Application of Phytogenic Liquid Supplementation in Soil Microbiome Restoration in Queensland Pasture Dieback
Abstract
1. Introduction
2. Materials and Methods
3. Results
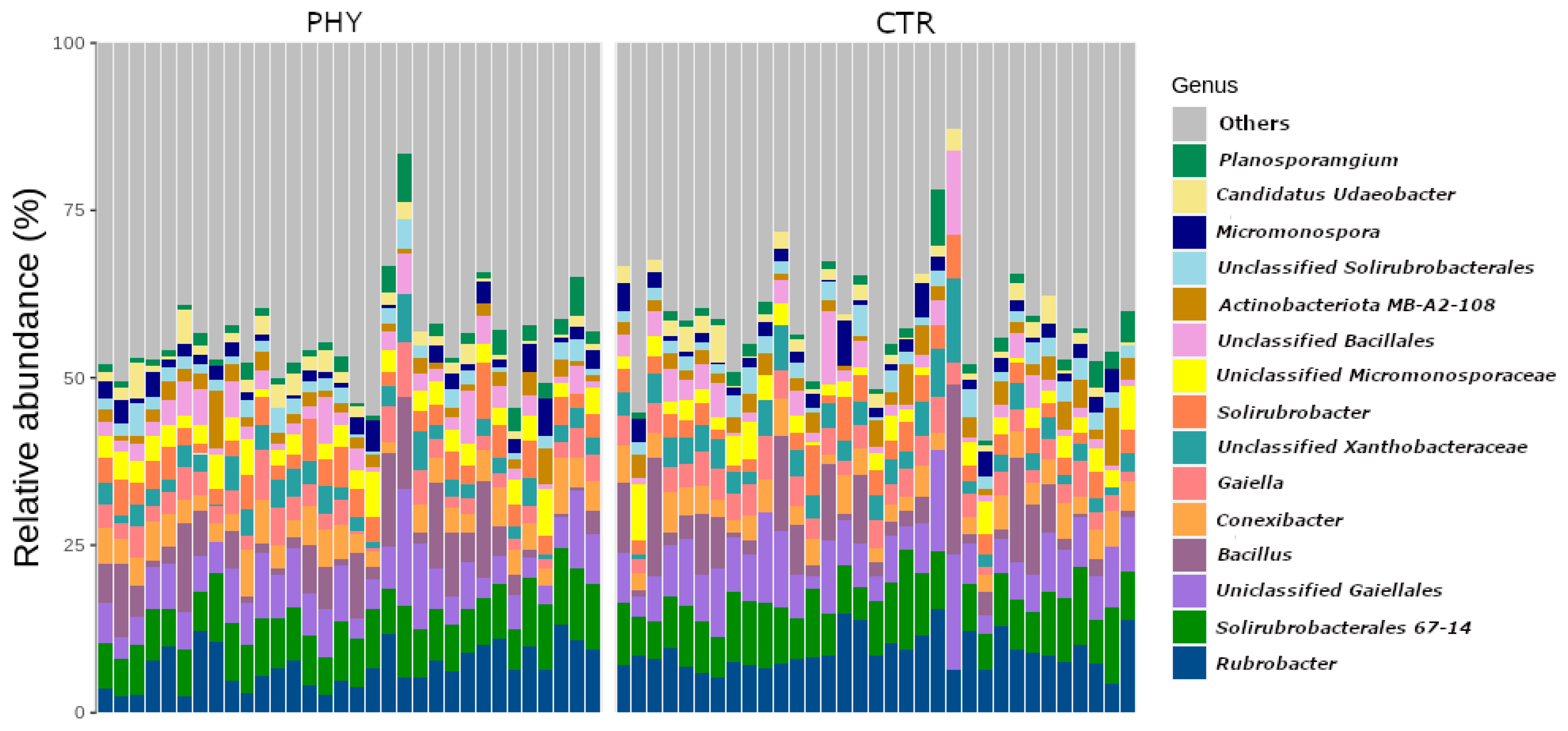
3.1. Soil Community Structure
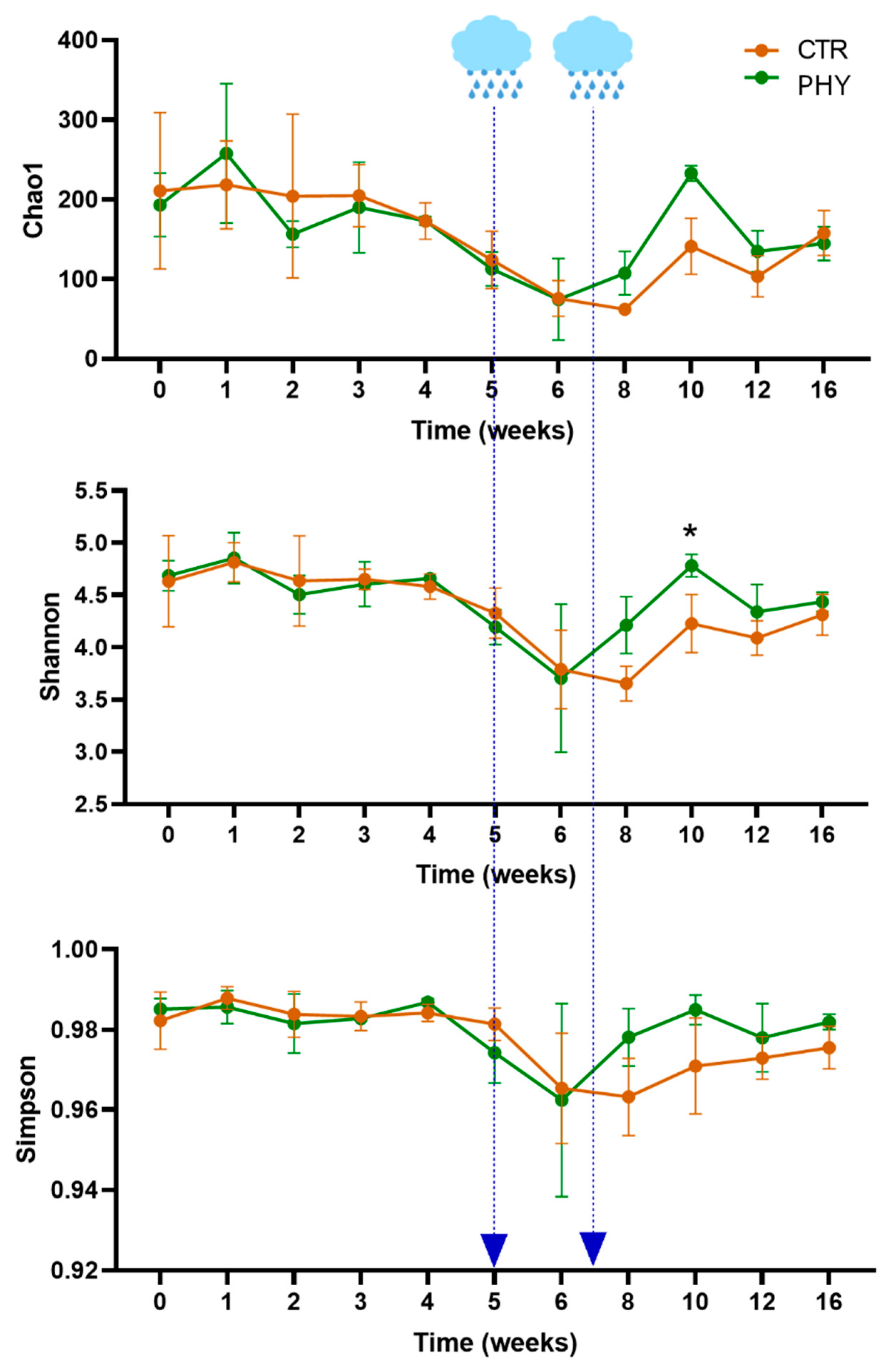
3.2. Alpha Diversity
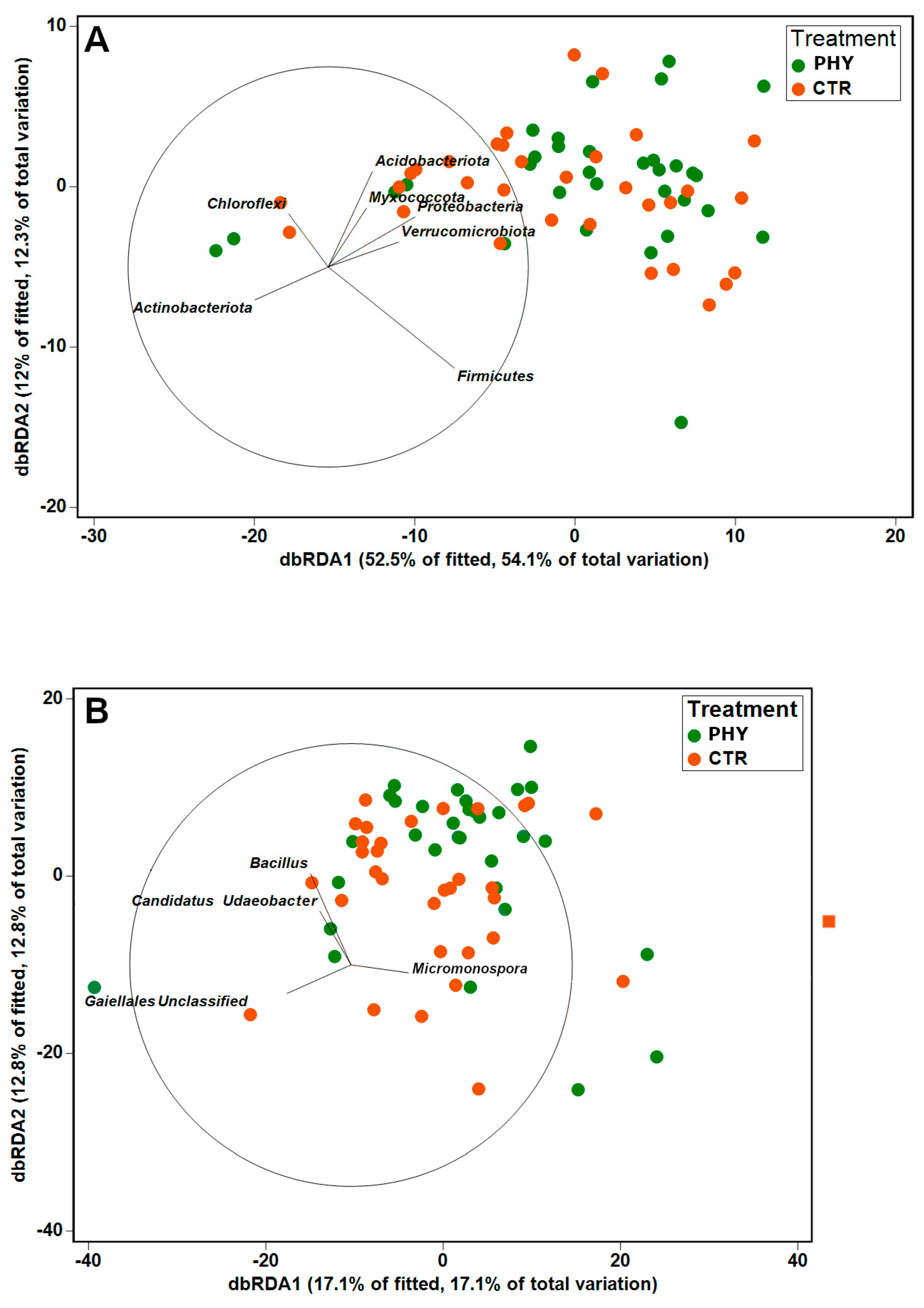
3.3. Spatial and Temporal Alterations in Beta Diversity
3.4. Temporal Correlation
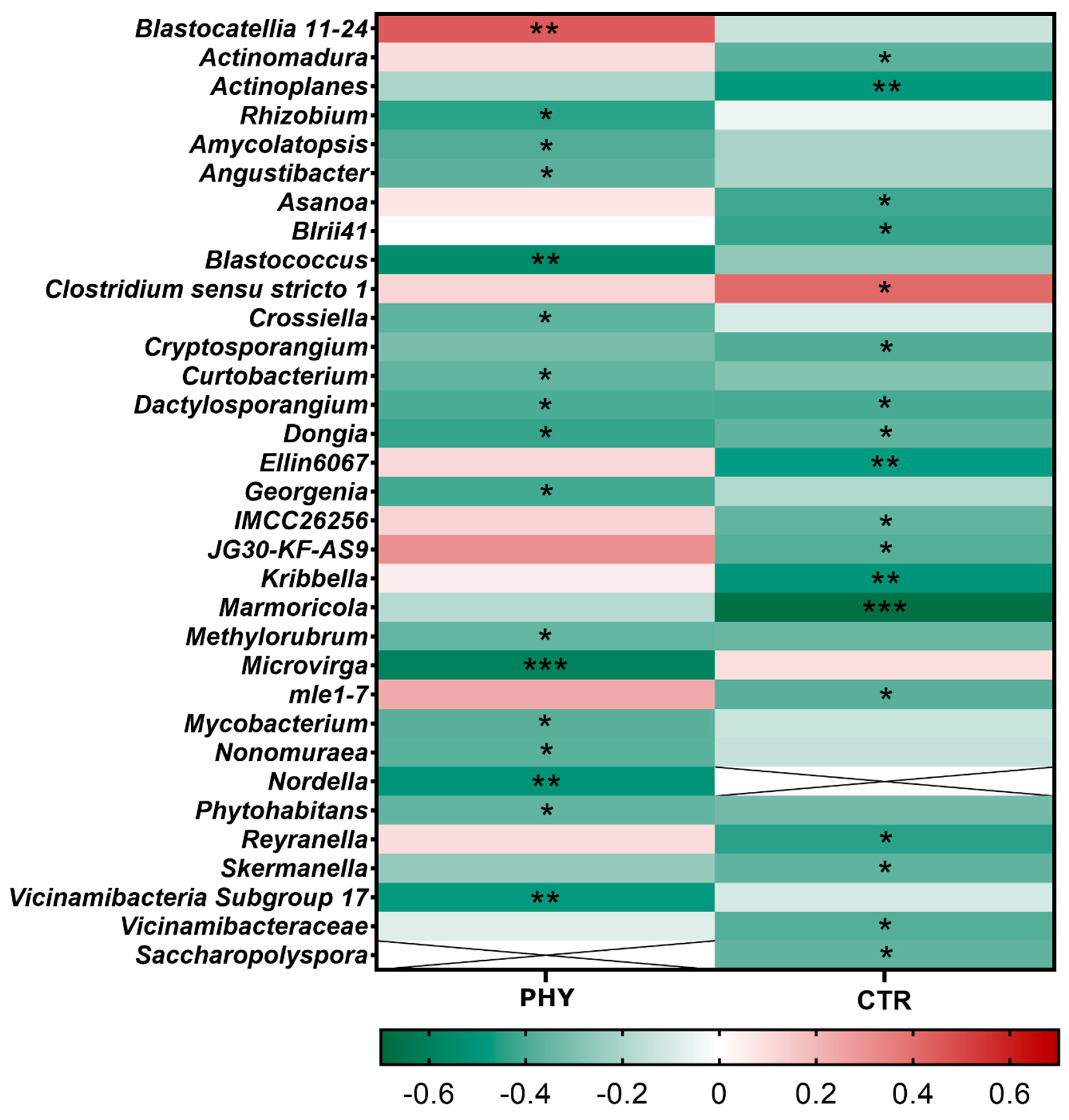
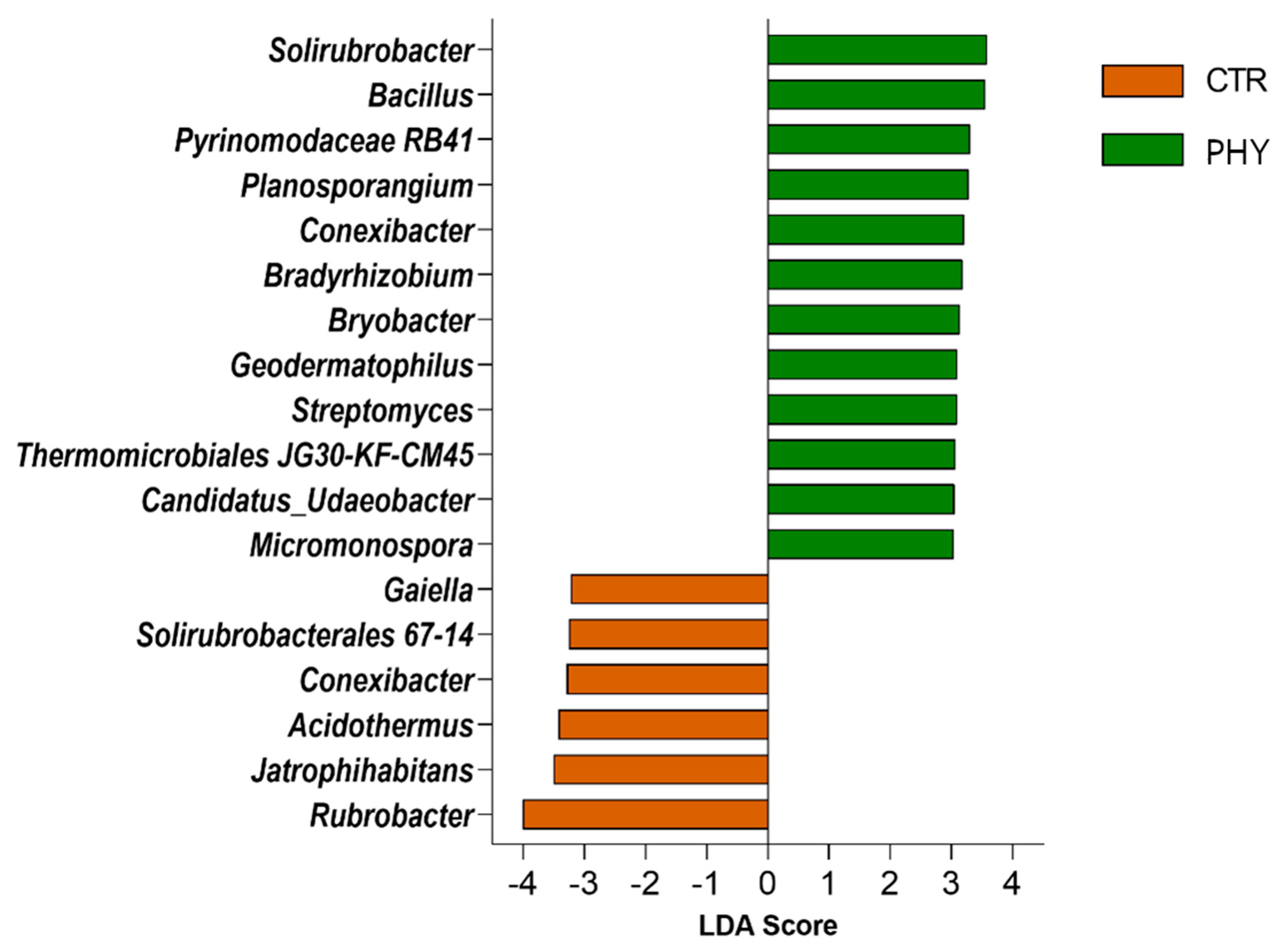
3.5. Taxa Responding to Phytogenic Liquid Application
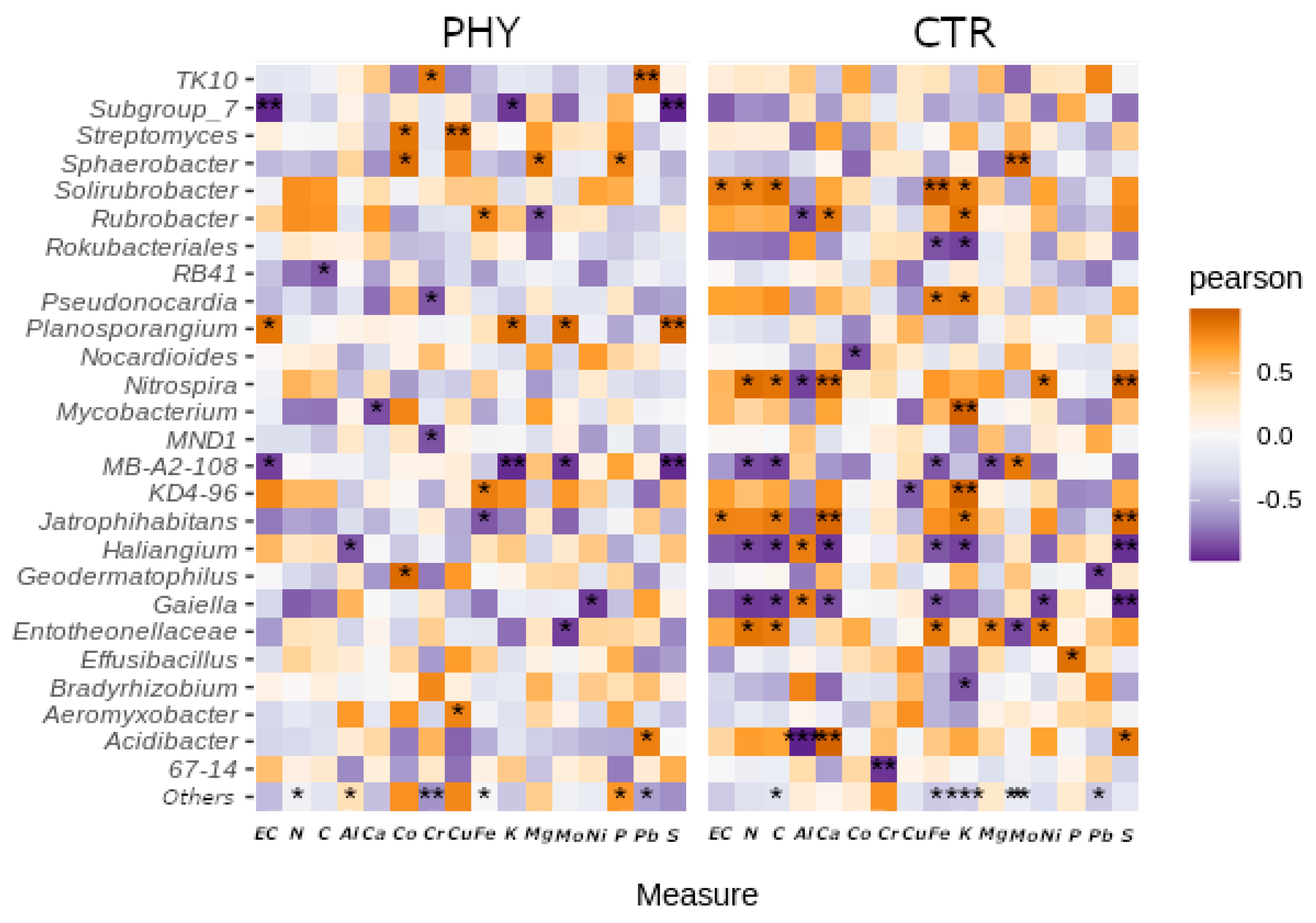
3.6. Interactions with Minerals
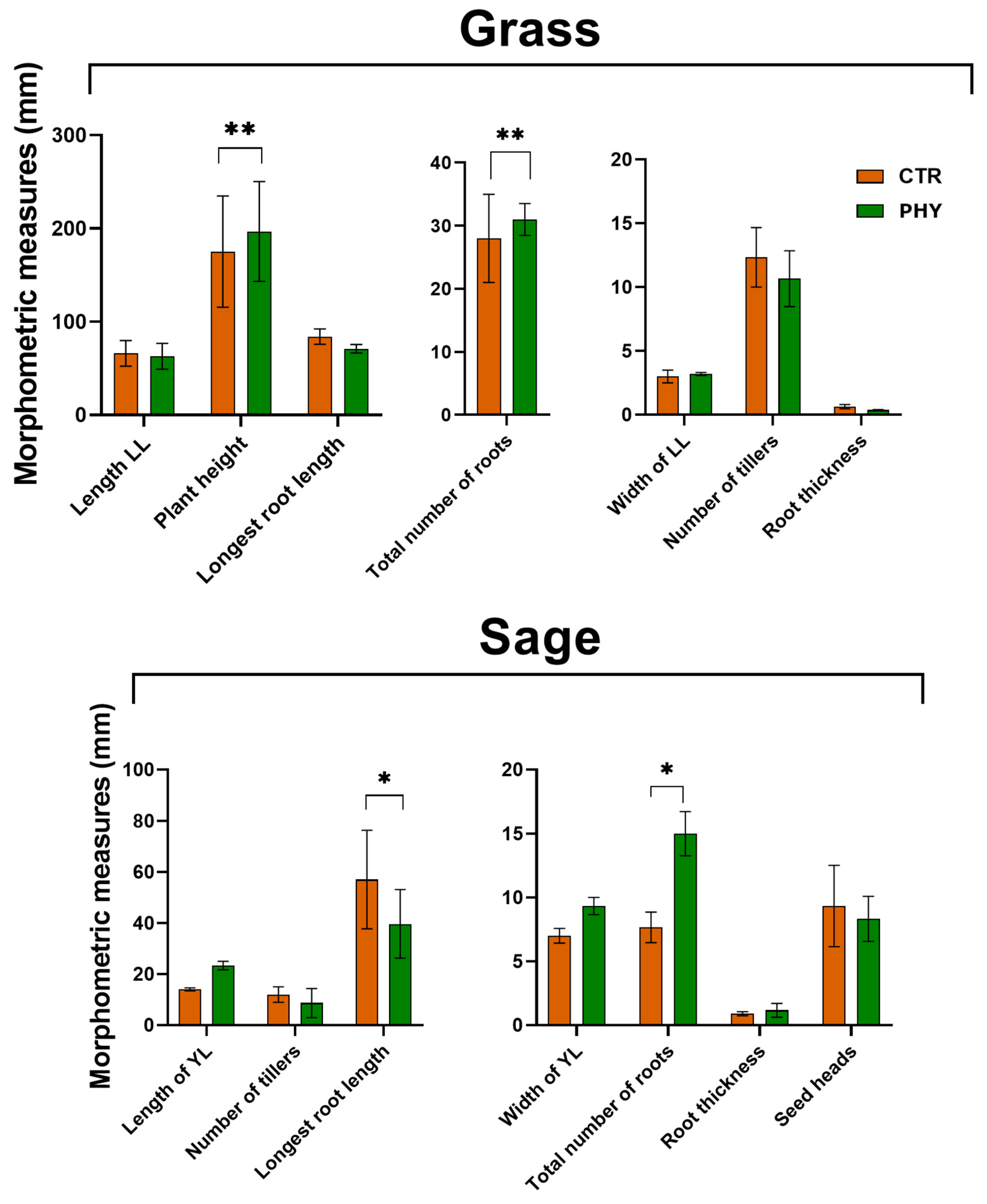
3.7. Plant Morphometrics
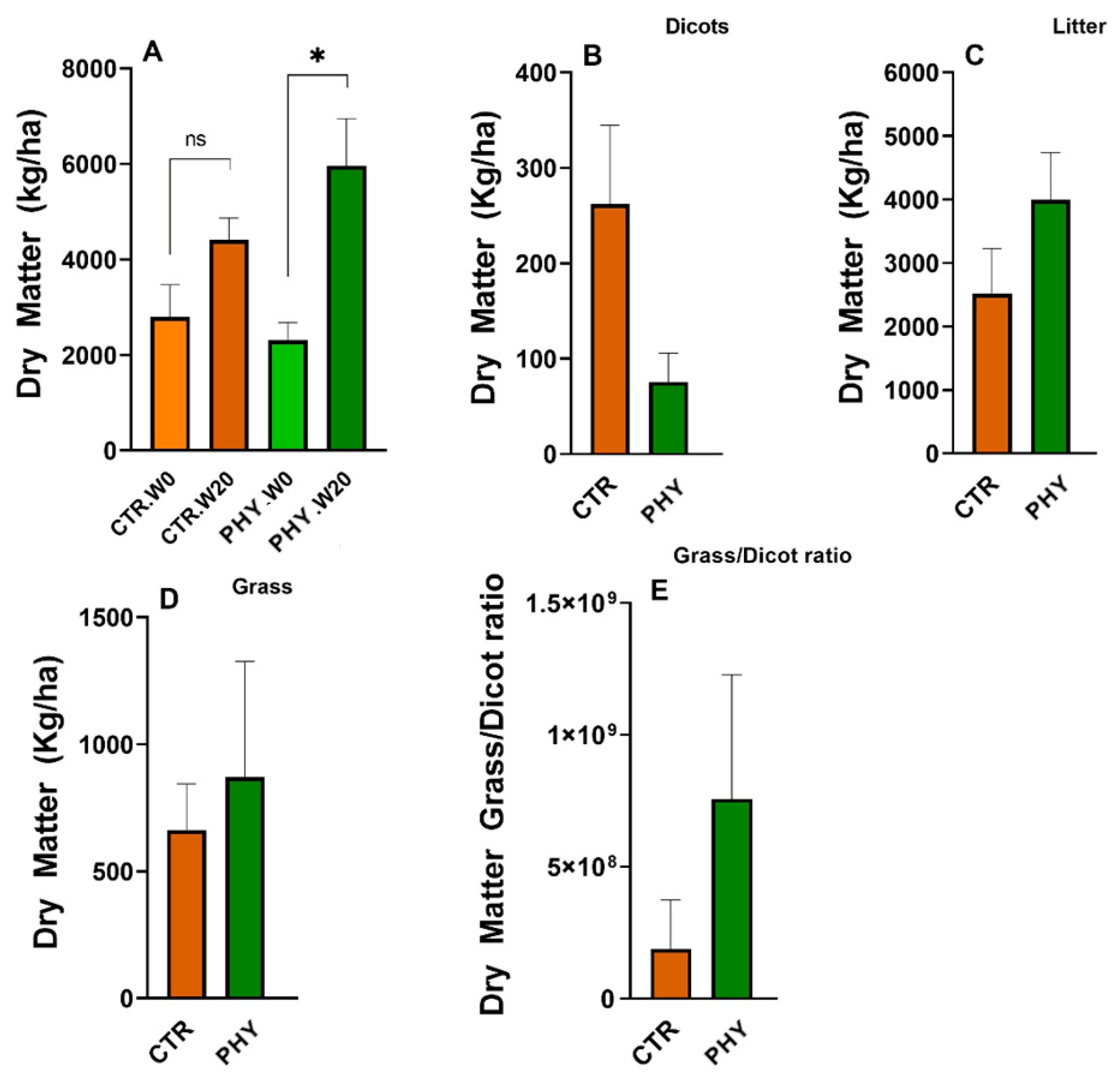
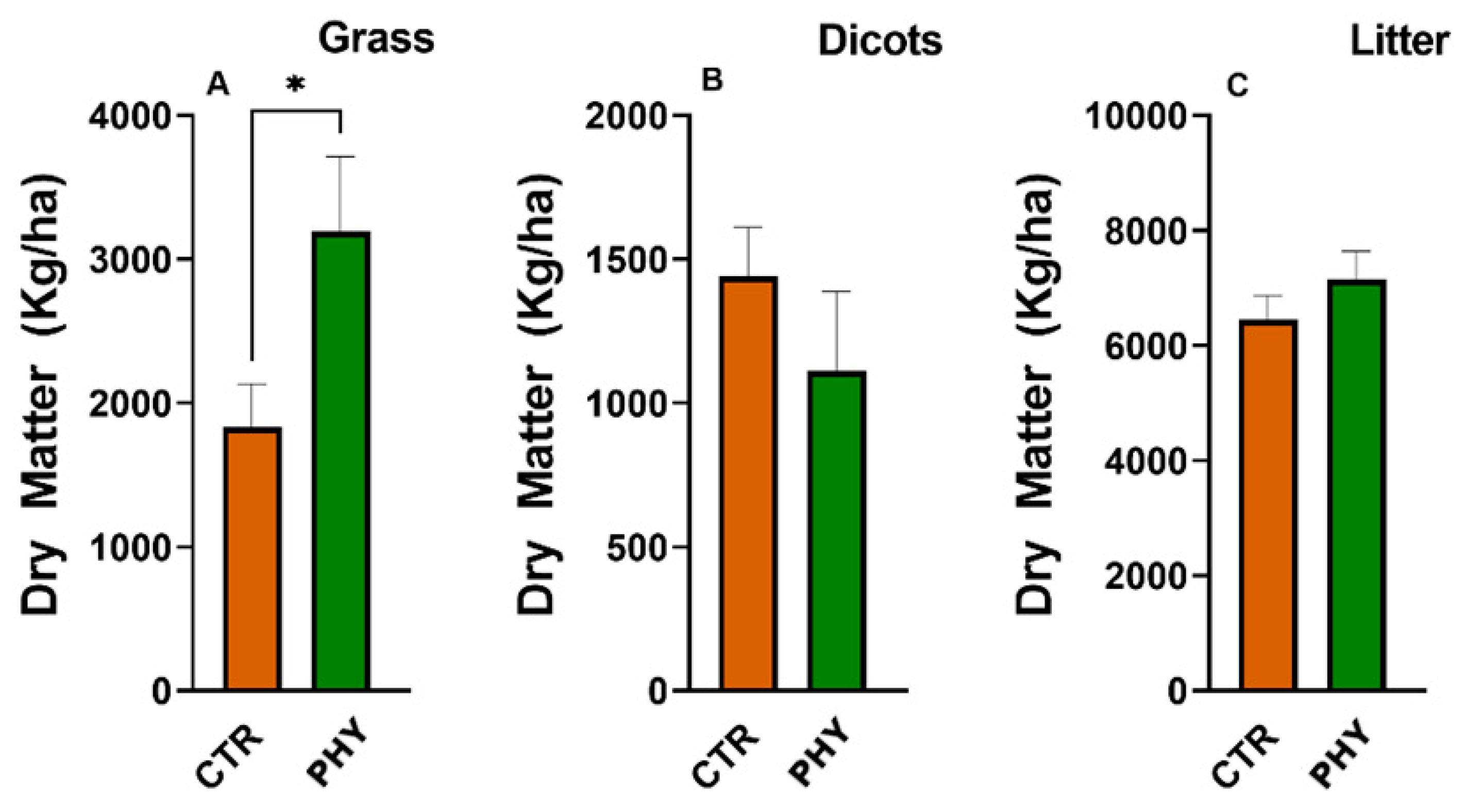
3.8. Dry Matter
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Yajun, W.; Chongchong, G.; Tianjing, C.; Jinshou, L.; Yan, X.; Dafang, F. Adaptability of enhanced bioretention cell for nitrogen and phosphorus removal under two antibiotics stress. Ecotoxicol. Environ. Saf. 2022, 230, 113114. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.J.; Milson, J.; Hall, C. Pasture Recovery, Land Condition and Some Other Observations after the Monsoon Flooding, Chill Event in North-West Queensland in Jan–Mar 2019. 2020. Available online: https://era.daf.qld.gov.au/id/eprint/7443/ (accessed on 1 December 2022).
- Makiela, S.; Harrower, K.M. Overview of the current status of buffel grass dieback. Australas. Plant Dis. Notes 2008, 3, 12. [Google Scholar] [CrossRef]
- Bell, A.W.; Charmley, E.; Hunter, R.A.; Archer, J.A. The Australasian beef industries—Challenges and opportunities in the 21st century. Anim. Front. 2011, 1, 10–19. [Google Scholar] [CrossRef]
- Australia, M.L. Industry Fast Facts. 2020. Available online: https://www.mla.com.au/prices-markets/market-news/2020/2020-industry-fast-facts-released/ (accessed on 1 December 2022).
- Australia, M.L. State of the Industry Report. 2020. Available online: https://www.mla.com.au/news-and-events/industry-news/state-of-the-industry-report-2020-released/ (accessed on 2 September 2020).
- Andreote, F.D.; e Silva, M.d.C.P. Microbial communities associated with plants: Learning from nature to apply it in agriculture. Curr. Opin. Microbiol. 2017, 37, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Millard, P.; Singh, B.K. Does grassland vegetation drive soil microbial diversity? Nutr. Cycl. Agroecosyst. 2010, 88, 147–158. [Google Scholar] [CrossRef]
- Gwinn, K.D. Chapter 7—Bioactive Natural Products in Plant Disease Control. In Studies in Natural Products Chemistry; University of Tennessee: Knoxville, TN, USA, 2018; Volume 56, pp. 229–246. [Google Scholar]
- Liu, Q.; Qiao, K.; Zhang, S. Potential of a small molecule carvacrol in management of vegetable diseases. Molecules 2019, 24, 1932. [Google Scholar] [CrossRef]
- Gurjar, M.S.; Ali, S.; Akhtar, M.; Singh, K.S. Efficacy of plant extracts in plant disease management. Agric. Sci. 2012, 3, 19046. [Google Scholar] [CrossRef]
- Imran, M.; Aslam, M.; Alsagaby, S.A.; Saeed, F.; Ahmad, I.; Afzaal, M.; Arshad, M.U.; Abdelgawad, M.A.; El-Ghorab, A.H.; Khames, A.; et al. Therapeutic application of carvacrol: A comprehensive review. Food Sci. Nutr. 2022, 10, 3544–3561. [Google Scholar] [CrossRef]
- Saghrouchni, H.; El Barnossi, A.; Salamatullah, A.M.; Bourhia, M.; Alzahrani, A.; Alkaltham, M.S.; Alyahya, H.K.; Tahiri, N.E.H.; Imtara, H.; Var, I. Carvacrol: A promising environmentally friendly agent to fight seeds damping-off diseases induced by fungal species. Agronomy 2021, 11, 985. [Google Scholar] [CrossRef]
- Javed, H.; Meeran, M.F.N.; Jha, N.K.; Ojha, S. Carvacrol, a Plant Metabolite Targeting Viral Protease (M(pro)) and ACE2 in Host Cells Can Be a Possible Candidate for COVID-19. Front. Plant Sci. 2021, 11, 601335. [Google Scholar] [CrossRef] [PubMed]
- Azizi, Z.; Majlessi, N.; Choopani, S.; Naghdi, N. Neuroprotective effects of carvacrol against Alzheimer’s disease and other neurodegenerative diseases: A review. Avicenna J. Phytomed. 2022, 12, 371–387. [Google Scholar] [CrossRef]
- Costa, M.F.; Durco, A.O.; Rabelo, T.K.; Barreto, R.S.S.; Guimaraes, A.G. Effects of Carvacrol, Thymol and essential oils containing such monoterpenes on wound healing: A systematic review. J. Pharm. Pharmacol. 2019, 71, 141–155. [Google Scholar] [CrossRef]
- de Carvalho, F.O.; Silva, E.R.; Gomes, I.A.; Santana, H.S.R.; do Nascimento Santos, D.; de Oliveira Souza, G.P.; de Jesus Silva, D.; Monteiro, J.C.M.; de Albuquerque Junior, R.L.C.; de Souza Araujo, A.A.; et al. Anti-inflammatory and antioxidant activity of carvacrol in the respiratory system: A systematic review and meta-analysis. Phytother. Res. 2020, 34, 2214–2229. [Google Scholar] [CrossRef]
- Sampaio, L.A.; Pina, L.T.S.; Serafini, M.R.; Tavares, D.D.S.; Guimaraes, A.G. Antitumor Effects of Carvacrol and Thymol: A Systematic Review. Front. Pharmacol. 2021, 12, 702487. [Google Scholar] [CrossRef]
- Silva, E.R.; de Carvalho, F.O.; Teixeira, L.G.B.; Santos, N.G.L.; Felipe, F.A.; Santana, H.S.R.; Shanmugam, S.; Quintans Junior, L.J.; de Souza Araujo, A.A.; Nunes, P.S. Pharmacological Effects of Carvacrol in In vitro Studies: A Review. Curr. Pharm. Des. 2018, 24, 3454–3465. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, M.Y.; Kujawska, M.; Nikbakhtzadeh, M.; Hassanshahi, A.; Ramezanpour, S.; Kamiab, Z.; Bazmandegan, G. Carvacrol as a Potential Neuroprotective Agent for Neurological Diseases: A Systematic Review Article. CNS Neurol. Disord. Drug Targets 2021, 20, 942–953. [Google Scholar] [CrossRef]
- Adams, S.; Kunz, B.; Weidenbörner, M. Mycelial deformations of Cladosporium herbarum due to the application of eugenol or carvacrol. J. Essent. Oil Res. 1996, 8, 535–540. [Google Scholar] [CrossRef]
- Final Report on the Safety Assessment of Sodium p -Chloro- m -Cresol, p -Chloro- m -Cresol, Chlorothymol, Mixed Cresols, m -Cresol, o -Cresol, p -Cresol, Isopropyl Cresols, Thymol, o -Cymen-5-ol, and Carvacrol1. Int. J. Toxicol. 2006, 25, 29–127. [CrossRef]
- Abbaszadeh, S.; Sharifzadeh, A.; Shokri, H.; Khosravi, A.R.; Abbaszadeh, A. Antifungal efficacy of thymol, carvacrol, eugenol and menthol as alternative agents to control the growth of food-relevant fungi. J. Mycol. Med. 2014, 24, e51–e56. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.F.; dos Santos, A.R.; Coelho Trevisan, D.A.; Ribeiro, A.B.; Zanetti Campanerut-Sá, P.A.; Kukolj, C.; de Souza, E.M.; Cardoso, R.F.; Estivalet Svidzinski, T.I.; de Abreu Filho, B.A.; et al. Cinnamaldehyde induces changes in the protein profile of Salmonella Typhimurium biofilm. Res. Microbiol. 2018, 169, 33–43. [Google Scholar] [CrossRef]
- Ka, S.-M.; Chao, L.K.; Lin, J.-C.; Chen, S.-T.; Li, W.-T.; Lin, C.-N.; Cheng, J.-C.; Jheng, H.-L.; Chen, A.; Hua, K.-F. A low toxicity synthetic cinnamaldehyde derivative ameliorates renal inflammation in mice by inhibiting NLRP3 inflammasome and its related signaling pathways. Free. Radic. Biol. Med. 2016, 91, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Pu, C.; Zhou, P.; Wang, P.; Liang, D.; Wang, Q.; Hu, Y.; Li, B.; Hao, X. Cinnamaldehyde prevents endothelial dysfunction induced by high glucose by activating Nrf2. Cell. Physiol. Biochem. 2015, 36, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.K.; Hua, K.-F.; Hsu, H.-Y.; Cheng, S.-S.; Lin, I.-F.; Chen, C.-J.; Chen, S.-T.; Chang, S.-T. Cinnamaldehyde inhibits pro-inflammatory cytokines secretion from monocytes/macrophages through suppression of intracellular signaling. Food Chem. Toxicol. 2008, 46, 220–231. [Google Scholar] [CrossRef]
- Doyle, A.A.; Stephens, J.C. A review of cinnamaldehyde and its derivatives as antibacterial agents. Fitoterapia 2019, 139, 104405. [Google Scholar] [CrossRef]
- Tahjib-Ul-Arif, M.; Zahan, M.I.; Karim, M.M.; Imran, S.; Hunter, C.T.; Islam, M.S.; Mia, M.A.; Hannan, M.A.; Rhaman, M.S.; Hossain, M.A.; et al. Citric Acid-Mediated Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2021, 22, 7235. [Google Scholar] [CrossRef]
- Jiao, H.; Xu, W.; Hu, Y.; Tian, R.; Wang, Z. Citric Acid in Rice Root Exudates Enhanced the Colonization and Plant Growth-Promoting Ability of Bacillus altitudinis LZP02. Microbiol. Spectr. 2022, 10, e0100222. [Google Scholar] [CrossRef]
- Mallhi, Z.I.; Rizwan, M.; Mansha, A.; Ali, Q.; Asim, S.; Ali, S.; Hussain, A.; Alrokayan, S.H.; Khan, H.A.; Alam, P.; et al. Citric Acid Enhances Plant Growth, Photosynthesis, and Phytoextraction of Lead by Alleviating the Oxidative Stress in Castor Beans. Plants 2019, 8, 525. [Google Scholar] [CrossRef]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to software and statistical methods. In PRIMER-E; PRIMER-E Ltd.: Plymouth, UK, 2008; p. 218. [Google Scholar]
- Whitton, M.M.; Ren, X.; Yu, S.J.; Irving, A.D.; Trotter, T.; Bajagai, Y.S.; Stanley, D. Sea Minerals Reduce Dysbiosis, Improve Pasture Productivity and Plant Morphometrics in Pasture Dieback Affected Soils. Sustainability 2022, 14, 14873. [Google Scholar] [CrossRef]
- Whitton, M.M.; Ren, X.; Yu, S.J.; Trotter, T.; Stanley, D.; Bajagai, Y.S. Remediation of Pasture Dieback Using Plant Growth Promotant. Agronomy 2022, 12, 3153. [Google Scholar] [CrossRef]
- Ranjani, A.; Dhanasekaran, D.; Gopinath, P.M. An Introduction to Actinobacteria; Chapter Actinobacteria—Basics and Biotechnological Applications; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Mao, W.; Wang, C.; Yin, S. Functional potential differences between Firmicutes and Proteobacteria in response to manure amendment in a reclaimed soil. Can. J. Microbiol. 2020, 66, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Islam, Z.F.; Cordero, P.R.; Feng, J.; Chen, Y.-J.; Bay, S.K.; Jirapanjawat, T.; Gleadow, R.M.; Carere, C.R.; Stott, M.B.; Chiri, E. Two Chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide. ISME J. 2019, 13, 1801–1813. [Google Scholar] [CrossRef] [PubMed]
- Sciences, Q.D.o.E.a. Climate Change in Australia. Available online: https://www.climatechangeinaustralia.gov.au/ (accessed on 1 November 2022).
- de Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, K.; Kumar, R.; Vijayanand, K.; Bhojani, G.; Kubavat, D. Structural And Functional Changes In Soil Microbes By Foliar Drift Spray of Seaweed Extract As Revealed By Metagenomics. Arch. Microbiol. 2021, 204, 72. [Google Scholar] [CrossRef]
- Egas, C.; Barroso, C.; Froufe, H.J.C.; Pacheco, J.; Albuquerque, L.; da Costa, M.S. Complete genome sequence of the Radiation-Resistant bacterium Rubrobacter radiotolerans RSPS-4. Stand. Genomic. Sci. 2014, 9, 1062–1075. [Google Scholar] [CrossRef]
- Peng, M.; Jia, H.; Wang, Q. The effect of land use on bacterial communities in saline–alkali soil. Curr. Microbiol. 2017, 74, 325–333. [Google Scholar] [CrossRef]
- Goswami, A.; Adkins-Jablonsky, S.; Barreto Filho, M.M.; Schilling, M.; Dawson, A.; Heiser, S.; O’Connor, A.; Walker, M.; Roberts, Q.; Morris, J. Heavy metal pollution impacts soil bacterial community structure and antimicrobial resistance at the Birmingham 35th Avenue Superfund Site. bioRxiv 2022. [Google Scholar] [CrossRef]
- Barabote, R.D.; Xie, G.; Leu, D.H.; Normand, P.; Necsulea, A.; Daubin, V.; Medigue, C.; Adney, W.S.; Xu, X.C.; Lapidus, A.; et al. Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations. Genome Res. 2009, 19, 1033–1043. [Google Scholar] [CrossRef]
- Wei, L.; Ouyang, S.; Wang, Y.; Shen, X.; Zhang, L. Solirubrobacter phytolaccae sp. nov., an endophytic bacterium isolated from roots of Phytolacca acinosa Roxb. Int. J. Syst. Evol. Microbiol. 2014, 64, 858–862. [Google Scholar] [CrossRef]
- Mahapatra, S.; Yadav, R.; Ramakrishna, W. Bacillus subtilis impact on plant growth, soil health and environment: Dr. Jekyll and Mr. Hyde. J. Appl. Microbiol. 2022, 132, 3543–3562. [Google Scholar] [CrossRef]
- Cao, Y.R.; Wang, Q.; Jin, R.X.; Jiang, Y.; Lai, H.X.; He, W.X.; Xu, L.H.; Jiang, C.L. Planosporangium mesophilum sp. nov., isolated from rhizosphere soil of Bletilla striata. Int. J. Syst. Evol. Microbiol. 2011, 61, 1330–1333. [Google Scholar] [CrossRef]
- Seki, T.; Matsumoto, A.; Shimada, R.; Inahashi, Y.; Omura, S.; Takahashi, Y. Conexibacter arvalis sp. nov., isolated from a cultivated field soil sample. Int. J. Syst. Evol. Microbiol. 2012, 62, 2400–2404. [Google Scholar] [CrossRef]
- Seki, T.; Matsumoto, A.; Ōmura, S.; Takahashi, Y. Distribution and isolation of strains belonging to the order Solirubrobacterales. J. Antibiot. 2015, 68, 763–766. [Google Scholar] [CrossRef]
- Loh, J.; Stacey, G. Nodulation gene regulation in Bradyrhizobium japonicum: A unique integration of global regulatory circuits. Appl. Environ. Microbiol. 2003, 69, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Meakin, G.E.; Jepson, B.J.; Richardson, D.J.; Bedmar, E.J.; Delgado, M.J. The role of Bradyrhizobium japonicum nitric oxide reductase in nitric oxide detoxification in soya bean root nodules. Biochem. Soc. Trans. 2006, 34, 195–196. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Bhattacharyya, S.; Kumar, R.; Kumar, A.; Ibanez, F.; Wang, J.; Guo, B.; Sudini, H.K.; Gopalakrishnan, S.; DasGupta, M.; et al. Molecular Basis of Root Nodule Symbiosis between Bradyrhizobium and ‘Crack-Entry’ Legume Groundnut (Arachis hypogaea L.). Plants 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Lee, H.G.; Kim, H.S.; Ahn, C.Y.; Oh, H.M. Geodermatophilus soli sp. nov. and Geodermatophilus terrae sp. nov., two actinobacteria isolated from grass soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 2625–2629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Chen, J.; Liu, H.Y.; Zhang, Y.Q.; Li, W.J.; Yu, L.Y. Geodermatophilus ruber sp. nov., isolated from rhizosphere soil of a medicinal plant. Int. J. Syst. Evol. Microbiol. 2011, 61, 190–193. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Zhang, X.; Huang, J.; Zhao, Y.; Zhao, Y.; Liu, J.; Huang, C.; Wang, J.; Hu, Y.; et al. Geodermatophilus daqingensis sp. nov., isolated from petroleum-contaminated soil. Antonie Van Leeuwenhoek 2017, 110, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.S.; Babalola, O.O. Streptomyces: Implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019, 103, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Whang, K.S. Micromonospora fulva sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 1746–1751. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, X.X.; Qu, Z.; Wang, H.L.; Lin, H.P.; Xie, Q.Y.; Ruan, J.S.; Hong, K. Micromonospora rhizosphaerae sp. nov., isolated from mangrove rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Thawai, C.; Kittiwongwattana, C.; Thanaboripat, D.; Laosinwattana, C.; Koohakan, P.; Parinthawong, N. Micromonospora soli sp. nov., isolated from rice rhizosphere soil. Antonie Van Leeuwenhoek 2016, 109, 449–456. [Google Scholar] [CrossRef]
| Distance | Significance of | p-Value | Significance |
|---|---|---|---|
| Weighted UniFrac | Treatment | 0.247 | ns |
| Time | 0.001 | *** | |
| Plot | 0.009 | ** | |
| Unweighted UniFrac | Treatment | 0.241 | ns |
| Time | 0.001 | *** | |
| Plot | 0.012 | * | |
| Phylum level Bray–Curtis | Treatment | 0.0273 | * |
| Time | 0.002 | ** | |
| Plot | 0.466 | ns | |
| Genus level Bray–Curtis | Treatment | 0.384 | ns |
| Time | 0.001 | *** | |
| Plot | 0.001 | *** |
| Phylum | Contribution (%) |
|---|---|
| Firmicutes | 19.38 |
| Proteobacteria | 10.14 |
| Actinobacteriota | 9.59 |
| Verrucomicrobiota | 9.32 |
| Acidobacteriota | 7.53 |
| Myxococcota | 6.44 |
| Chloroflexi | 6.42 |
| Gemmatimonadota | 5.73 |
| Genus | Contribution (%) |
| Bacillus | 7.19 |
| Rubrobacter | 4.96 |
| Solirubrobacterales 67-14 | 3.06 |
| Solirubrobacter | 2.74 |
| Acidothermus | 2.39 |
| Conexibacter | 2.31 |
| Gaiella | 2.28 |
| MB-A2-108 | 2.26 |
| Micromonospora | 2.21 |
| Planosporangium | 2.11 |
| Candidatus Udaeobacter | 1.89 |
| Geodermatophilus | 1.77 |
| Jatrophihabitans | 1.45 |
| Pseudonocardia | 1.25 |
| Chloroflexi TK10 | 1.22 |
| Chloroflexi JG30-KF-CM45 | 1.21 |
| Streptomyces | 1.07 |
| Pyrinomonadaceae RB41 | 1 |
| Rhodoplanes | 0.93 |
| IMCC26256 | 0.9 |
| Actinoallomurus | 0.81 |
| Actinoplanes | 0.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, X.; Whitton, M.M.; Yu, S.J.; Trotter, T.; Bajagai, Y.S.; Stanley, D. Application of Phytogenic Liquid Supplementation in Soil Microbiome Restoration in Queensland Pasture Dieback. Microorganisms 2023, 11, 561. https://doi.org/10.3390/microorganisms11030561
Ren X, Whitton MM, Yu SJ, Trotter T, Bajagai YS, Stanley D. Application of Phytogenic Liquid Supplementation in Soil Microbiome Restoration in Queensland Pasture Dieback. Microorganisms. 2023; 11(3):561. https://doi.org/10.3390/microorganisms11030561
Chicago/Turabian StyleRen, Xipeng, Maria M. Whitton, Sung J. Yu, Tieneke Trotter, Yadav S. Bajagai, and Dragana Stanley. 2023. "Application of Phytogenic Liquid Supplementation in Soil Microbiome Restoration in Queensland Pasture Dieback" Microorganisms 11, no. 3: 561. https://doi.org/10.3390/microorganisms11030561
APA StyleRen, X., Whitton, M. M., Yu, S. J., Trotter, T., Bajagai, Y. S., & Stanley, D. (2023). Application of Phytogenic Liquid Supplementation in Soil Microbiome Restoration in Queensland Pasture Dieback. Microorganisms, 11(3), 561. https://doi.org/10.3390/microorganisms11030561










