Genomic Characterization of Multidrug-Resistant Extended Spectrum β-Lactamase-Producing Klebsiella pneumoniae from Clinical Samples of a Tertiary Hospital in South Kivu Province, Eastern Democratic Republic of Congo
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Clearance
2.2. Bacterial Strains
2.3. Antimicrobial Susceptibility Testing
2.4. Bacterial DNA Extraction
2.5. Library Preparation and Next Generation Sequencing
2.6. Genome Assembly, Annotation, and Alignment
3. Results
3.1. Bacterial Strains
3.2. Antimicrobial Susceptibility Testing
3.3. Phylogenomic Analysis
3.4. Genetic Determinants of Virulence
3.4.1. Detection of Virulence Factors Genes
3.4.2. Replicon Typing
3.4.3. Identification of the Capsule Type
3.4.4. Detection of AMR Genes
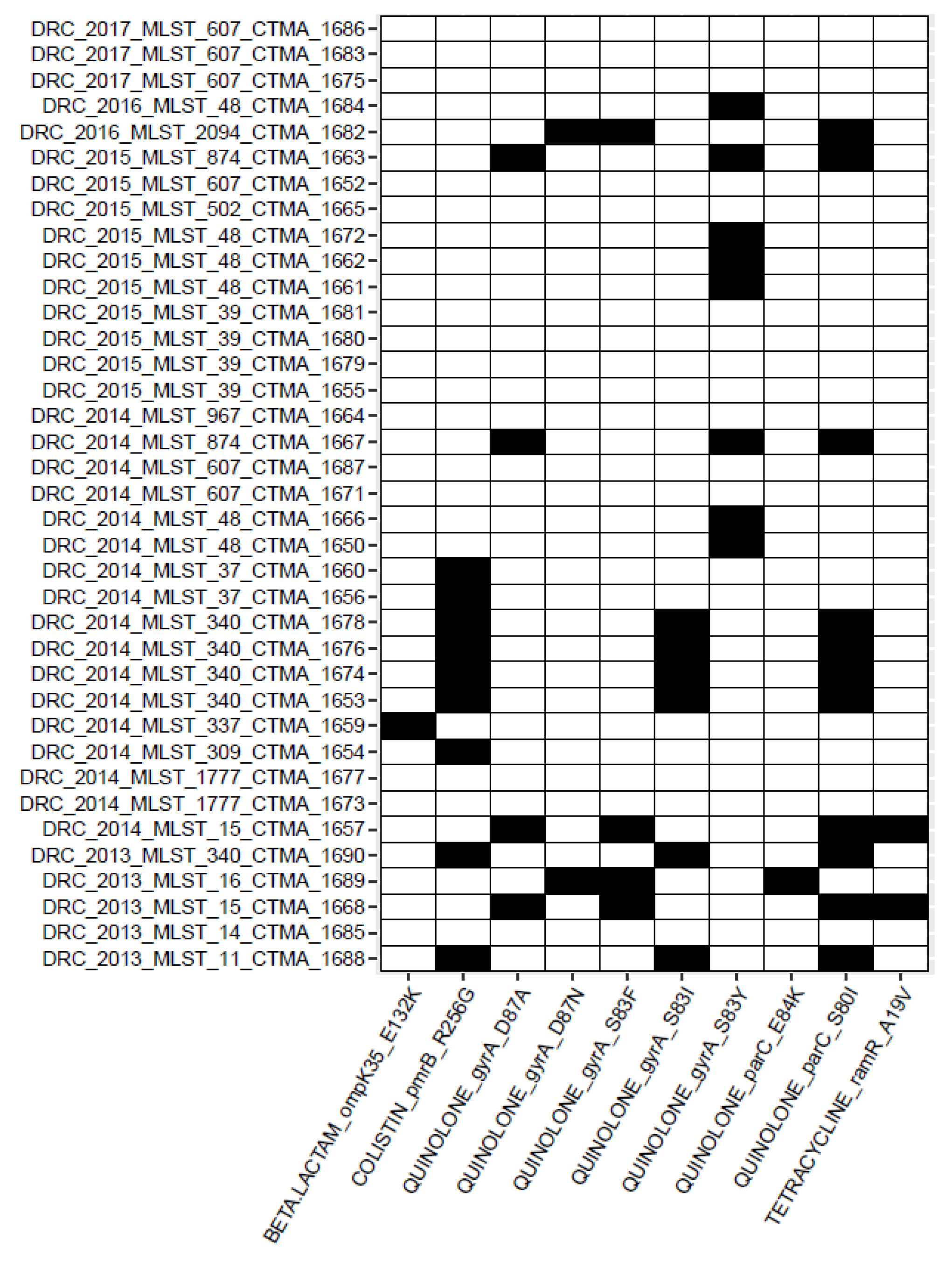
3.4.5. Detection of AMR Point Mutations
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tseng, W.P.; Chen, Y.C.; Chen, S.Y.; Chen, S.Y.; Chang, S.C. Risk for subsequent infection and mortality after hospitalization among patients with multidrug-resistant gram-negative bacteria colonization or infection. Antimicrob. Resist. Infect. Control 2018, 7, 93. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, M.; Ramachandran, B.; Barabadi, H. The prevalence and drug resistance pattern of extended spectrum β-lactamases (ESBLs) producing Enterobacteriaceae in Africa. Microb. Pathog. 2018, 114, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Casellas, J.M. Antibacterial drug resistance in Latin America: Consequences for infectious disease control. Rev. Panam Salud Publica 2011, 30, 519–528. [Google Scholar] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Pitout, J.D.D. Enterobacteriaceae that produce extended-spectrum β-lactamases and AmpC β-lactamases in the community: The tip of the iceberg? Curr. Pharm. Des. 2013, 19, 257–263. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef]
- Aibinu, I.; Odugbemi, T.; Koenig, W.; Ghebremedhin, B. Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin. Microbiol. Infect. 2012, 18, E49–E51. [Google Scholar] [CrossRef]
- Irenge, L.M.; Ambroise, J.; Bearzatto, B.; Durant, J.F.; Chirimwami, R.B.; Gala, J.L. Whole-genome sequences of multi-drug-resistant Escherichia coli in South-Kivu Province, Democratic Republic of Congo: Characterization of phylogenomic changes, virulence and resistance genes. BMC Infect. Dis. 2019, 19, 137. [Google Scholar] [CrossRef]
- Martischang, R.; Francois, P.; Cherkaoui, A.; Gaia, N.; Renzi, G.; Agostinho, A.; Perez, M.; Graf, C.E.; Harbarth, S. Epidemi-ology of ESBL-producing Escherichia coli from repeated prevalence studies over 11 years in a long-term-care facility. Antimicrob. Resist. Infect. Control 2021, 10, 148. [Google Scholar] [CrossRef]
- Eger, E.; Heiden, S.E.; Korolew, K.; Bayingana, C.; Ndoli, J.M.; Sendegeya, A.; Gahutu, J.B.; Kurz, M.S.E.; Mockenhaupt, F.P.; Muller, J.; et al. Circulation of Extended-Spectrum β-Lactamase-Producing Escherichia coli of Pandemic Sequence Types 131, 648, and 410 Among Hospitalized Patients, Caregivers, and the Community in Rwanda. Front. Microbiol. 2021, 12, 662575. [Google Scholar] [CrossRef]
- Tegha, G.; Ciccone, E.J.; Krysiak, R.; Kaphatika, J.; Chikaonda, T.; Ndhlovu, I.; van Duin, D.; Hoffman, I.; Juliano, J.J.; Wang, J. Genomic epidemiology of Escherichia coli isolates from a tertiary referral center in Lilongwe, Malawi. Microb. Genom. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Seni, J.; Peirano, G.; Mshana, S.E.; Pitout, J.D.D.; DeVinney, R. The importance of Escherichia coli clonal complex 10 and ST131 among Tanzanian patients on antimicrobial resistance surveillance programs. Eur. J. Clin. Microbiol. Infect. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Caldes, C.; Romero-Alvarez, D.; Valdez-Velez, V.; Morales, R.D.; Montalvo-Hernandez, A.; Gomes-Dias, C.; Calopina, M. Extended-Spectrum β-Lactamases Producing Escherichia coli in South America: A Systematic Review with a One Health Perspective. Infect. Drug Resist. 2022, 15, 5759–5779. [Google Scholar] [CrossRef] [PubMed]
- Onduru, O.G.; Mkakosya, R.S.; Aboud, S.; Rumisha, S.F. Genetic Determinants of Resistance among ESBL-Producing Enterobacteriaceae in Community and Hospital Settings in East, Central, and Southern Africa: A Systematic Review and Meta-Analysis of Prevalence. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 5153237. [Google Scholar] [CrossRef]
- Irenge, L.M.; Kabego, L.; Vandenberg, O.; Chirimwami, R.B.; Gala, J.L. Antimicrobial resistance in urinary isolates from inpatients and outpatients at a tertiary care hospital in South-Kivu Province (Democratic Republic of Congo). BMC Res. Notes 2014, 7, 374. [Google Scholar] [CrossRef] [PubMed]
- Irenge, L.M.; Kabego, L.; Kinunu, F.B.; Itongwa, M.; Mitangala, P.N.; Gala, J.L.; Chirimwami, R.B. Antimicrobial resistance of bacteria isolated from patients with bloodstream infections at a tertiary care hospital in the Democratic Republic of the Congo. S. Afr. Med. J. 2015, 105, 752–755. [Google Scholar] [CrossRef]
- Bitew, A.; Tsige, E. High Prevalence of Multidrug-Resistant and Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: A Cross-Sectional Study at Arsho Advanced Medical Laboratory, Addis Ababa, Ethiopia. J. Trop. Med. 2020, 2020, 6167234. [Google Scholar] [CrossRef]
- Moglad, E.H. Antibiotics Profile, Prevalence of Extended-Spectrum β-Lactamase (ESBL), and Multidrug-Resistant Enterobacteriaceae from Different Clinical Samples in Khartoum State, Sudan. Int. J. Microbiol. 2020, 2020, 8898430. [Google Scholar] [CrossRef]
- Osman, E.A.; Yokoyama, M.; Altayb, H.N.; Cantillon, D.; Wille, J.; Seifert, H.; Higgins, P.G.; Al Hassan, L. Klebsiella pneumonia in Sudan: Multidrug Resistance, Polyclonal Dissemination, and Virulence. Antibiotics 2023, 12, 233. [Google Scholar] [CrossRef]
- Muraya, A.; Kyany’a, C.; Kiyaga, S.; Smith, H.J.; Kibet, C.; Martin, M.J.; Kimani, J.; Musila, L. Antimicrobial Resistance and Virulence Characteristics of Klebsiella pneumoniae Isolates in Kenya by Whole-Genome Sequencing. Pathogens 2022, 11, 545. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae Population Genomics and Antimicrobial-Resistant Clones. Trends Microbiol. 2016, 24, 944–956. [Google Scholar] [CrossRef]
- Mulinganya, G.M.; Claeys, M.; Balolebwami, S.Z.; Bamuleke, B.A.; Mongane, J.I.; Boelens, J.; Delanghe, J.; De Vos, D.; Kam-bale, R.M.; Maheshe, G.B.; et al. Etiology of Early-Onset Neonatal Sepsis and Antibiotic Resistance in Bukavu, Democratic Republic of the Congo. Clin. Infect. Dis. 2021, 73, e976–e980. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. v.0.11.9. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 27 January 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, D581–D591. [Google Scholar] [CrossRef]
- Gardner, S.N.; Hall, B.G. When whole-genome alignments just won’t work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 2013, 8, e81760. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Wick, R.R.; Gorrie, C.; Jenney, A.; Follador, R.; Thomson, N.R.; Holt, K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016, 2, e000102. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.; Garcia-Fernandez, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef] [PubMed]
- Moubareck, C.A.; Mouftah, S.F.; Pal, T.; Ghazawi, A.; Halat, D.H.; Nabi, A.; AlSharhan, M.A.; AlDeesi, Z.O.; Peters, C.C.; Celiloglu, H.; et al. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int. J. Antimicrob. Agents 2018, 52, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Martins, W.; Nicolas, M.F.; Yu, Y.; Li, M.; Dantas, P.; Sands, K.; Portal, E.; Almeida, L.G.P.; Vasconcelos, A.T.R.; Medeiros, E.A.; et al. Clinical and Molecular Description of a High-Copy IncQ1 KPC-2 Plasmid Harbored by the International ST15 Klebsiella pneumoniae Clone. mSphere 2020, 5, e00756-20. [Google Scholar] [CrossRef]
- Karampatakis, T.; Zarras, C.; Pappa, S.; Vagdatli, E.; Iosifidis, E.; Roilides, E.; Papa, A. Emergence of ST39 carbapenem-resistant Klebsiella pneumoniae producing VIM-1 and KPC-2. Microb. Pathog. 2022, 162, 105373. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Bi, R.; Cao, X.; Qian, H.; Hu, R.; Ma, P. Clonal dissemination of KPC-2-producing Klebsiella pneumoniae ST11 and ST48 clone among multiple departments in a tertiary teaching hospital in Jiangsu Province, China. Ann. Transl. Med. 2019, 7, 716. [Google Scholar] [CrossRef]
- Tolentino, F.M.; Bueno, M.F.C.; Franscisco, G.R.; Barcelos, D.D.P.; Lobo, S.M.; Tomaz, F.; da Silva, N.S.; de Andrade, L.N.; Casella, T.; Darini, A.; et al. Endemicity of the High-Risk Clone Klebsiella pneumoniae ST340 Coproducing QnrB, CTX-M-15, and KPC-2 in a Brazilian Hospital. Microb. Drug. Resist. 2019, 25, 528–537. [Google Scholar] [CrossRef]
- Peirano, G.; Bradford, P.A.; Kazmierczak, K.M.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D. Importance of Clonal Complex 258 and IncF(K2-like) Plasmids among a Global Collection of Klebsiella pneumoniae with bla(KPC). Antimicrob. Agents Chemother. 2017, 61, e02610–e02616. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef] [PubMed]
- Remya, P.A.; Shanthi, M.; Sekar, U. Characterisation of virulence genes associated with pathogenicity in Klebsiella pneumoniae. Indian J. Med. Microbiol. 2019, 37, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Corvec, S.; Rapoport, M.; Mugnier, P.; Petroni, A.; Pasteran, F.; Faccone, D.; Galas, M.; Drugeon, H.; Cattoir, V.; et al. Identification of the novel narrow-spectrum β-lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob. Agents Chemother. 2007, 51, 2179–2184. [Google Scholar] [CrossRef] [PubMed]
- Agyepong, N.; Govinden, U.; Owusu-Ofori, A.; Amoako, D.G.; Allam, M.; Janice, J.; Pedersen, T.; Sundsfjord, A.; Essack, S. Genomic characterization of multidrug-resistant ESBL-producing Klebsiella pneumoniae isolated from a Ghanaian teaching hospital. Int. J. Infect. Dis. 2019, 85, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Afolayan, A.O.; Oaikhena, A.O.; Aboderin, A.O.; Olabisi, O.F.; Amupitan, A.A.; Abiri, O.V.; Ogunleye, V.O.; Odih, E.E.; Adeyemo, A.T.; Adeyemo, A.T.; et al. Clones and Clusters of Antimicrobial-Resistant Klebsiella From Southwestern Nigeria. Clin. Infect. Dis. 2021, 73, S308–S315. [Google Scholar] [CrossRef]
- Can, F.; Menekse, S.; Ispir, P.; Atac, N.; Albayrak, O.; Demir, T.; Karaaslan, D.C.; Karahan, S.N.; Kapmaz, M.; Kurt Azap, O.; et al. Impact of the ST101 clone on fatality among patients with colistin-resistant Klebsiella pneumoniae infection. J. Antimicrob. Chemother. 2018, 73, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D.D. Emerging Antimicrobial-Resistant High-Risk Klebsiella pneumoniae Clones ST307 and ST147. Antimicrob. Agents Chemother. 2020, 64, e01148-20. [Google Scholar] [CrossRef]
- Marsh, J.W.; Mustapha, M.M.; Griffith, M.P.; Evans, D.R.; Ezeonwuka, C.; Pasculle, A.W.; Shutt, K.A.; Sundermann, A.; Ayres, A.M.; Shields, R.K.; et al. Evolution of Outbreak-Causing Carbapenem-Resistant Klebsiella pneumoniae ST258 at a Tertiary Care Hospital over 8 Years. mBio 2019, 10, e01945-19. [Google Scholar] [CrossRef]
- Villa, L.; Feudi, C.; Fortini, D.; Brisse, S.; Passet, V.; Bonura, C.; Endimiani, A.; Mammina, C.; Ocampo, A.M.; Jimenez, J.N.; et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genom. 2017, 3, e000110. [Google Scholar] [CrossRef]
- Loconsole, D.; Accogli, M.; De Robertis, A.L.; Capozzi, L.; Bianco, A.; Morea, A.; Mallamaci, R.; Quarto, M.; Parisi, A.; Chironna, M. Emerging high-risk ST101 and ST307 carbapenem-resistant Klebsiella pneumoniae clones from bloodstream infections in Southern Italy. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 24. [Google Scholar] [CrossRef] [PubMed]
- Peona, V.; Weissensteiner, M.H.; Suh, A. How complete are “complete” genome assemblies? An avian perspective. Mol. Ecol. Resour. 2018, 18, 1188–1195. [Google Scholar] [CrossRef] [PubMed]


| Isolate | Tissue | ST | AMP | AMX | AMC | FEP | CTX | CAZ | CRO | IPM | MERO | CIP | AKN | GENT | TET | COL | NITRO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTMA-1678 | Bloodstream | 340 | >256 | >256 | >256 | 48 | >256 | >256 | >32 | 0.094 | 0.75 | >32 | 12 | 96 | >256 | 0.25 | >512 |
| CTMA-1674 | Bloodstream | 340 | >256 | >256 | >256 | 64 | >256 | >256 | >32 | 0.094 | 0.094 | >32 | 6 | >256 | >256 | 0.25 | >512 |
| CTMA-1676 | Bloodstream | 340 | >256 | >256 | >256 | 64 | >256 | >256 | >32 | 0.094 | 0.047 | >32 | 8 | 64 | >256 | 0.25 | >512 |
| CTMA-1653 | Bloodstream | 340 | >256 | >256 | >256 | 32 | >256 | >256 | >32 | 0.5 | 0.47 | >32 | 8 | 64 | >256 | 0.25 | >512 |
| CTMA-1690 | UTI | 340 | >256 | >256 | >256 | 64 | >256 | 192 | >32 | 0.19 | 0.094 | >32 | 8 | 128 | >256 | <0.25 | >512 |
| CTMA-1668 | UTI | 15 | >256 | >256 | >256 | 48 | >256 | >256 | >32 | 0.25 | 0.032 | >32 | 6 | 96 | >256 | 0.25 | 48 |
| CTMA-1660 | UTI | 37 | >256 | >256 | 16 | 0.25 | 12 | 12 | >32 | 0.125 | 0.023 | 1 | 24 | >256 | >256 | <0.25 | 3 |
| CTMA-1656 | UTI | 37 | >256 | >256 | >256 | 0.75 | 24 | 24 | >32 | 0.19 | 0.23 | 1 | 32 | >256 | >256 | 0.25 | 6 |
| CTMA-1654 | UTI | 309 | >256 | >256 | >256 | 12 | >256 | >256 | >32 | 0.125 | 0.032 | >32 | 24 | >256 | >256 | 0.25 | 64 |
| CTMA-1689 | UTI | 16 | >256 | >256 | >256 | 48 | >256 | >256 | >32 | 0.25 | 0.002 | >32 | 8 | 96 | >256 | 0.25 | >512 |
| CTMA-1682 | UTI | 2094 | >256 | >256 | >256 | 64 | >256 | >256 | >32 | 0.023 | 0.125 | >32 | 12 | 48 | 4 | 0.5 | 32 |
| CTMA-1677 | Bloodstream | 1777 | >256 | >256 | >256 | 12 | 64 | 16 | >32 | 0.094 | 0.064 | 1.5 | 3 | 96 | >256 | 0.25 | >512 |
| CTMA-1673 | Bloodstream | 1777 | >256 | >256 | 12 | 12 | >256 | >256 | >32 | 0.25 | 0.047 | 1.5 | 3 | 96 | >256 | 0.25 | 128 |
| CTMA-1665 | Peritoneal fluid | 502 | >256 | >256 | >256 | 24 | >256 | >256 | >32 | 0.002 | 0.23 | 4 | 24 | 128 | >256 | <0.25 | 12 |
| CTMA-1680 | Bloodstream | 39 | >256 | >256 | >256 | 24 | 96 | >256 | >32 | 0.125 | 0.032 | 3 | 48 | >256 | >256 | 0.25 | 96 |
| CTMA-1679 | Bloodstream | 39 | >256 | >256 | >256 | 24 | >256 | >256 | >32 | 0.125 | 0.064 | 2 | 48 | >256 | >256 | 0.25 | 32 |
| CTMA-1681 | Bloodstream | 39 | >256 | >256 | >256 | 32 | >256 | 32 | >32 | 0.125 | 0.032 | 2 | 64 | >256 | >256 | 0.5 | 96 |
| CTMA-1655 | UTI | 39 | >256 | >256 | >256 | 32 | >256 | >256 | >32 | 0.19 | 0.32 | 2 | 24 | >256 | >256 | 0.25 | 6 |
| CTMA-1664 | UTI | 967 | >256 | >256 | 8 | 8 | >256 | 24 | >32 | 0.064 | 0.47 | 1.5 | 8 | 24 | >256 | 0.25 | 64 |
| CTMA-1659 | UTI | 337 | >256 | >256 | 8 | 24 | >256 | 16 | >32 | 0.125 | 0.05 | 0.75 | 2 | 0.75 | >256 | 0.25 | 24 |
| CTMA-1661 | Soft tissue | 48 | >256 | >256 | >256 | 16 | >256 | >256 | >32 | 0.094 | 0.023 | 6 | 6 | 32 | >256 | <0.25 | 128 |
| CTMA-1666 | Bloodstream | 48 | >256 | >256 | 64 | 8 | >256 | >256 | >32 | 0.023 | 0.125 | 1.5 | 8 | 24 | 64 | <0.25 | 96 |
| CTMA-1672 | Bloodstream | 48 | >256 | >256 | 1.5 | 16 | >256 | >256 | >32 | 0.19 | 0.047 | >32 | 16 | 96 | >256 | 0.25 | >512 |
| CTMA-1684 | Bloodstream | 48 | >256 | >256 | >256 | 16 | >256 | >256 | >32 | 0.094 | 0.047 | 8 | 12 | 32 | 16 | 0.5 | 128 |
| CTMA-1650 | Bloodstream | 48 | >256 | >256 | >256 | 16 | >256 | >256 | >32 | 0.094 | 0.023 | >32 | 16 | 96 | >256 | 0.25 | >512 |
| CTMA-1662 | Soft tissue | 48 | >256 | >256 | 64 | 24 | >256 | >256 | >32 | 0.094 | 0.023 | 6 | 6 | 32 | >256 | <0.25 | 128 |
| CTMA-1667 | Bloodstream | 874 | >256 | >256 | >256 | 1.5 | >256 | >256 | >32 | 0.047 | 0.094 | 0.5 | 38 | 0.094 | >256 | 0.25 | 16 |
| CTMA-1663 | Bloodstream | 874 | >256 | >256 | 48 | 24 | >256 | >256 | >32 | 0.5 | 0.047 | >32 | 8 | 96 | >256 | 0.25 | 192 |
| CTMA-1683 | Bloodstream | 607 | >256 | >256 | >256 | 16 | >256 | >256 | >32 | 0.256 | 0.125 | 2 | 4 | 128 | >256 | 0.25 | >512 |
| CTMA-1687 | UTI | 607 | >256 | >256 | >256 | >256 | >256 | >256 | >32 | 0.094 | 0.064 | 1.000 | 3 | >256 | >256 | 0.5 | >512 |
| CTMA-1686 | UTI | 607 | >256 | >256 | >256 | >256 | >256 | >256 | >32 | 0.19 | 0.023 | 0.094 | 6 | 192 | 1.5 | 0.5 | >512 |
| CTMA-1671 | Bloodstream | 607 | >256 | >256 | >256 | 1.5 | >256 | >256 | >32 | 0.094 | 0.047 | >32.000 | 12 | 128 | >256 | 0.25 | >512 |
| CTMA-1675 | Bloodstream | 607 | >256 | >256 | >256 | 48 | >256 | >256 | >32 | 0.125 | 0.047 | 0.125 | 4 | >256 | >256 | 0.25 | 192 |
| CTMA-1652 | UTI | 607 | >256 | >256 | >256 | >256 | >256 | >256 | >32 | 0.125 | 0.47 | 0.125 | 4 | >256 | >256 | 0.25 | >512 |
| CTMA-1685 | UTI | 14 | >256 | >256 | >256 | 6 | >256 | >256 | >32 | 0.19 | 0.023 | 1.5 | 6 | 96 | 128 | 0.5 | >512 |
| CTMA-1657 | UTI | 15 | >256 | >256 | >256 | 16 | >256 | >256 | >32 | 0.125 | 0.06 | >32 | 8 | 64 | >256 | 0.25 | 64 |
| CTMA-1688 | Bloodstream | 11 | >256 | >256 | >256 | 48 | >256 | >256 | >32 | 0.256 | 0.023 | 1.500 | 6 | 64 | 2 | 0.25 | 48 |
| Antibiotics | Resistance (R) n = 37 (%) | Susceptible (S) n = 37 (%) | Area of Technical Uncertainty (ATU) n = 37 (%) |
|---|---|---|---|
| Ampicillin | 37 (100) | 0 (0) | 0 (0) |
| Amoxicillin | 37 (100) | 0 (0) | 0 (0) |
| Amoxicillin-clavulanate | 35 (94.6) | 2 (5.4) | 0 (0) |
| Cefepime | 34 (91.9) | 3 (8.1) | 0 (0) |
| Cefotaxime | 37 (100) | 0 (0) | 0 (0) |
| Ceftazidime | 37 (100) | 0 (0) | 0 (0) |
| Ceftriaxone | 37 (100) | 0 (0) | 0 (0) |
| Imipenem | 0 (0) | 37 (100) | 0 (0) |
| Meropenem | 0 (0) | 37 (100) | 0 (0) |
| Ciprofloxacin | 35 (94.6) | 1 (2.7) | 1 (2.7) |
| Amikacin | 15 (40.5) | 22 (59.5) | 0 (0) |
| Gentamicin | 35 (94.6) | 2 (5.4) | 0 (0) |
| Tetracycline | 34 (91.9) | 3 (8.1) | 0 (0) |
| Colistin | 0 (0) | 37 (100) | 0 (0) |
| Nitrofurantoin | 24 (64.9) | 13 (35.1) | 0 (0) |
| Year of Isolation | Strain | Origin | ST | Replicon ST | K_Type | O Type | Yersiniabactin | (Hyper) Virulence Genes | Virulence Score |
|---|---|---|---|---|---|---|---|---|---|
| 2014 | CTMA_1678 | Bloodstream | ST340 | F1:A−:B− | unknown (KL107) | unknown (O4) | ybt 9; ICEKp3 (truncated) | - | 1 |
| 2014 | CTMA_1674 | Bloodstream | ST340 | F1:A13:B− | unknown (K15) | O4 | ybt 9; ICEKp3 (truncated) | - | 1 |
| 2014 | CTMA_1676 | Bloodstream | ST340 | F1:A13:B− | unknown (K15) | O4 | ybt 9; ICEKp3 (truncated) | - | 1 |
| 2014 | CTMA_1653 | Bloodstream | ST340 | F1:A13:B− | unknown (KL107) | unknown (O4) | ybt 9; ICEKp3 (truncated) | - | 1 |
| 2013 | CTMA_1690 | UTI | ST340 | F−:A13:B− | unknown (K15) | O4 | ybt 14; ICEKp5 | - | 1 |
| 2013 | CTMA_1688 | UTI | ST11 | F13:A13:B− | unknown (KL105) | O2afg | ybt 9; ICEKp3 | - | 1 |
| 2014 | CTMA_1660 | UTI | ST37 | F−:A13:B− | K14 | O3b | ybt 15; ICEKp11 | - | 1 |
| 2014 | CTMA_1656 | UTI | ST37 | F−:A13:B− | K14 | O3b | ybt 15; ICEKp11 | - | 1 |
| 2014 | CTMA_1654 | UTI | ST309 | ND | unknown (K42) | unknown (O4) | - | - | 0 |
| 2013 | CTMA_1689 | UTI | ST16 | F12:A26:B− | unknown (K51) | O3b | ybt 9; ICEKp3 | - | 1 |
| 2016 | CTMA_1682 | UTI | ST2094 | F−:A13:B− | unknown (KL107) | unknown (OL101) | - | - | 0 |
| 2014 | CTMA_1677 | Bloodstream | ST1777 | F13:A13:B− | unknown (KL111) | O3b | - | - | 0 |
| 2014 | CTMA_1673 | Bloodstream | ST1777 | F13:A13:B− | unknown (KL111) | O3b | - | - | 0 |
| 2015 | CTMA_1665 | Soft tissue | ST502 | F13:A−:B− | unknown (K15) | unknown (O4) | - | - | 0 |
| 2015 | CTMA_1680 | Bloodstream | ST39 | F2:A13:B− | unknown (K2) | O2a | ybt 16; ICEKp12 | - | 1 |
| 2015 | CTMA_1679 | Bloodstream | ST39 | F2:A13:B− | unknown (K2) | O1 | ybt 16; ICEKp12 | - | 1 |
| 2015 | CTMA_1681 | Bloodstream | ST39 | F2:A13:B− | unknown (K2) | O1 | ybt 16; ICEKp12 | - | 1 |
| 2015 | CTMA_1655 | UTI | ST39 | F2:A13:B− | K2 | O1 | ybt 16; ICEKp12 | - | 1 |
| 2014 | CTMA_1664 | UTI | ST967 | F2:A−:B− | unknown (K18) | O2afg | - | - | 0 |
| 2014 | CTMA_1659 | UTI | ST337 | ND | unknown (KL109) | O2afg | - | - | 0 |
| 2015 | CTMA_1661 | Peritoneal fluid | ST48 | F13:A−:B− | unknown (K62) | O1 | ybt 14; ICEKp5 | - | 1 |
| 2014 | CTMA_1666 | Bloodstream | ST48 | F13:A−:B− | unknown (K62) | O2a | ybt 14; ICEKp5 | - | 1 |
| 2015 | CTMA_1672 | Bloodstream | ST48 | F13:A−:B− | unknown (K62) | unknown (O2a) | ybt 14; ICEKp5 | - | 1 |
| 2016 | CTMA_1684 | Bloodstream | ST48 | F13:A−:B− | unknown (K62) | O1 | ybt 14; ICEKp5 | - | 1 |
| 2014 | CTMA_1650 | Bloodstream | ST48 | F13:A−:B− | K62 | O1 | ybt 14; ICEKp5 | - | 1 |
| 2015 | CTMA_1662 | Peritoneal fluid | ST48 | F13:A−:B− | unknown (K62) | O1 | ybt 14; ICEKp5 | - | 1 |
| 2014 | CTMA_1667 | Bloodstream | ST874 | ND | unknown (K45) | O1 | ybt 14; ICEKp12 | - | 1 |
| 2015 | CTMA_1663 | Bloodstream | ST874 | ND | unknown (K45) | unknown (O1) | ybt 14; ICEKp12 | - | 1 |
| 2017 | CTMA_1683 | Bloodstream | ST607 | F13:A13:B− | unknown (K25) | O1 | ybt 14; ICEKp5 | - | 1 |
| 2014 | CTMA_1687 | UTI | ST607 | F13:A13:B− | unknown (K25) | O1 | - | - | 0 |
| 2017 | CTMA_1686 | UTI | ST607 | F13:A13:B− | unknown (K25) | O1 | ybt 14; ICEKp5 | - | 1 |
| 2014 | CTMA_1671 | Bloodstream | ST607 | F13:A13:B− | unknown (K25) | O2a | ybt 14; ICEKp5 | - | 1 |
| 2017 | CTMA_1675 | Bloodstream | ST607 | F13:A13:B− | unknown (K25) | O1 | ybt 14; ICEKp5 | - | 1 |
| 2015 | CTMA_1652 | UTI | ST607 | F13:A13:B− | unknown (K25) | O2a | ybt 14; ICEKp5 | - | 1 |
| 2013 | CTMA_1685 | UTI | ST14 | F9:A−:B− | K2 | O1 | - | 0 | |
| 2014 | CTMA_1657 | UTI | ST15 | F8:A13:B− | unknown (KL112) | O1 | - | - | 0 |
| 2013 | CTMA_1668 | Bloodstream | ST15 | F−:A13:B− | unknown (K48) | O2a | - | - | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irenge, L.M.; Ambroise, J.; Bearzatto, B.; Durant, J.-F.; Bonjean, M.; Gala, J.-L. Genomic Characterization of Multidrug-Resistant Extended Spectrum β-Lactamase-Producing Klebsiella pneumoniae from Clinical Samples of a Tertiary Hospital in South Kivu Province, Eastern Democratic Republic of Congo. Microorganisms 2023, 11, 525. https://doi.org/10.3390/microorganisms11020525
Irenge LM, Ambroise J, Bearzatto B, Durant J-F, Bonjean M, Gala J-L. Genomic Characterization of Multidrug-Resistant Extended Spectrum β-Lactamase-Producing Klebsiella pneumoniae from Clinical Samples of a Tertiary Hospital in South Kivu Province, Eastern Democratic Republic of Congo. Microorganisms. 2023; 11(2):525. https://doi.org/10.3390/microorganisms11020525
Chicago/Turabian StyleIrenge, Leonid M., Jérôme Ambroise, Bertrand Bearzatto, Jean-François Durant, Maxime Bonjean, and Jean-Luc Gala. 2023. "Genomic Characterization of Multidrug-Resistant Extended Spectrum β-Lactamase-Producing Klebsiella pneumoniae from Clinical Samples of a Tertiary Hospital in South Kivu Province, Eastern Democratic Republic of Congo" Microorganisms 11, no. 2: 525. https://doi.org/10.3390/microorganisms11020525
APA StyleIrenge, L. M., Ambroise, J., Bearzatto, B., Durant, J.-F., Bonjean, M., & Gala, J.-L. (2023). Genomic Characterization of Multidrug-Resistant Extended Spectrum β-Lactamase-Producing Klebsiella pneumoniae from Clinical Samples of a Tertiary Hospital in South Kivu Province, Eastern Democratic Republic of Congo. Microorganisms, 11(2), 525. https://doi.org/10.3390/microorganisms11020525







