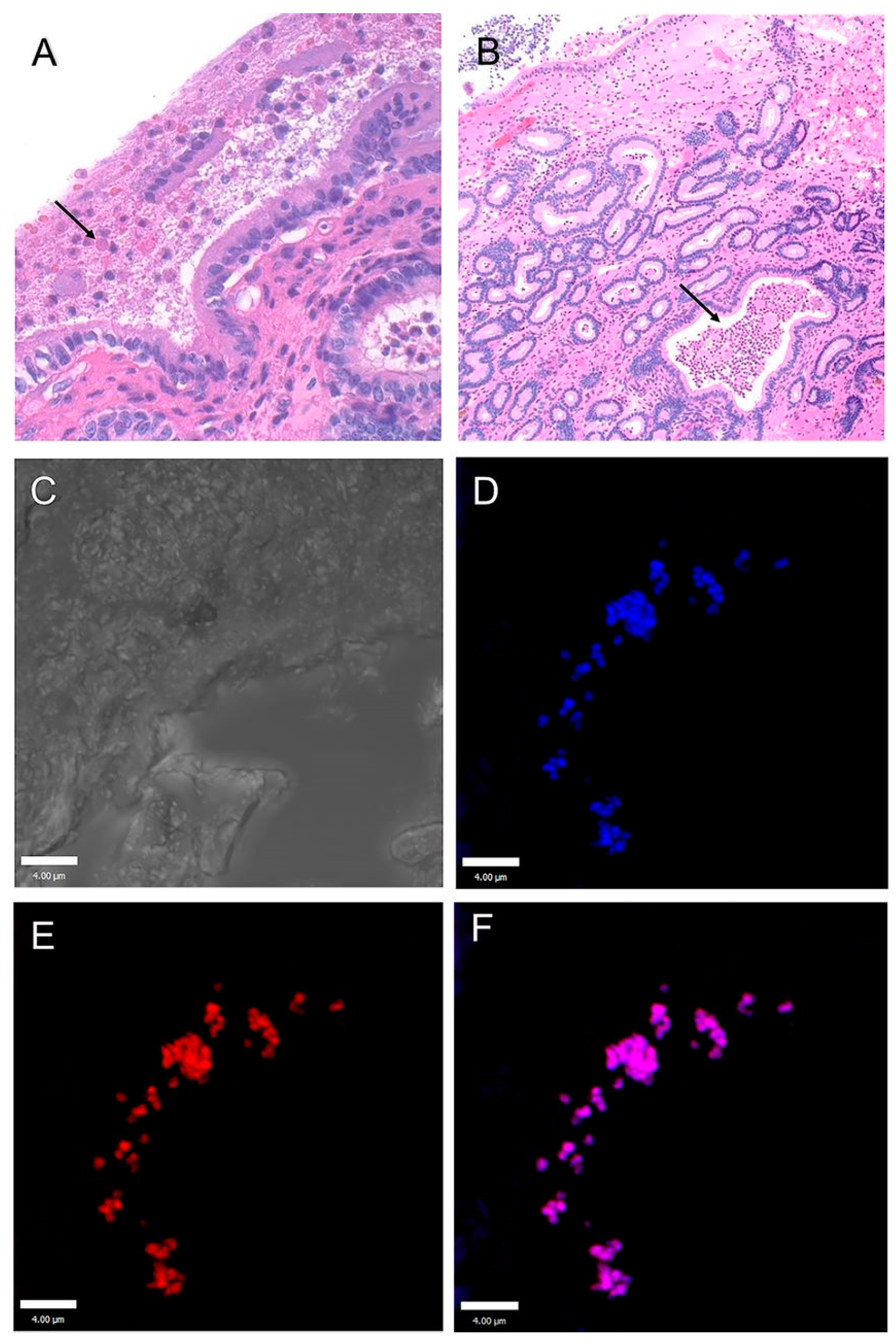
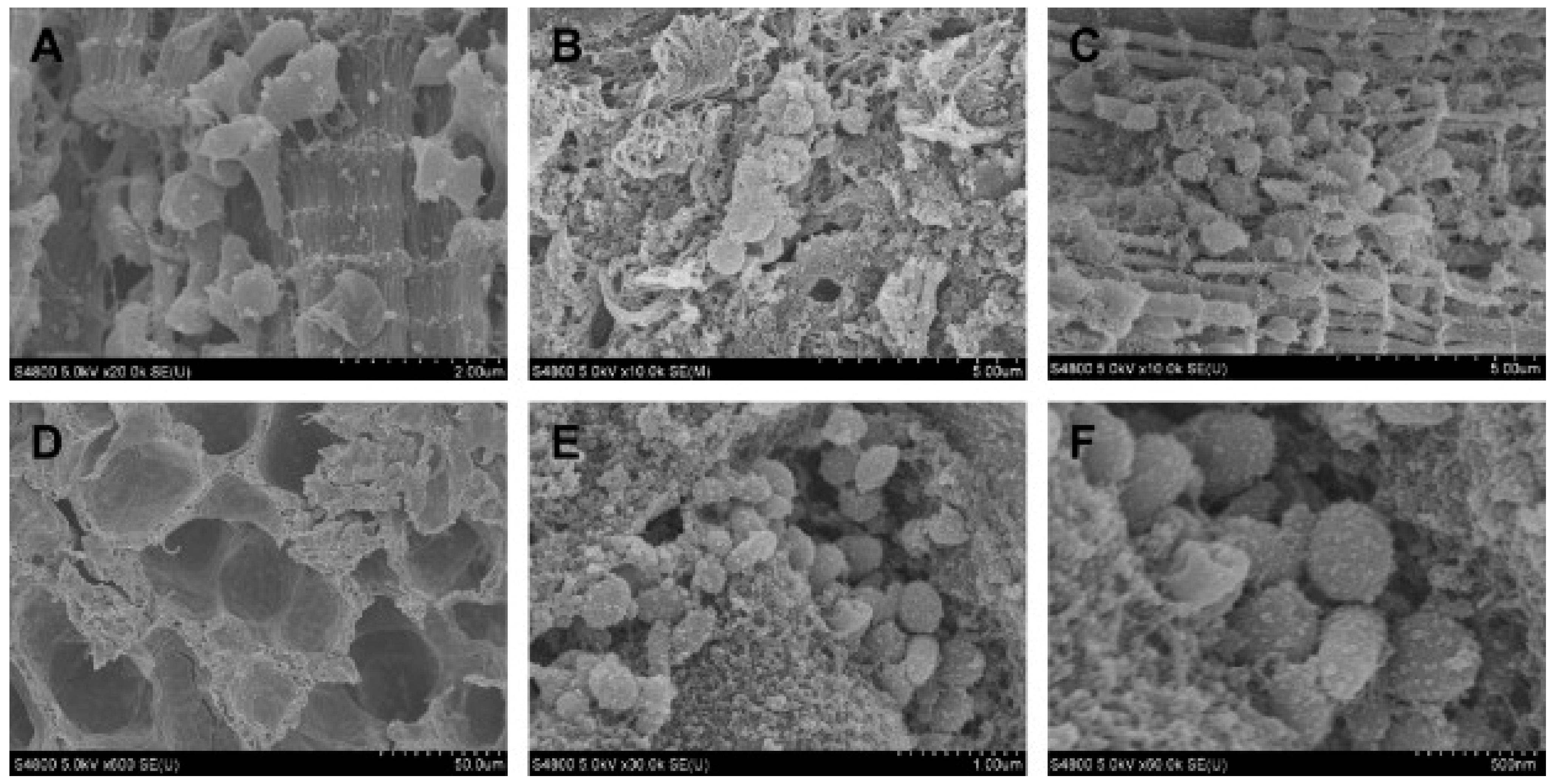
6.1. Studies on Naturally Occurring Wound Infections
6.2. Laboratory Studies on Wound Infections
7. Reproductive System
7.1. Endometritis
7.1.1. Endometritis in Dogs
7.1.2. Endometritis in Mares
7.2. Endometritis in Cows
7.3. Mastitis
Mastitis in Cows
8. Respiratory System
8.1. Bovine Respiratory Disease Complex (BRDC)
8.2. Porcine Enzootic Pneumonia
8.3. Porcine Pleuropneumonia
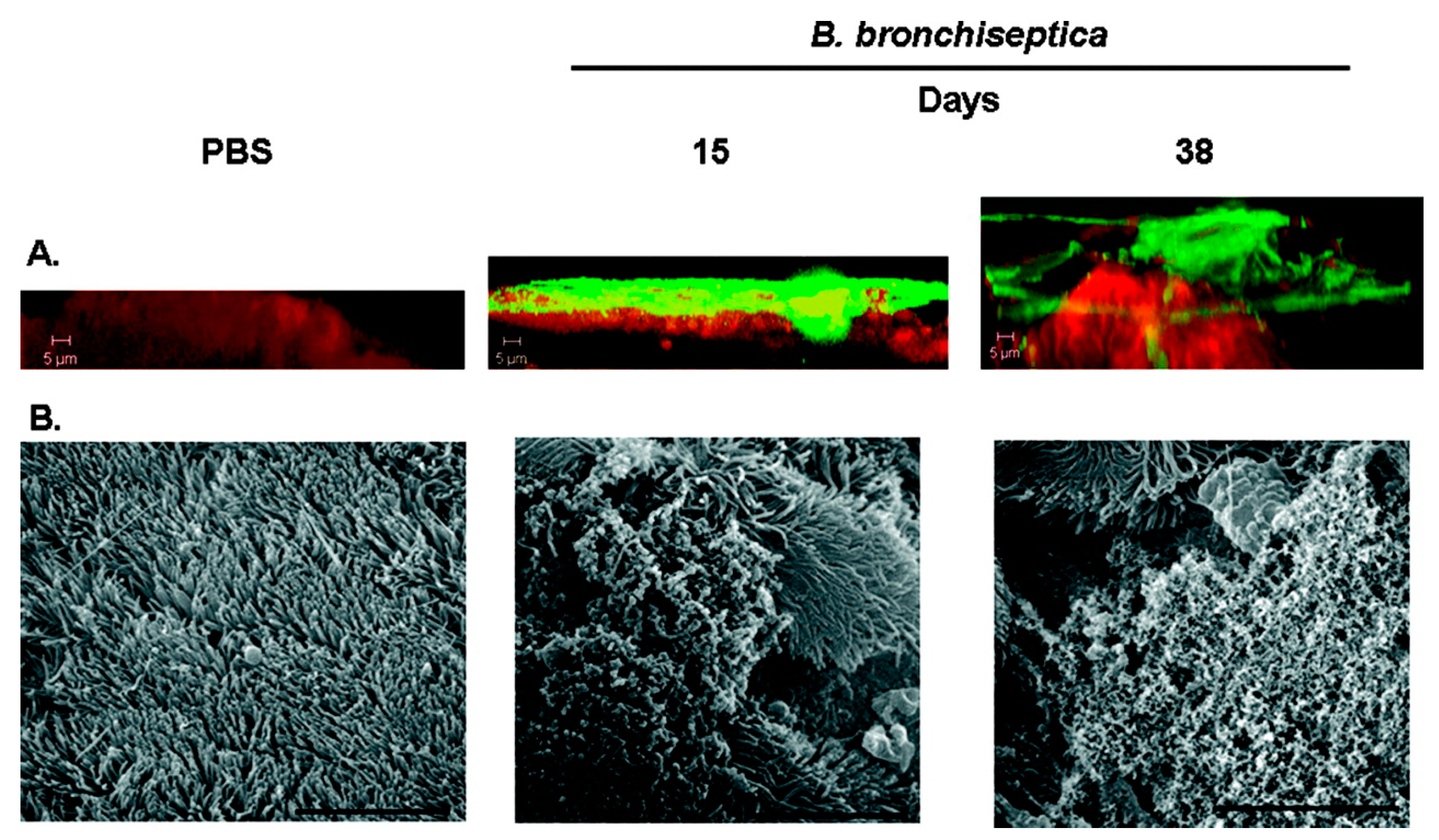
8.4. Bordetella Bronchiseptica Infections
9. Skeletal System
10. Urinary System
Urinary Tract Infections
11. Visual System
Equine Recurrent Uveitis
12. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013, 121, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W. The etiology and persistence of cryptic bacterial infections: A hypothesis. Rev. Infect. Dis. 1984, 6 (Suppl. S3), S608–S616. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Stoodley, P. Evolving concepts in biofilm infections. Cell Microbiol. 2009, 11, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N. A short history of microbial biofilms and biofilm infections. APMIS 2017, 125, 272–275. [Google Scholar] [CrossRef]
- Vestby, L.K.; Gronseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Alhede, M.; Alhede, M.; Eickhardt-Sorensen, S.R.; Moser, C.; Kuhl, M.; Jensen, P.O.; Hoiby, N. The in vivo biofilm. Trends Microbiol. 2013, 21, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Anju, V.T.; Busi, S.; Imchen, M.; Kumavath, R.; Mohan, M.S.; Salim, S.A.; Subhaswaraj, P.; Dyavaiah, M. Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies. Antibiotics 2022, 11, 1731. [Google Scholar] [CrossRef]
- Peters, B.M.; Jabra-Rizk, M.A.; Scheper, M.A.; Leid, J.G.; Costerton, J.W.; Shirtliff, M.E. Microbial interactions and differential protein expression in Staphylococcus aureus-Candida albicans dual-species biofilms. FEMS Immunol. Med. Microbiol. 2010, 59, 493–503. [Google Scholar] [CrossRef]
- Tolker-Nielsen, T. Biofilm Development. Microbiol. Spectr. 2015, 3, 51–66. [Google Scholar] [CrossRef]
- Solano, C.; Echeverz, M.; Lasa, I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014, 18, 96–104. [Google Scholar] [CrossRef]
- Songtanin, B.; Peterson, C.J.; Molehin, A.J.; Nugent, K. Biofilms and Benign Colonic Diseases. Int. J. Mol. Sci. 2022, 23, 14259. [Google Scholar] [CrossRef] [PubMed]
- Uruen, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as Promoters of Bacterial Antibiotic Resistance and Tolerance. Antibiotics 2020, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Rybtke, M.; Hultqvist, L.D.; Givskov, M.; Tolker-Nielsen, T. Pseudomonas aeruginosa Biofilm Infections: Community Structure, Antimicrobial Tolerance and Immune Response. J. Mol. Biol. 2015, 427, 3628–3645. [Google Scholar] [CrossRef] [PubMed]
- Zare, M.; Vehreschild, M.; Wagenlehner, F. Management of uncomplicated recurrent urinary tract infections. BJU Int. 2022, 129, 668–678. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Hoiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef]
- Totani, T.; Nishiuchi, Y.; Tateishi, Y.; Yoshida, Y.; Kitanaka, H.; Niki, M.; Kaneko, Y.; Matsumoto, S. Effects of nutritional and ambient oxygen condition on biofilm formation in Mycobacterium avium subsp. hominissuis via altered glycolipid expression. Sci. Rep. 2017, 7, 41775. [Google Scholar] [CrossRef]
- Chan, W.Y.; Hickey, E.E.; Page, S.W.; Trott, D.J.; Hill, P.B. Biofilm production by pathogens associated with canine otitis externa, and the antibiofilm activity of ionophores and antimicrobial adjuvants. J. Vet. Pharmacol. Ther. 2019, 42, 682–692. [Google Scholar] [CrossRef]
- Parnell-Turner, H.; Griffin, C.E.; Rosenkrantz, W.S.; Kelly Keating, M.; Bidot, W.A. Evaluation of the use of paired modified Wright’s and periodic acid Schiff stains to identify microbial aggregates on cytological smears of dogs with microbial otitis externa and suspected biofilm. Vet. Dermatol. 2021, 32, 448-e122. [Google Scholar] [CrossRef]
- Sandal, I.; Shao, J.Q.; Annadata, S.; Apicella, M.A.; Boye, M.; Jensen, T.K.; Saunders, G.K.; Inzana, T.J. Histophilus somni biofilm formation in cardiopulmonary tissue of the bovine host following respiratory challenge. Microbes Infect. 2009, 11, 254–263. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, D.; Allen, T.; Hunter, R.; Corbeil, L.B. Diagnostic Exercise: Myocarditis Due to Histophilus somni in Feedlot and Backgrounded Cattle. Vet. Pathol. 2009, 46, 1015–1017. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, D.; Sondgeroth, K.S. Histophilosis as a Natural Disease; Springer International Publishing: Cham, Switzerland, 2015; pp. 15–48. [Google Scholar]
- Elswaifi, S.F.; Scarratt, W.K.; Inzana, T.J. The role of lipooligosaccharide phosphorylcholine in colonization and pathogenesis of Histophilus somni in cattle. Vet. Res. 2012, 43, 49. [Google Scholar] [CrossRef] [PubMed]
- Isiaku, A.I.; Sabri, M.Y.; Ina-Salwany, M.Y.; Hassan, M.D.; Tanko, P.N.; Bello, M.B. Biofilm is associated with chronic streptococcal meningoencephalitis in fish. Microb. Pathog. 2017, 102, 59–68. [Google Scholar] [CrossRef] [PubMed]
- van der Mee-Marquet, N.; Fourny, L.; Arnault, L.; Domelier, A.S.; Salloum, M.; Lartigue, M.F.; Quentin, R. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J. Clin. Microbiol. 2008, 46, 2906–2911. [Google Scholar] [CrossRef]
- Bowater, R.O.; Forbes-Faulkner, J.; Anderson, I.G.; Condon, K.; Robinson, B.; Kong, F.; Gilbert, G.L.; Reynolds, A.; Hyland, S.; McPherson, G.; et al. Natural outbreak of Streptococcus agalactiae (GBS) infection in wild giant Queensland grouper, Epinephelus lanceolatus (Bloch), and other wild fish in northern Queensland, Australia. J. Fish Dis. 2012, 35, 173–186. [Google Scholar] [CrossRef]
- Delannoy, C.M.; Crumlish, M.; Fontaine, M.C.; Pollock, J.; Foster, G.; Dagleish, M.P.; Turnbull, J.F.; Zadoks, R.N. Human Streptococcus agalactiae strains in aquatic mammals and fish. BMC Microbiol. 2013, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Iregui, C.A.; Comas, J.; Vasquez, G.M.; Verjan, N. Experimental early pathogenesis of Streptococcus agalactiae infection in red tilapia Oreochromis spp. J. Fish Dis. 2016, 39, 205–215. [Google Scholar] [CrossRef]
- Motta, J.P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334. [Google Scholar] [CrossRef]
- Hathroubi, S.; Servetas, S.L.; Windham, I.; Merrell, D.S.; Ottemann, K.M. Helicobacter pylori Biofilm Formation and Its Potential Role in Pathogenesis. Microbiol. Mol. Biol. Rev. 2018, 82, e00001-18. [Google Scholar] [CrossRef]
- Miller, A.L.; Pasternak, J.A.; Medeiros, N.J.; Nicastro, L.K.; Tursi, S.A.; Hansen, E.G.; Krochak, R.; Sokaribo, A.S.; MacKenzie, K.D.; Palmer, M.B.; et al. In vivo synthesis of bacterial amyloid curli contributes to joint inflammation during S. Typhimurium infection. PLoS Pathog. 2020, 16, e1008591. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, E.; Bjarnsholt, T.; Jacobsen, S. Biofilm and Equine Limb Wounds. Animals 2021, 11, 2825. [Google Scholar] [CrossRef] [PubMed]
- Westgate, S.J.; Percival, S.L.; Knottenbelt, D.C.; Clegg, P.D.; Cochrane, C.A. Microbiology of equine wounds and evidence of bacterial biofilms. Vet. Microbiol. 2011, 150, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Freeman, K.; Woods, E.; Welsby, S.; Percival, S.L.; Cochrane, C.A. Biofilm evidence and the microbial diversity of horse wounds. Can. J. Microbiol. 2009, 55, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Van Hecke, L.L.; Hermans, K.; Haspeslagh, M.; Chiers, K.; Pint, E.; Boyen, F.; Martens, A.M. A quantitative swab is a good non-invasive alternative to a quantitative biopsy for quantifying bacterial load in wounds healing by second intention in horses. Vet. J. 2017, 225, 63–68. [Google Scholar] [CrossRef]
- Konig, L.; Klopfleisch, R.; Kershaw, O.; Gruber, A.D. Prevalence of biofilms on surgical suture segments in wounds of dogs, cats, and horses. Vet. Pathol. 2015, 52, 295–297. [Google Scholar] [CrossRef]
- Konig, L.M.; Klopfleisch, R.; Hoper, D.; Gruber, A.D. Next Generation Sequencing Analysis of Biofilms from Three Dogs with Postoperative Surgical Site Infection. Int. Sch. Res. Notices 2014, 2014, 282971. [Google Scholar] [CrossRef]
- Swanson, E.A.; Freeman, L.J.; Seleem, M.N.; Snyder, P.W. Biofilm-infected wounds in a dog. J. Am. Vet. Med. Assoc. 2014, 244, 699–707. [Google Scholar] [CrossRef]
- Theoret, C.L. Wound repair in the horse: Problems and proposed innovative solutions. Clin. Tech. Equine Pract. 2004, 3, 134–140. [Google Scholar] [CrossRef]
- Roberts, A.E.; Kragh, K.N.; Bjarnsholt, T.; Diggle, S.P. The Limitations of In Vitro Experimentation in Understanding Biofilms and Chronic Infection. J. Mol. Biol. 2015, 427, 3646–3661. [Google Scholar] [CrossRef]
- Jorgensen, E.; Bay, L.; Bjarnsholt, T.; Bundgaard, L.; Sorensen, M.A.; Jacobsen, S. The occurrence of biofilm in an equine experimental wound model of healing by secondary intention. Vet. Microbiol. 2017, 204, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Singh, P.K. Bacterial biofilms: An emerging link to disease pathogenesis. Annu. Rev. Microbiol. 2003, 57, 677–701. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.K.; Johansen, A.S.B.; Jensen, H.E. Porcine Models of Biofilm Infections with Focus on Pathomorphology. Front. Microbiol. 2017, 8, 1961. [Google Scholar] [CrossRef]
- Maslova, E.; Eisaiankhongi, L.; Sjöberg, F.; McCarthy, R.R. Burns and biofilms: Priority pathogens and in vivo models. Npj Biofilms Microbiomes 2021, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Elliot, S.; Wikramanayake, T.C.; Jozic, I.; Tomic-Canic, M. A Modeling Conundrum: Murine Models for Cutaneous Wound Healing. J. Investig. Dermatol. 2018, 138, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Thaarup, I.C.; Iversen, A.K.S.; Lichtenberg, M.; Bjarnsholt, T.; Jakobsen, T.H. Biofilm Survival Strategies in Chronic Wounds. Microorganisms 2022, 10, 775. [Google Scholar] [CrossRef] [PubMed]
- Dalton, T.; Dowd, S.E.; Wolcott, R.D.; Sun, Y.; Watters, C.; Griswold, J.A.; Rumbaugh, K.P. An In Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. PLoS ONE 2011, 6, e27317. [Google Scholar] [CrossRef]
- Watters, C.; DeLeon, K.; Trivedi, U.; Griswold, J.A.; Lyte, M.; Hampel, K.J.; Wargo, M.J.; Rumbaugh, K.P. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med. Microbiol. Immunol. 2013, 202, 131–141. [Google Scholar] [CrossRef]
- Gurjala, A.N.; Geringer, M.R.; Seth, A.K.; Hong, S.J.; Smeltzer, M.S.; Galiano, R.D.; Leung, K.P.; Mustoe, T.A. Development of a novel, highly quantitative in vivo model for the study of biofilm-impaired cutaneous wound healing. Wound Repair Regen. 2011, 19, 400–410. [Google Scholar] [CrossRef]
- Metcalf, D.; Bowler, P. Biofilm delays wound healing: A review of the evidence. Burn. Trauma 2013, 1, 5. [Google Scholar] [CrossRef]
- Pastar, I.; Nusbaum, A.G.; Gil, J.; Patel, S.B.; Chen, J.; Valdes, J.; Stojadinovic, O.; Plano, L.R.; Tomic-Canic, M.; Davis, S.C. Interactions of Methicillin Resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in Polymicrobial Wound Infection. PLoS ONE 2013, 8, e56846. [Google Scholar] [CrossRef] [PubMed]
- Moreno, I.; Franasiak, J.M. Endometrial microbiota-new player in town. Fertil Steril 2017, 108, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Adnane, M.; Chapwanya, A. A Review of the Diversity of the Genital Tract Microbiome and Implications for Fertility of Cattle. Animals 2022, 12, 460. [Google Scholar] [CrossRef] [PubMed]
- Holyoak, G.R.; Premathilake, H.U.; Lyman, C.C.; Sones, J.L.; Gunn, A.; Wieneke, X.; DeSilva, U. The healthy equine uterus harbors a distinct core microbiome plus a rich and diverse microbiome that varies with geographical location. Sci. Rep. 2022, 12, 14790. [Google Scholar] [CrossRef]
- Lyman, C.C.; Holyoak, G.R.; Meinkoth, K.; Wieneke, X.; Chillemi, K.A.; DeSilva, U. Canine endometrial and vaginal microbiomes reveal distinct and complex ecosystems. PLoS ONE 2019, 14, e0210157. [Google Scholar] [CrossRef]
- Wen, C.; Li, Q.; Lan, F.; Li, X.; Li, G.; Yan, Y.; Wu, G.; Yang, N.; Sun, C. Microbiota continuum along the chicken oviduct and its association with host genetics and egg formation. Poult. Sci. 2021, 100, 101104. [Google Scholar] [CrossRef]
- Swidsinski, A.; Verstraelen, H.; Loening-Baucke, V.; Swidsinski, S.; Mendling, W.; Halwani, Z. Presence of a polymicrobial endometrial biofilm in patients with bacterial vaginosis. PLoS ONE 2013, 8, e53997. [Google Scholar] [CrossRef]
- Rocha, M.F.G.; Paiva, D.D.Q.; Amando, B.R.; Melgarejo, C.M.A.; Freitas, A.S.; Gomes, F.I.F.; Ocadaque, C.J.; Costa, C.L.; Guedes, G.M.M.; Lima-Neto, R.G.; et al. Antimicrobial susceptibility and production of virulence factors by bacteria recovered from bitches with pyometra. Reprod. Domest. Anim. 2022, 57, 1063–1073. [Google Scholar] [CrossRef]
- Fiamengo, T.E.; Runcan, E.E.; Premanandan, C.; Blawut, B.; Coutinho da Silva, M.A. Evaluation of Biofilm Production by Escherichia coli Isolated from Clinical Cases of Canine Pyometra. Top. Companion Anim. Med. 2020, 39, 100429. [Google Scholar] [CrossRef]
- LeBlanc, M.M.; Causey, R.C. Clinical and subclinical endometritis in the mare: Both threats to fertility. Reprod. Domest. Anim. 2009, 44 (Suppl. S3), 10–22. [Google Scholar] [CrossRef]
- LeBlanc, M.M. Advances in the diagnosis and treatment of chronic infectious and post-mating-induced endometritis in the mare. Reprod. Domest. Anim. 2010, 45 (Suppl. S2), 21–27. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.A.; McCue, P.M.; Borlee, G.I.; Loncar, K.D.; Hennet, M.L.; Borlee, B.R. In Vitro Efficacy of Nonantibiotic Treatments on Biofilm Disruption of Gram-Negative Pathogens and an In Vivo Model of Infectious Endometritis Utilizing Isolates from the Equine Uterus. J. Clin. Microbiol. 2016, 54, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.A.; McCue, P.M.; Borlee, G.I.; Glapa, K.E.; Martin, K.H.; Mangalea, M.R.; Hennet, M.L.; Wolfe, L.M.; Broeckling, C.D.; Borlee, B.R. Model of Chronic Equine Endometritis Involving a Pseudomonas aeruginosa Biofilm. Infect. Immun. 2017, 85, e00332-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.L.; Liu, M.C.; Xu, J.; An, L.G.; Wang, J.F.; Zhu, Y.H. Uterine Microbiota of Dairy Cows with Clinical and Subclinical Endometritis. Front. Microbiol. 2018, 9, 2691. [Google Scholar] [CrossRef] [PubMed]
- Pascottini, O.B.; Van Schyndel, S.J.; Spricigo, J.F.W.; Rousseau, J.; Weese, J.S.; LeBlanc, S.J. Dynamics of uterine microbiota in postpartum dairy cows with clinical or subclinical endometritis. Sci. Rep. 2020, 10, 12353. [Google Scholar] [CrossRef] [PubMed]
- Madoz, L.V.; Giuliodori, M.J.; Migliorisi, A.L.; Jaureguiberry, M.; de la Sota, R.L. Endometrial cytology, biopsy, and bacteriology for the diagnosis of subclinical endometritis in grazing dairy cows. J. Dairy Sci. 2014, 97, 195–201. [Google Scholar] [CrossRef]
- Ahmadi, M.; Derakhshandeh, A.; Shirian, S.; Daneshbod, Y.; Ansari-Lari, M.; Nazifi, S. Detection of bacterial biofilm in uterine of repeat breeder dairy cows. Asian Pac. J. Reprod. 2017, 6, 136–139. [Google Scholar] [CrossRef]
- Rzewuska, M.; Kwiecien, E.; Chrobak-Chmiel, D.; Kizerwetter-Swida, M.; Stefanska, I.; Gierynska, M. Pathogenicity and Virulence of Trueperella pyogenes: A Review. Int. J. Mol. Sci. 2019, 20, 2737. [Google Scholar] [CrossRef]
- Hensen, S.M.; Pavicic, M.J.; Lohuis, J.A.; de Hoog, J.A.; Poutrel, B. Location of Staphylococcus aureus within the experimentally infected bovine udder and the expression of capsular polysaccharide type 5 in situ. J. Dairy Sci. 2000, 83, 1966–1975. [Google Scholar] [CrossRef]
- Schonborn, S.; Kromker, V. Detection of the biofilm component polysaccharide intercellular adhesin in Staphylococcus aureus infected cow udders. Vet. Microbiol. 2016, 196, 126–128. [Google Scholar] [CrossRef]
- Gogoi-Tiwari, J.; Williams, V.; Waryah, C.B.; Eto, K.Y.; Tau, M.; Costantino, P.; Tiwari, H.K.; Mukkur, T. Comparative studies of the immunogenicity and protective potential of biofilm vs planktonic Staphylococcus aureus vaccine against bovine mastitis using non-invasive mouse mastitis as a model system. Biofouling 2015, 31, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.M.; Prenafeta, A.; Valle, J.; Penades, J.; Rota, C.; Solano, C.; Marco, J.; Grillo, M.J.; Lasa, I.; Irache, J.M.; et al. Protection from Staphylococcus aureus mastitis associated with poly-N-acetyl beta-1,6 glucosamine specific antibody production using biofilm-embedded bacteria. Vaccine 2009, 27, 2379–2386. [Google Scholar] [CrossRef] [PubMed]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.; Penades, J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, E.; Melville, P.A.; Saidenberg, A.B.; Laes, M.A.; Gonsales, F.F.; Salaberry, S.R.; Gregori, F.; Brandao, P.E.; dos Santos, F.G.; Lincopan, N.E.; et al. Occurrence of genes coding for MSCRAMM and biofilm-associated protein Bap in Staphylococcus spp. isolated from bovine subclinical mastitis and relationship with somatic cell counts. Microb. Pathog. 2015, 89, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hebert, A.; Sayasith, K.; Senechal, S.; Dubreuil, P.; Lagace, J. Demonstration of intracellular Staphylococcus aureus in bovine mastitis alveolar cells and macrophages isolated from naturally infected cow milk. FEMS Microbiol. Lett. 2000, 193, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Grunert, T.; Stessl, B.; Wolf, F.; Sordelli, D.O.; Buzzola, F.R.; Ehling-Schulz, M. Distinct phenotypic traits of Staphylococcus aureus are associated with persistent, contagious bovine intramammary infections. Sci. Rep. 2018, 8, 15968. [Google Scholar] [CrossRef]
- Tan, N.C.; Foreman, A.; Jardeleza, C.; Douglas, R.; Vreugde, S.; Wormald, P.J. Intracellular Staphylococcus aureus: The Trojan horse of recalcitrant chronic rhinosinusitis? Int. Forum Allergy Rhinol. 2013, 3, 261–266. [Google Scholar] [CrossRef]
- Ha, K.R.; Psaltis, A.J.; Tan, L.; Wormald, P.J. A sheep model for the study of biofilms in rhinosinusitis. Am. J. Rhinol. 2007, 21, 339–345. [Google Scholar] [CrossRef]
- Petruzzi, B.; Dickerman, A.; Lahmers, K.; Scarratt, W.K.; Inzana, T.J. Polymicrobial Biofilm Interaction between Histophilus somni and Pasteurella multocida. Front. Microbiol. 2020, 11, 1561. [Google Scholar] [CrossRef]
- Maes, D.; Verdonck, M.; Deluyker, H.; de Kruif, A. Enzootic pneumonia in pigs. Vet. Q. 1996, 18, 104–109. [Google Scholar] [CrossRef]
- Tajima, M.; Yagihashi, T. Interaction of Mycoplasma hyopneumoniae with the porcine respiratory epithelium as observed by electron microscopy. Infect. Immun. 1982, 37, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Raymond, B.B.A.; Jenkins, C.; Turnbull, L.; Whitchurch, C.B.; Djordjevic, S.P. Extracellular DNA release from the genome-reduced pathogen Mycoplasma hyopneumoniae is essential for biofilm formation on abiotic surfaces. Sci. Rep. 2018, 8, 10373. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, Y.D.N.; Labrie, J.; Chenier, S.; Jacques, M. Actinobacillus pleuropneumoniae grows as aggregates in the lung of pigs: Is it time to refine our in vitro biofilm assays? Microb. Biotechnol. 2017, 10, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Cattelan, N.; Dubey, P.; Arnal, L.; Yantorno, O.M.; Deora, R. Bordetella biofilms: A lifestyle leading to persistent infections. Pathog. Dis. 2016, 74, ftv108. [Google Scholar] [CrossRef] [PubMed]
- Sloan, G.P.; Love, C.F.; Sukumar, N.; Mishra, M.; Deora, R. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J. Bacteriol. 2007, 189, 8270–8276. [Google Scholar] [CrossRef] [PubMed]
- Conover, M.S.; Mishra, M.; Deora, R. Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PLoS ONE 2011, 6, e16861. [Google Scholar] [CrossRef] [PubMed]
- Gieling, F.; Peters, S.; Erichsen, C.; Richards, R.G.; Zeiter, S.; Moriarty, T.F. Bacterial osteomyelitis in veterinary orthopaedics: Pathophysiology, clinical presentation and advances in treatment across multiple species. Vet. J. 2019, 250, 44–54. [Google Scholar] [CrossRef]
- Meroni, G.; Tsikopoulos, A.; Tsikopoulos, K.; Allemanno, F.; Martino, P.A.; Soares Filipe, J.F. A Journey into Animal Models of Human Osteomyelitis: A Review. Microorganisms 2022, 10, 1135. [Google Scholar] [CrossRef]
- Patel, M.; Rojavin, Y.; Jamali, A.A.; Wasielewski, S.J.; Salgado, C.J. Animal models for the study of osteomyelitis. Semin. Plast Surg. 2009, 23, 148–154. [Google Scholar] [CrossRef]
- Johansen, L.K.; Koch, J.; Frees, D.; Aalbaek, B.; Nielsen, O.L.; Leifsson, P.S.; Iburg, T.M.; Svalastoga, E.; Buelund, L.E.; Bjarnsholt, T.; et al. Pathology and biofilm formation in a porcine model of staphylococcal osteomyelitis. J. Comp. Pathol. 2012, 147, 343–353. [Google Scholar] [CrossRef]
- Bortolami, A.; Zendri, F.; Maciuca, E.I.; Wattret, A.; Ellis, C.; Schmidt, V.; Pinchbeck, G.; Timofte, D. Diversity, Virulence, and Clinical Significance of Extended-Spectrum β-Lactamase- and pAmpC-Producing Escherichia coli from Companion Animals. Front. Microbiol. 2019, 10, 1260. [Google Scholar] [CrossRef] [PubMed]
- Clark, H.; Lasarev, M.; Wood, M. Risk factors of enterococcal bacteriuria in cats: A retrospective study. Can. Vet. J. 2023, 64, 40–44. [Google Scholar]
- Wood, M.W.; Lepold, A.; Tesfamichael, D.; Lasarev, M.R. Risk factors for enterococcal bacteriuria in dogs: A retrospective study. J. Vet. Intern. Med. 2020, 34, 2447–2453. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.F.; Litster, A.L.; Platell, J.L.; Trott, D.J. Canine bacterial urinary tract infections: New developments in old pathogens. Vet. J. 2011, 190, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Ballash, G.A.; Mollenkopf, D.F.; Diaz-Campos, D.; Van Balen, J.C.; Cianciolo, R.E.; Wittum, T.E. Pathogenomics and clinical recurrence influence biofilm capacity of Escherichia coli isolated from canine urinary tract infections. PLoS ONE 2022, 17, e0270461. [Google Scholar] [CrossRef]
- Dorsch, R.; Teichmann-Knorrn, S.; Sjetne Lund, H. Urinary tract infection and subclinical bacteriuria in cats: A clinical update. J. Feline Med. Surg. 2019, 21, 1023–1038. [Google Scholar] [CrossRef]
- Litster, A.; Thompson, M.; Moss, S.; Trott, D. Feline bacterial urinary tract infections: An update on an evolving clinical problem. Vet. J. 2011, 187, 18–22. [Google Scholar] [CrossRef]
- Kern, Z.T.; Jacob, M.E.; Gilbertie, J.M.; Vaden, S.L.; Lyle, S.K. Characteristics of Dogs with Biofilm-Forming Escherichia coli Urinary Tract Infections. J. Vet. Intern. Med. 2018, 32, 1645–1651. [Google Scholar] [CrossRef]
- Gilbertie, J.M.; Levent, G.; Norman, K.N.; Vinasco, J.; Scott, H.M.; Jacob, M.E. Comprehensive phenotypic and genotypic characterization and comparison of virulence, biofilm, and antimicrobial resistance in urinary Escherichia coli isolated from canines. Vet. Microbiol. 2020, 249, 108822. [Google Scholar] [CrossRef]
- Allbaugh, R.A. Equine recurrent uveitis: A review of clinical assessment and management. Equine Vet. Educ. 2017, 29, 279–288. [Google Scholar] [CrossRef]
- Brandes, K.; Wollanke, B.; Niedermaier, G.; Brem, S.; Gerhards, H. Recurrent uveitis in horses: Vitreal examinations with ultrastructural detection of leptospires. J. Vet. Med. A 2007, 54, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, K.; Kenngott, R.; Settles, M.; Gerhards, H.; Maierl, J.; Wollanke, B. In Vivo Biofilm Formation of Pathogenic Leptospira spp. in the Vitreous Humor of Horses with Recurrent Uveitis. Microorganisms 2021, 9, 1915. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N.; Bjarnsholt, T.; Moser, C.; Bassi, G.L.; Coenye, T.; Donelli, G.; Hall-Stoodley, L.; Hola, V.; Imbert, C.; Kirketerp-Moller, K.; et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 2015, 21 (Suppl. S1), S1–S25. [Google Scholar] [CrossRef] [PubMed]
- Jacques, M.; Malouin, F. One Health-One Biofilm. Vet. Res. 2022, 53, 51. [Google Scholar] [CrossRef] [PubMed]
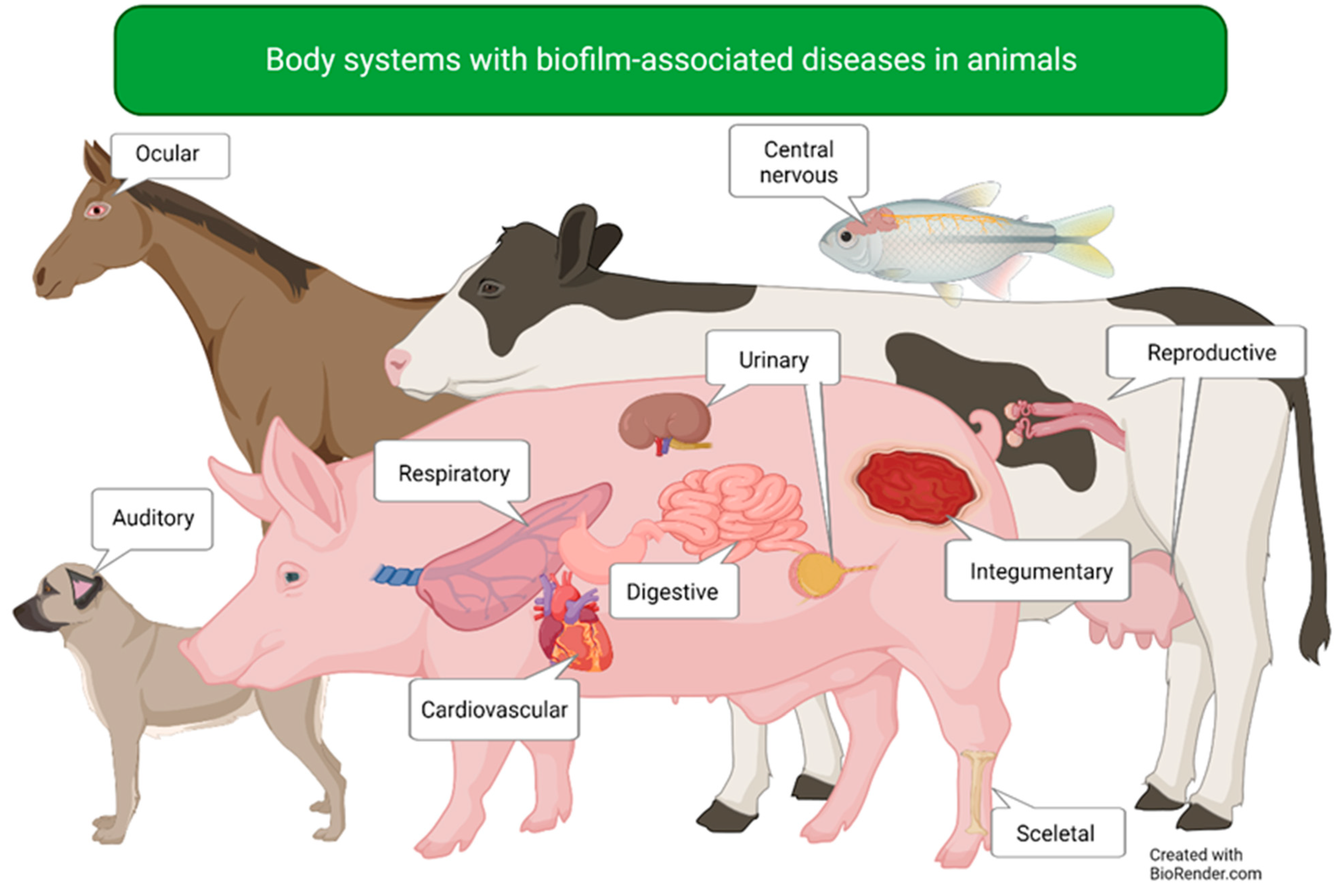
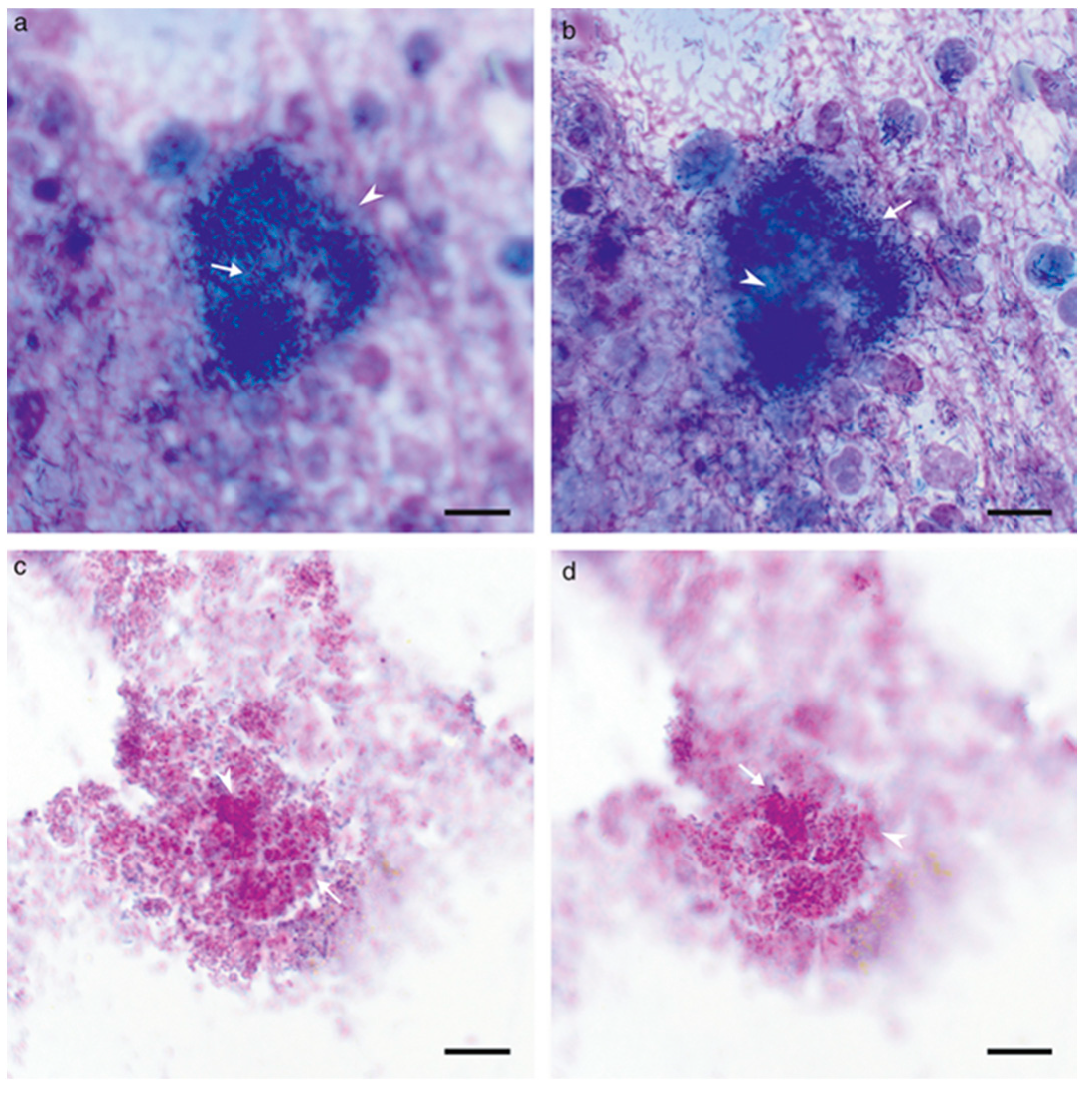
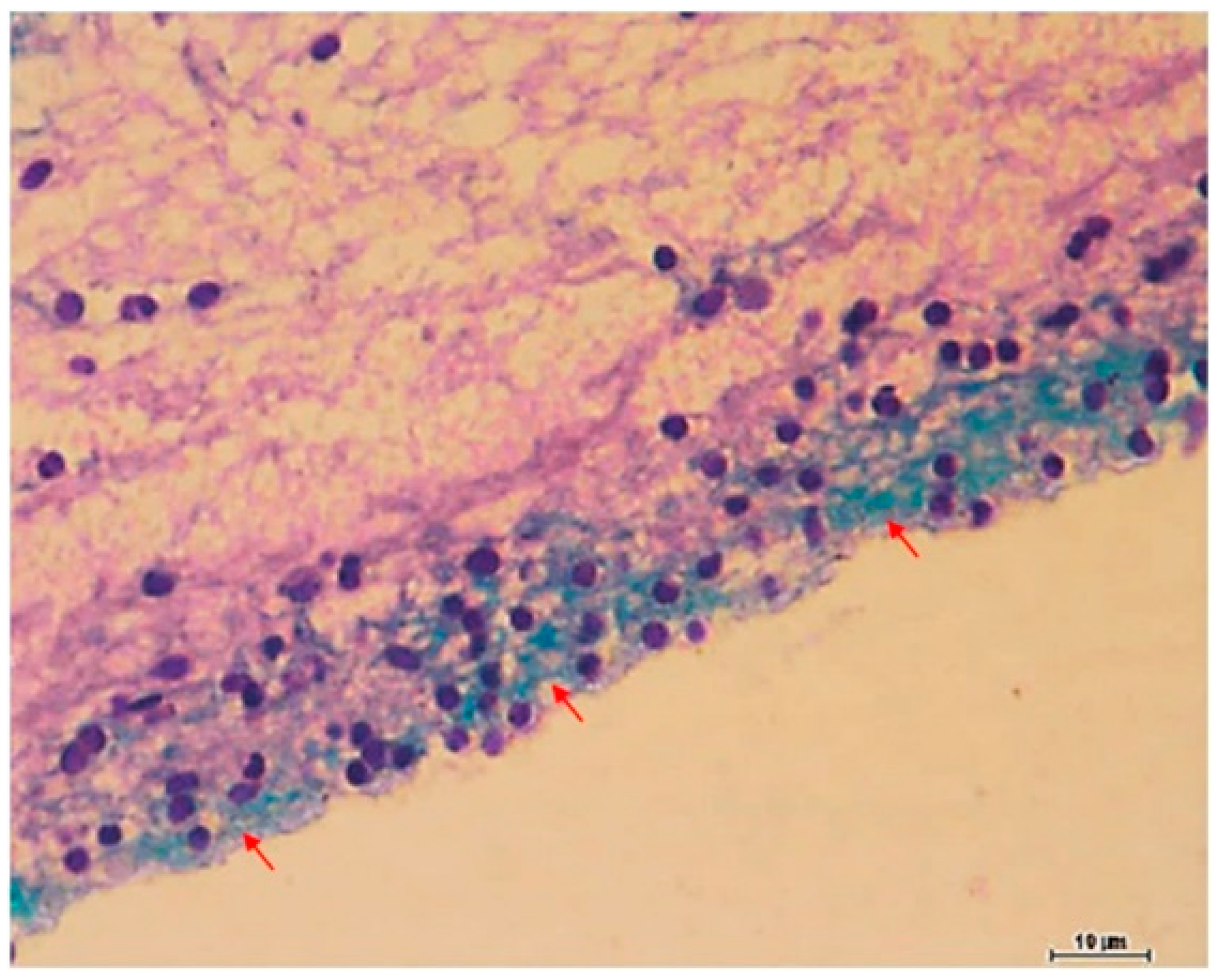
| Body System | Affected Organs | Disease |
|---|---|---|
| Auditory | Ear | Canine otitis externa |
| Cardiovascular | Heart | Bovine myocarditis |
| Central nervous | Brain | Piscine meningoencephalitis |
| Digestive | Intestines | Salmonella infection |
| Integumentary | Skin and underlying tissue | Wound infections |
| Reproductive | Uterus | Endometritis |
| Mammary glands | Mastitis | |
| Respiratory | Lower airways | Bovine respiratory disease |
| Lower airways | Porcine enzootic pneumonia | |
| Lower airways | Porcine pleuropneumonia | |
| Upper and lower airways | Bordetella bronchiseptica infection | |
| Skeletal | Bones | Osteomyelitis |
| Urinary | Urinary tract | Uropathogenic E. coli infection |
| Visual | Eye | Equine recurrent uveitis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nesse, L.L.; Osland, A.M.; Vestby, L.K. The Role of Biofilms in the Pathogenesis of Animal Bacterial Infections. Microorganisms 2023, 11, 608. https://doi.org/10.3390/microorganisms11030608
Nesse LL, Osland AM, Vestby LK. The Role of Biofilms in the Pathogenesis of Animal Bacterial Infections. Microorganisms. 2023; 11(3):608. https://doi.org/10.3390/microorganisms11030608
Chicago/Turabian StyleNesse, Live L., Ane Mohr Osland, and Lene K. Vestby. 2023. "The Role of Biofilms in the Pathogenesis of Animal Bacterial Infections" Microorganisms 11, no. 3: 608. https://doi.org/10.3390/microorganisms11030608
APA StyleNesse, L. L., Osland, A. M., & Vestby, L. K. (2023). The Role of Biofilms in the Pathogenesis of Animal Bacterial Infections. Microorganisms, 11(3), 608. https://doi.org/10.3390/microorganisms11030608