Presence of Polyketide Synthase (PKS) Gene and Counterpart Virulence Determinants in Klebsiella pneumoniae Strains Enhances Colorectal Cancer Progression In-Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of K. pneumoniae from Clinical Specimens
2.2. Maintenance of CRC Cell Lines
2.3. Genomic DNA Extraction
2.4. PCR Detection of Virulence Genes including PKS Colibactin Genotoxin
2.5. Antibiotic Susceptibility Testing (AST)
2.6. Biofilm Assay
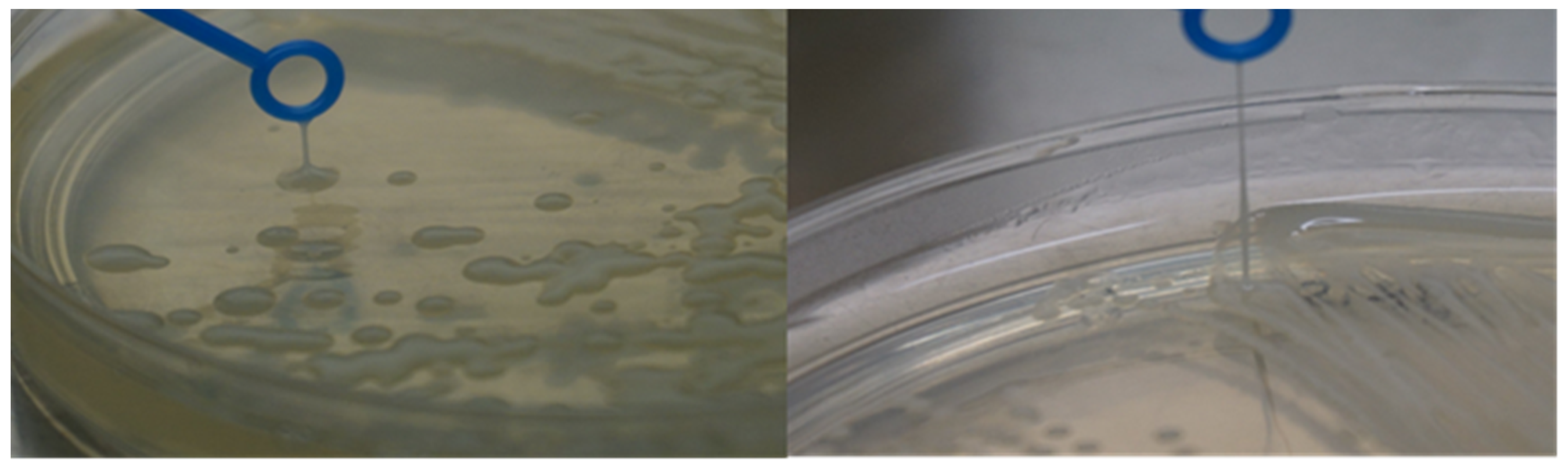
2.7. String Test
2.8. Solubilised Antigen Extraction (Crude Whole Cell Extract)
2.9. Enhancement of Cell Proliferation (In-Vitro)
2.10. Electrical Cell-Substrate Impedance Sensing (ECIS)
2.11. Statistical Analysis
3. Results
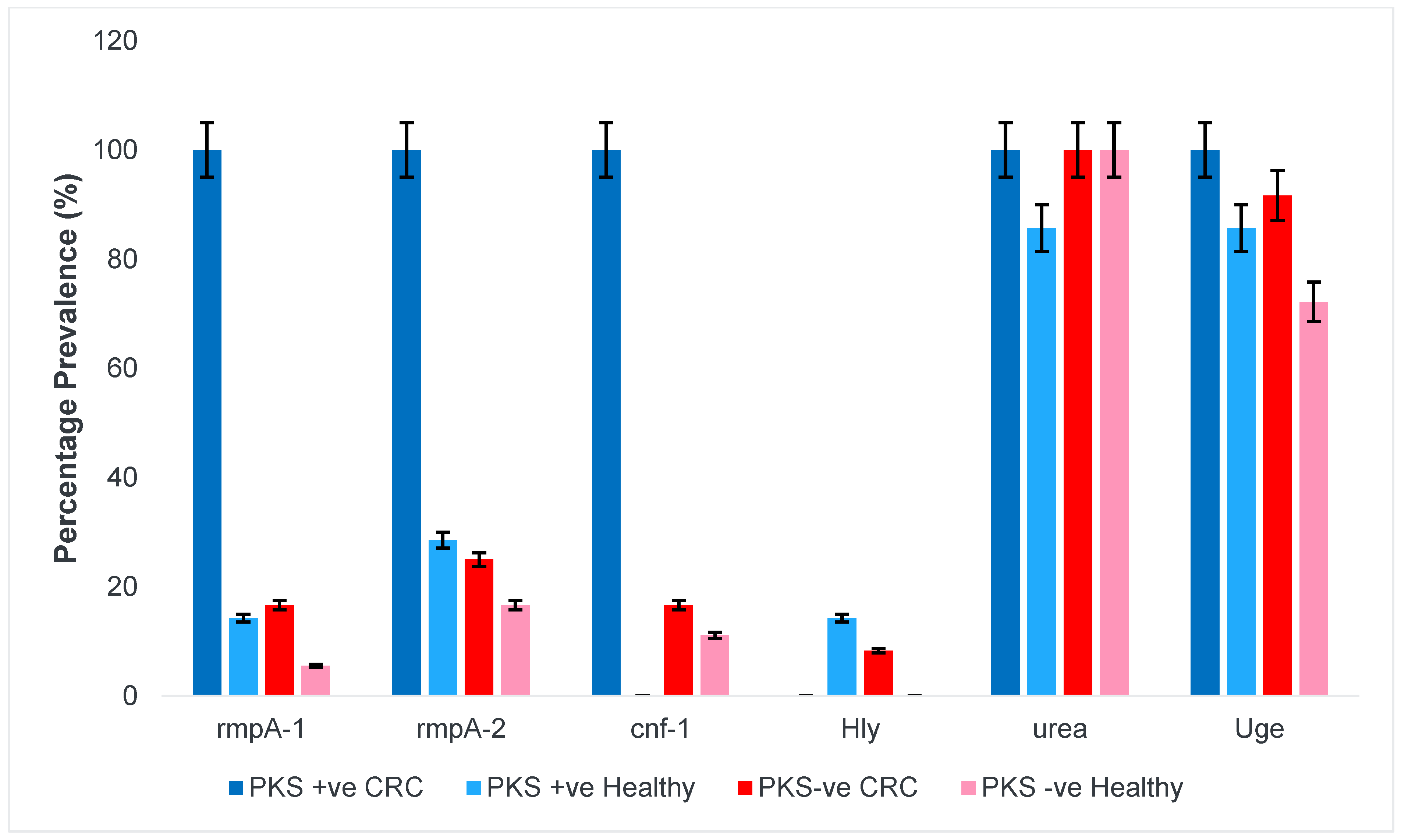
3.1. PKS-Positive K. pneumoniae Isolates Harboured More Virulence Genes Compared to PKS-Negative Isolates
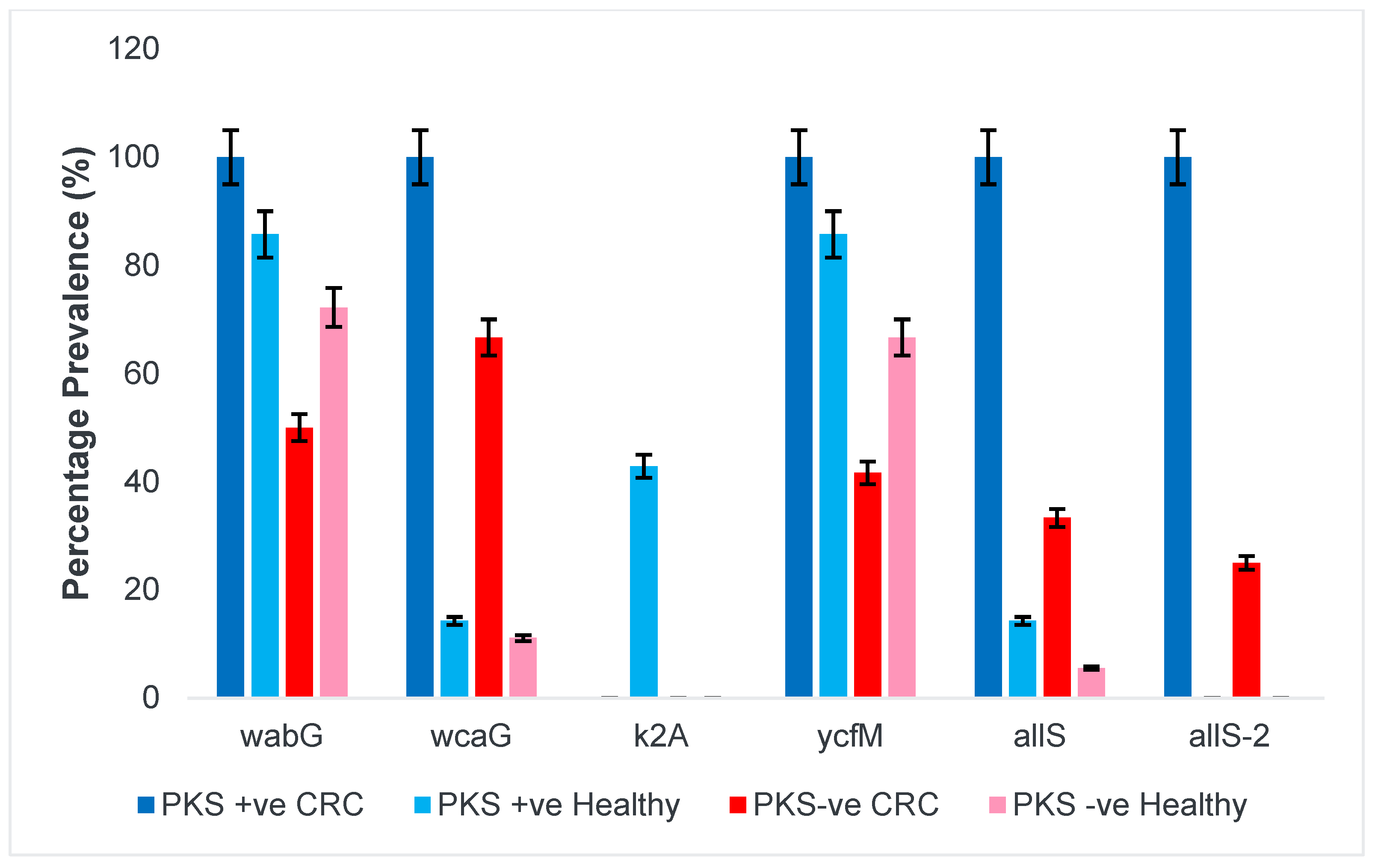
3.1.1. Presence of Colibactin (PKS) and Capsular Serotype
3.1.2. Siderophores and Iron Uptake Systems
3.1.3. Regulator of Mucoid Phenotype and Accessory Genes
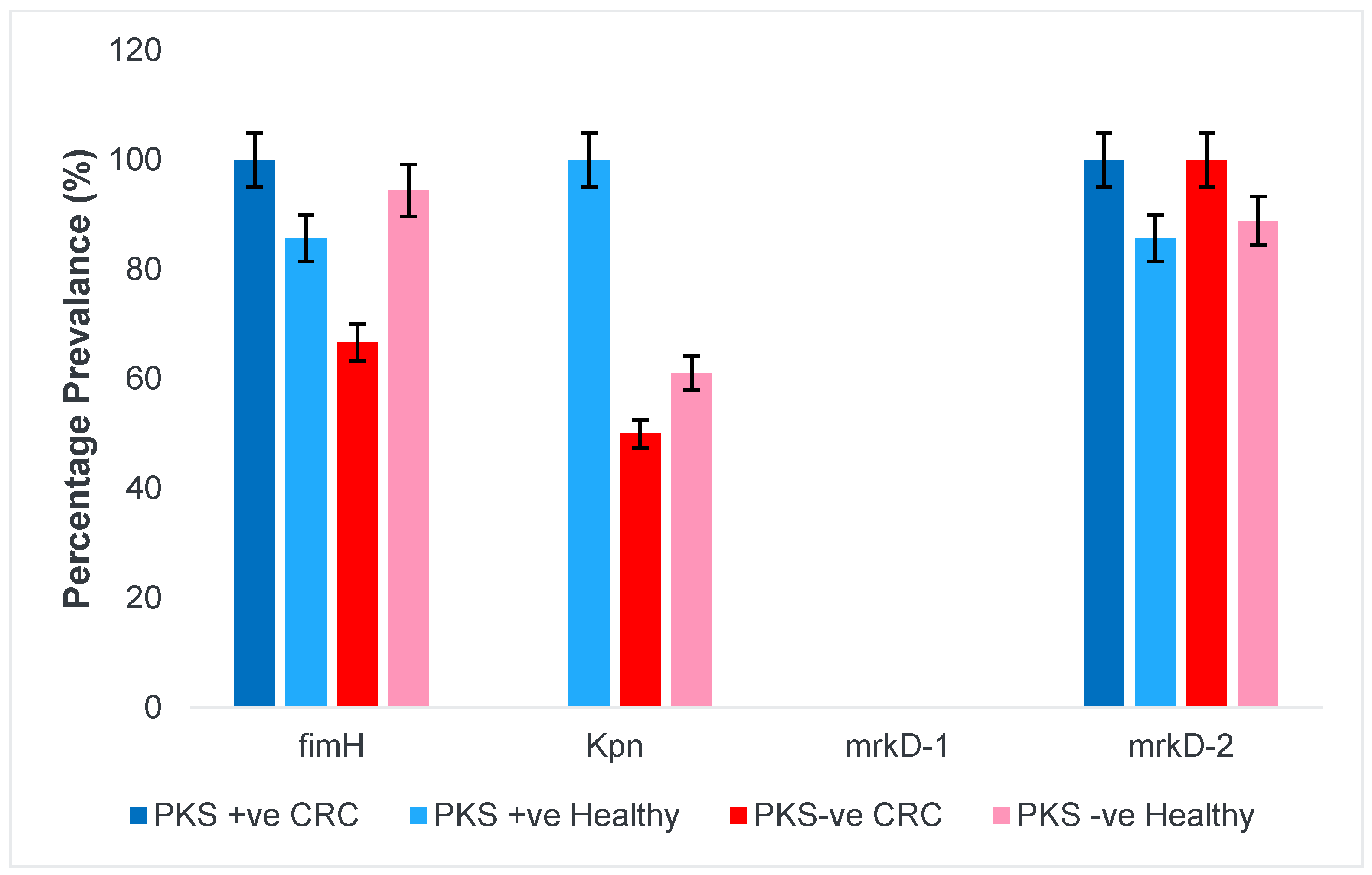
3.1.4. Fimbriae and Adhesins
3.1.5. Allantoin Metabolism and Lipopolysaccharide
3.2. Carbapenem Resistance Genes and Its Correlation with Antibiotic Susceptibility Testing (AST)
3.3. Presence of PKS Gene Improves Biofilm-Forming Capabilities of K. pneumoniae Isolates
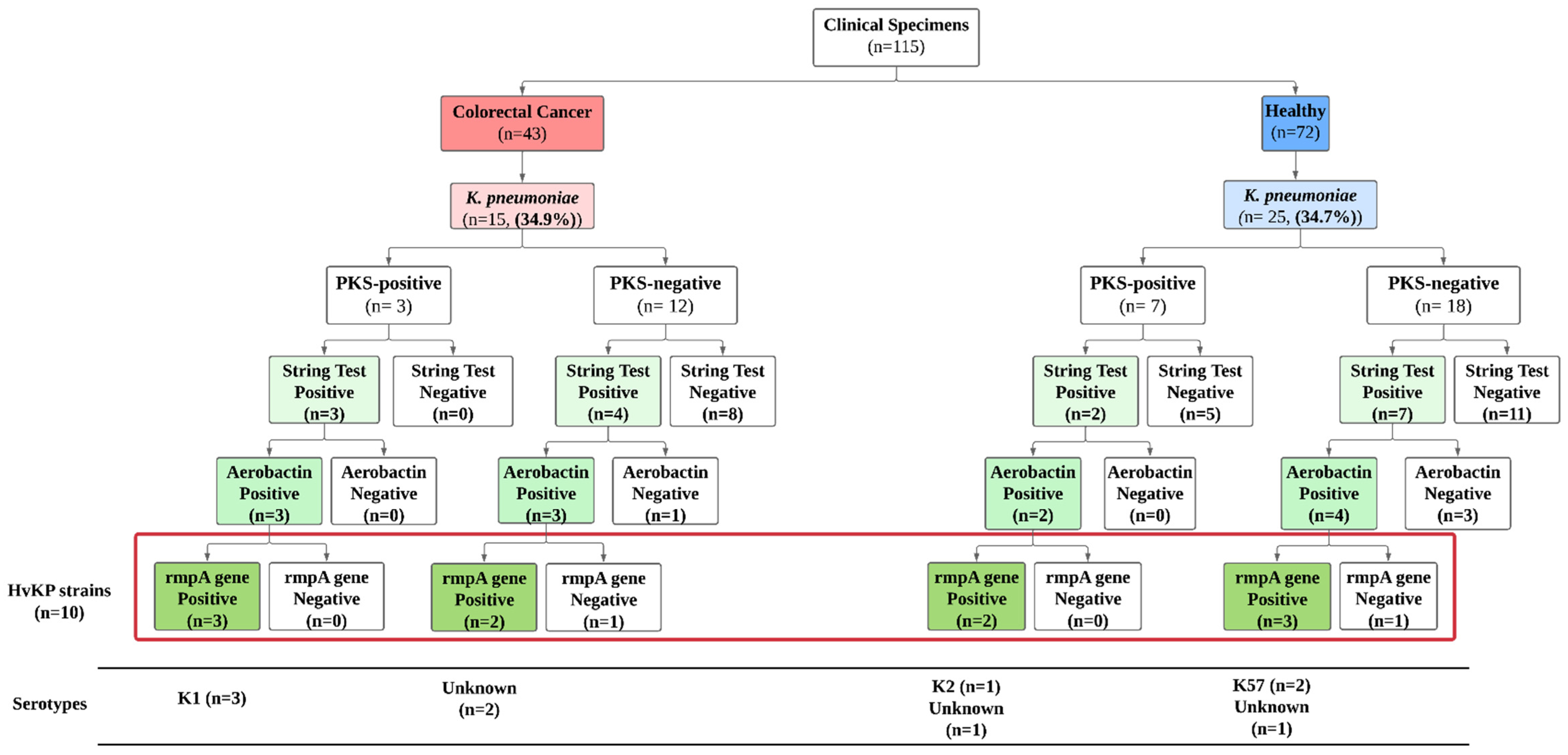
3.4. Prevalence of Hypervirulent K. pneumoniae (hvKp) Strains
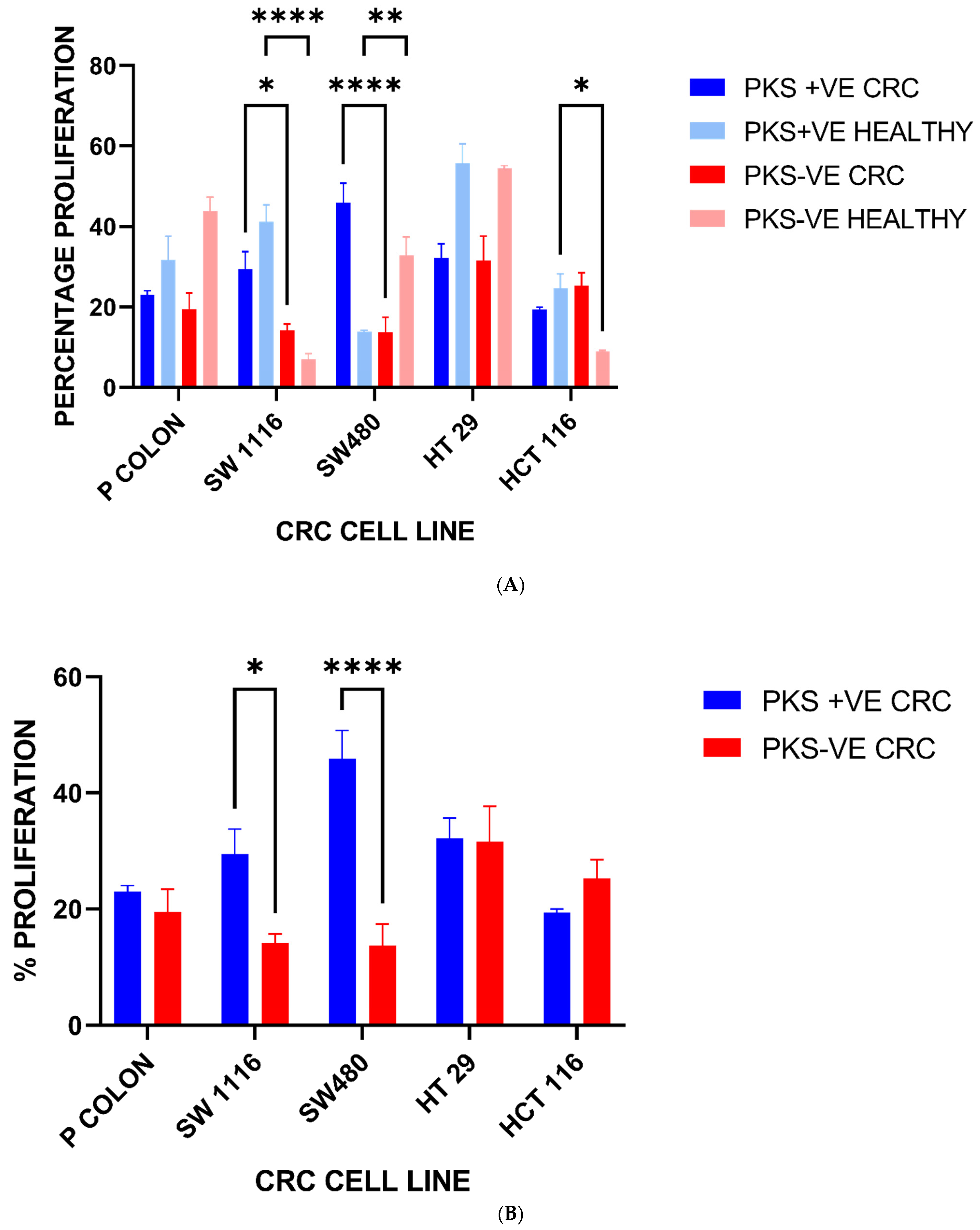
3.5. Colorectal Cancer Cell Line Proliferation Is Enhanced upon Exposure to K. pneumoniae Antigens
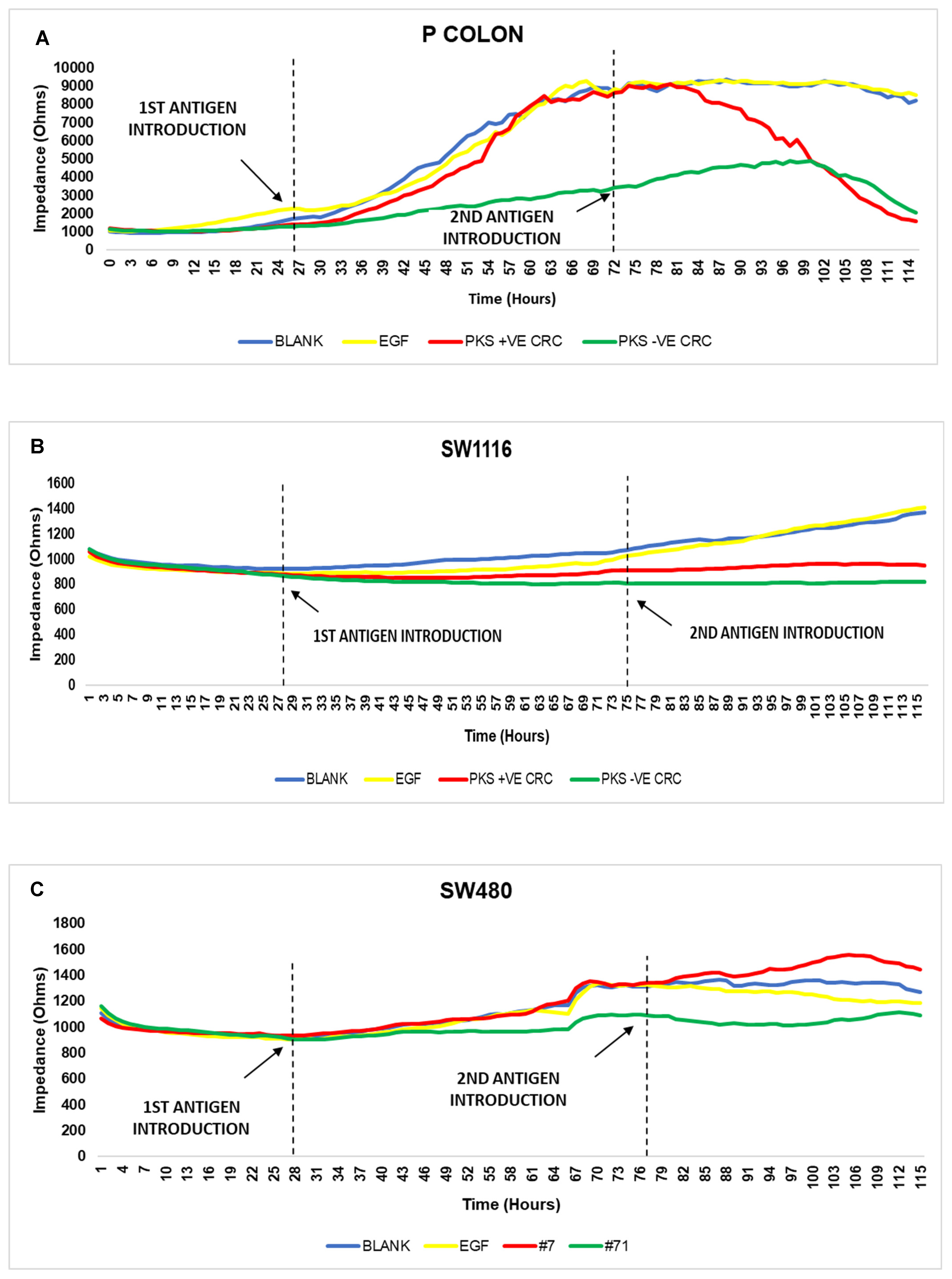
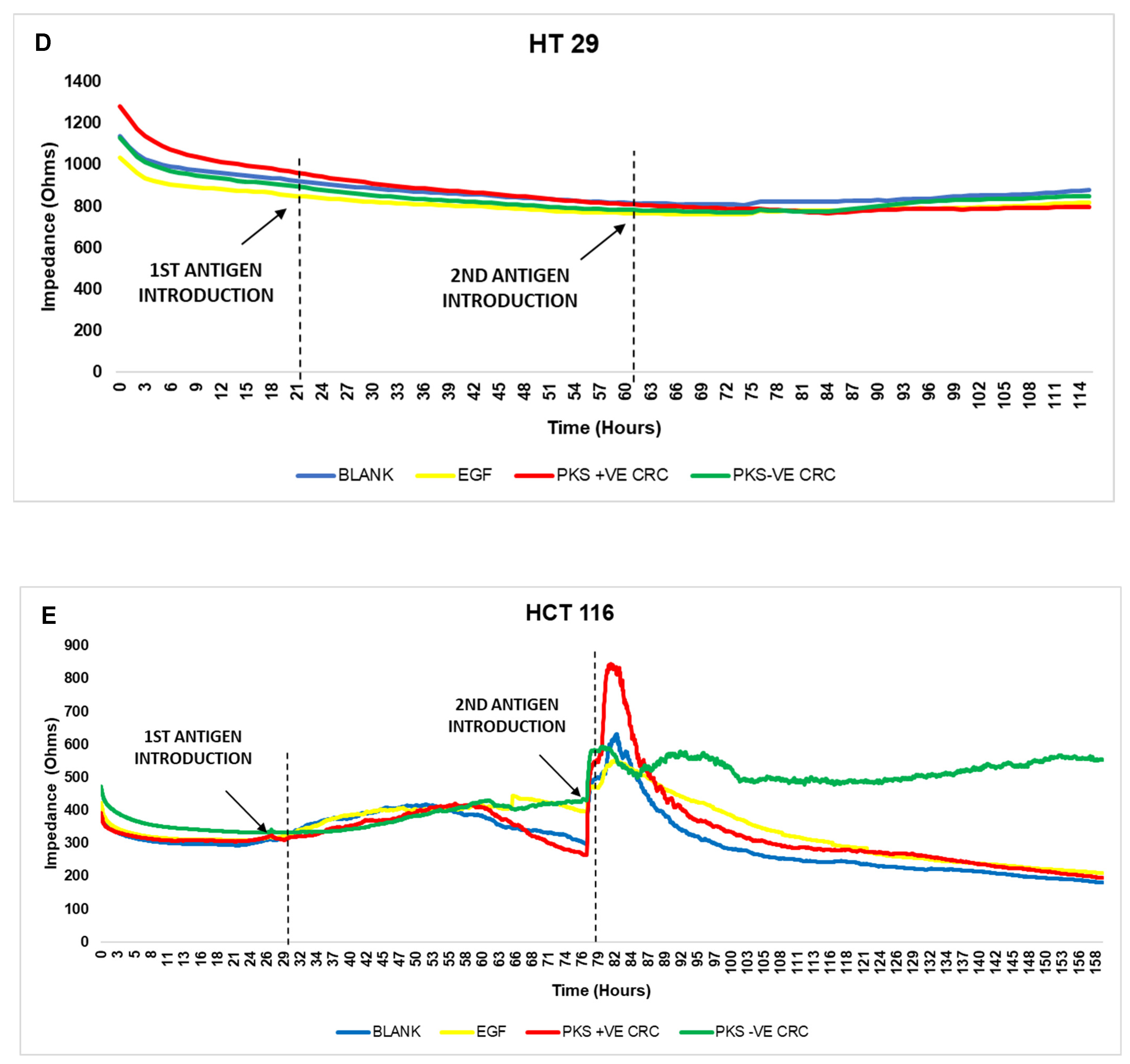
3.6. Persistent K. pneumoniae Infection Enhances Colorectal Cancer Progression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, Q.; Gu, D.; Wang, Q.; Hu, Y.; Shu, L.; Hu, J.; Zhang, R.; Chen, G.-X. Dynamic Colonization of Klebsiella pneumoniae Isolates in Gastrointestinal Tract of Intensive Care Patients. Front. Med. 2019, 10, 230. [Google Scholar]
- Martin, R.M.; Cao, J.; Brisse, S.; Passet, V.; Wu, W.; Zhao, L.; Malani, P.N.; Rao, K.; Bachman, M.A. Molecular Epidemiology of Colonizing and Infecting Isolates of Klebsiella. mSphere 2016, 1, e00261-16. [Google Scholar]
- Yu, W.; Ko, W.; Cheng, K.; Lee, H.; Ke, D.; Lee, C.; Ke, D.-S.; Lee, C.-C.; Fung, C.-P.; Chuang, Y.-C. Association between rmpA and magA Genes and Clinical Syndromes Caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 2006, 42, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Shon, A.S.; Beanan, J.M.; Olson, R.; Macdonald, U.; Pomakov, A.O.; Visitacion, M.P. Hypervirulent K. Pneumoniae Secretes More and More Active Iron-Acquisition Molecules than ‘“ Classical ”’ K. pneumoniae Thereby Enhancing its Virulence. PLoS ONE 2011, 6, e26734. [Google Scholar] [CrossRef] [PubMed]
- Giske, C.G.; Monnet, D.L.; Cars, O.; Carmeli, Y. Clinical and Economic Impact of Common Multidrug-Resistant Gram-Negative Bacilli. Antimicrob. Agents Chemother. 2008, 52, 813–821. [Google Scholar] [PubMed]
- Lederman, E.R.; Crum, N.F. Pyogenic Liver Abscess with a Focus on Klebsiella pneumoniae as a Primary Pathogen: An Emerging Disease with Unique Clinical Characteristics. Am. J. Gastroenterol. 2005, 100, 322–331. [Google Scholar] [PubMed]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrede, J.-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.C.; Lin, C.C.; Cheng, K.S.; Kao, J.T.; Chou, J.W.; Peng, C.Y.; Lai, S.; Chen, P.-C.; Sung, F. Increased Incidence of Gastrointestinal Cancers Among Patients With. Gastroenterology 2014, 146, 129–137.e1. [Google Scholar] [CrossRef] [PubMed]
- Mohan, B.P.; Meyyur Aravamudan, V.; Khan, S.R.; Chandan, S.; Ponnada, S.; Asokkumar, R.; Navaneethan, U.; Adler, D.G. Prevalence of colorectal cancer in cryptogenic pyogenic liver abscess patients. Do they need screening colonoscopy? A systematic review and meta-analysis. Dig. Liver Dis. 2019, 51, 1641–1645. [Google Scholar] [CrossRef]
- Lai, S.W. Pyogenic liver abscess and colorectal cancer. QJM 2019, 112, 559. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Kim, C.W.; Kim, D.R.; Cho, Y.W.; Cho, J.; Kim, W.J. Recurrent pyogenic liver abscess as a presenting manifestation of colorectal cancer. Korean J. Intern. Med. 2017, 32, 174–177. [Google Scholar] [PubMed]
- Huang, W.-K.; Chang, J.W.-C.; See, L.-C.; Tu, H.-T.; Chen, J.-S.; Liaw, C.-C.; Lin, Y.C.; Yang, T.S. Higher rate of colorectal cancer among patients with pyogenic liver abscess with Klebsiella pneumoniae than those without: An 11-year follow-up study. Color Dis. 2012, 14, e794–e801. [Google Scholar]
- Li, Z.; Su, Y.; Wang, X.; Yan, H.; Sun, M.; Shu, Z. Hepatic portal venous gas associated with colon cancer. Medicine 2017, 96, e9352. [Google Scholar] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility. Am. Soc. Microbiol. 2016, 3, 6–13. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; Available online: http://www.emeraldinsight.com/doi/10.1108/08876049410065598 (accessed on 28 August 2020).
- (EUCAST) TEC on AST. Breakpoint Tables for Interpretation of MICs and Zone Diameters [Internet]. 2020. 0–77p. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf (accessed on 28 August 2020).
- Singh, A.K.; Prakash, P.; Achra, A.; Singh, G.P.; Das, A.; Singh, R.K. Standardization and Classification of In vitro Biofilm Formation by Clinical Isolates of Staphylococcus aureus. J. Glob. Infect. Dis. 2017, 9, 93–101. [Google Scholar]
- Emery, B.D.D.; Furian, T.Q.; Pilatti, R.M.; Chitolina, G.Z.; Borges, K.A.; Salle, C.T.P.; Moraes, H.L.S. Evaluation of the biofilm formation capacity of Pasteurella multocida strains isolated from cases of fowl cholera and swine lungs and its relationship with pathogenicity 1. Pesqui. Vet. Bras. 2017, 37, 1041–1048. [Google Scholar]
- Magiorakos, A.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2011, 18, 268–281. [Google Scholar] [CrossRef]
- Liu, C.; Guo, J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: Antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann. Clin. Microbiol. Antimicrob. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, C.; Wang, Q.; Wang, X.; Chen, H.; Li, H.; Zhang, F.; Li, S.; Wang, R.; Wang, H. High Prevalence of Hypervirulent. Antimicrob. Agents Chemother. 2016, 60, 6115–6120. [Google Scholar]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam, M.M.C.; Duchêne, S.; Jenney, A.; Holt, K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019, 15, e1008114. [Google Scholar]
- Li, W.; Sun, G.; Yu, Y.; Li, N.; Chen, M.; Jin, R.; Jiao, Y.; Wu, H. Increasing occurrence of antimicrobial-resistant hypervirulent (Hypermucoviscous) klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 2014, 58, 225–232. [Google Scholar] [PubMed]
- Liu, C.; Shi, J.; Guo, J. High prevalence of hypervirulent klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect. Drug Resist. 2018, 11, 1031–1041. [Google Scholar]
- Shen, D.; Ma, G.; Li, C.; Jia, X.; Qin, C.; Yang, T.; Wang, L.; Jiang, X.; Ding, N.; Zhang, X.; et al. Emergence of a Multidrug-Resistant Hypervirulent Klebsiella pneumoniae Sequence Type 23 Strain with a Rare blaCTX-M-24-Harboring Virulence Plasmid. Antimicrob. Agents Chemother. 2019, 63, e02273-18. [Google Scholar] [PubMed]
- Fu, Y.; Xu, M.; Liu, Y.; Li, A.; Zhou, J. Virulence and genomic features of a bla CTX-M-3 and bla CTX-M-14 coharboring hypermucoviscous Klebsiella pneumoniae of serotype K2 and ST65. Infect. Drug Resist. 2019, 12, 145–159. [Google Scholar] [PubMed]
- Khaertynov, K.S.; Anokhin, V.A.; Rizvanov, A.A.; Davidyuk, Y.N.; Semyenova, D.R.; Lubin, S.A.; Skvortsova, N.N. Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Front. Med. 2018, 5, 225. [Google Scholar]
- Chen, T.; Dong, G.; Zhang, S.; Zhang, X.; Zhao, Y.; Cao, J.; Zhou, T.; Wu, Q. Effects of iron on the growth, biofilm formation and virulence of Klebsiella pneumoniae causing liver abscess. BMC Microbiol. 2020, 20, 36. [Google Scholar]
- Lam, M.M.C.; Wyres, K.L.; Duchêne, S.; Wick, R.R.; Judd, L.M.; Gan, Y.; Hoh, C.-H.; Archuleta, S.; Molton, J.S.; Kalimuddin, S.; et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 2018, 9, 2703. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.C.; Lee, C.Z.; Ma, L.C.; Fang, C.T.; Chang, S.C.; Wang, J.T. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect. Immun. 2004, 72, 3783–3792. [Google Scholar] [CrossRef]
- Strakova, N.; Korena, K.; Karpiskova, R. Klebsiella pneumoniae producing bacterial toxin colibactin as a risk of colorectal cancer development–A systematic review. Toxicon 2021, 197, 126–135. [Google Scholar] [CrossRef]



| Antimicrobial Category | Antimicrobial Agent |
|---|---|
| Aminoglycosides | Amikacin Gentamicin Kanamycin |
| Antipseudomonal penicillins + β-lactamase inhibitors | Piperacillin-tazobactam Ticarcilin-clavulanic acid |
| Carbapenems | Imipenem Meropenem |
| Non-extended spectrum cephalosporins (1st and 2nd generation cephalosporins) | Cefuroxime |
| Extended spectrum cephalosporins (3rd and 4th generation cephalosporins) | Cefotaxime Ceftazidime Cefepime |
| Cephamycins | Cefoxitin |
| Fluroquinolones | Ciprofloxacin |
| Folate Pathway Inhibitors | Trimethoprim-sulfamethoxazole |
| Glycylcyclines | Tigecycline |
| Monobactams | Aztreonam |
| Penicillins | Ampicillin Amoxycillin Piperacillin |
| Penicillins + β-lactamase inhibitors | Amoxicillin-clavulanic acid Ampicillin-sulbactam |
| Phenicols | Chloramphenicol |
| Polymyxins | Colistin Polymyxin B |
| Tetracyclines | Doxycycline Tetracycline |
| Quinolones | Levofloxacin Ofloxacin Norfloxacin |
| Virulence Gene | Colorectal Cancer (n = 15) | Healthy (n = 25) | Total | ||
|---|---|---|---|---|---|
| PKS +ve | PKS –ve | PKS +ve | PKS -ve | ||
| Colibactin (PKS) | 3 (20%) | 12 (80%) | 7 (28%) | 18 (72%) | 40 |
| K1 serotype | 3 | 0 | 0 | 0 | 3 |
| K2 serotype | 0 | 1 | 5 | 0 | 6 |
| K5 serotype | 0 | 0 | 1 | 2 | 3 |
| K20 serotype | 0 | 0 | 0 | 0 | 0 |
| K54 serotype | 0 | 0 | 0 | 0 | 0 |
| K57 serotype | 0 | 0 | 0 | 2 | 2 |
| Unknown * | 0 | 11 | 1 | 14 | 26 |
| PKS STATUS | SEROTYPE | wabG | fimH | ycfM | entB | hly | wcaG | ybtS | iutA1 | entB1 | Aerobactin | IucB | ureA | cnf1 | uge | iroN | kfuBC | iroNB | mrkD2 | rmpA2 | allS | kpn | allS2 | iutA | ybtA | rmpA1 | kfu | k2A | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKS STATUS | 1 | ||||||||||||||||||||||||||||
| SEROTYPE | 0.335 * | 1 | |||||||||||||||||||||||||||
| wabG | 0.294 | −0.123 | 1 | ||||||||||||||||||||||||||
| fimH | 0.095 | 0.265 | 0.163 | 1 | |||||||||||||||||||||||||
| ycfM | 0.320 * | −0.087 | 0.946 ** | 0.138 | 1 | ||||||||||||||||||||||||
| entB | 0.269 | −0.160 | 0.829 ** | 0.037 | 0.780 ** | 1 | |||||||||||||||||||||||
| hly | 0.118 | −0.145 | −0.089 | −0.547 ** | −0.076 | −0.352 * | 1 | ||||||||||||||||||||||
| wcaG | 0.028 | −0.250 | −0.173 | −0.284 | −0.241 | 0.011 | 0.076 | 1 | |||||||||||||||||||||
| ybtS | 0.620 ** | 0.336 * | 0.141 | 0.128 | 0.085 | 0.092 | 0.006 | 0.120 | 1 | ||||||||||||||||||||
| iutA1 | 0.569 ** | 0.386 * | 0.191 | 0.171 | 0.120 | 0.155 | 0.063 | 0.201 | 0.373 * | 1 | |||||||||||||||||||
| entB1 | 0.014 | 0.210 | 0.306 | 0.794 ** | 0.283 | 0.150 | −0.689 ** | −0.283 | 0.156 | 0.079 | 1 | ||||||||||||||||||
| Aerobactin | 0.346 * | 0.486 ** | 0.244 | 0.402 ** | 0.201 | 0.068 | −0.063 | 0.013 | 0.437 ** | 0.472 ** | 0.262 | 1 | |||||||||||||||||
| IucB | 0.454 ** | 0.469 ** | 0.191 | 0.314 * | 0.120 | 0.155 | −0.172 | 0.201 | 0.373 * | 0.895 ** | 0.250 | 0.577 ** | 1 | ||||||||||||||||
| ureA | −0.261 | 0.101 | 0.232 | 0.382 * | 0.220 | 0.246 | −0.698 ** | −0.220 | −0.162 | −0.208 | 0.481 ** | 0.208 | 0.120 | 1 | |||||||||||||||
| cnf1 | 0.257 | −0.164 | 0.203 | 0.030 | 0.225 | 0.046 | 0.174 | 0.035 | 0.012 | 0.009 | −0.046 | 0.246 | 0.009 | 0.078 | 1 | ||||||||||||||
| uge | 0.128 | −0.135 | 0.248 | −0.004 | 0.220 | 0.278 | −0.198 | 0.053 | 0.313 * | −0.059 | 0.069 | 0.059 | 0.075 | 0.348 * | 0.223 | 1 | |||||||||||||
| iroN | −0.170 | −0.180 | 0.191 | −0.149 | 0.202 | 0.181 | −0.064 | −0.005 | −0.274 | −0.213 | 0.092 | 0.019 | −0.213 | 0.044 | 0.334 * | 0.127 | 1 | ||||||||||||
| kfuBC | −0.088 | −0.208 | 0.049 | 0.007 | −0.024 | 0.011 | −0.163 | 0.458 ** | 0.223 | 0.201 | 0.237 | 0.120 | 0.201 | 0.114 | 0.035 | 0.190 | −0.005 | 1 | |||||||||||
| iroNB | −0.057 | 0.311 * | −0.227 | 0.154 | −0.203 | −0.252 | −0.084 | 0.046 | 0.382 * | −0.283 | 0.123 | 0.283 | −0.283 | 0.059 | −0.183 | 0.169 | −0.105 | 0.203 | 1 | ||||||||||
| mrkD2 | −0.041 | 0.180 | 0.010 | 0.149 | −0.005 | 0.025 | −0.371 * | 0.005 | 0.087 | 0.019 | 0.223 | 0.175 | 0.213 | 0.563 ** | 0.138 | 0.370 * | 0.079 | 0.202 | 0.105 | 1 | |||||||||
| rmpA2 | 0.336 * | 0.335 * | 0.093 | 0.266 | 0.124 | 0.060 | −0.146 | 0.215 | 0.230 | 0.624 ** | 0.212 | 0.489 ** | 0.736 ** | 0.102 | 0.224 | 0.007 | −0.181 | 0.215 | −0.240 | 0.181 | 1 | ||||||||
| allS | 0.297 | −0.224 | 0.021 | −0.408 ** | −0.070 | 0.116 | 0.399 ** | 0.549 ** | 0.127 | 0.276 | −0.579 ** | 0.078 | 0.158 | −0.278 | 0.437 ** | 0.107 | −0.160 | 0.190 | −0.212 | −0.059 | 0.259 | 1 | |||||||
| kpn | 0.146 | 0.266 | 0.100 | −0.048 | 0.162 | 0.255 | −0.051 | −0.162 | −0.220 | −0.015 | −0.095 | 0.015 | −0.015 | −0.126 | −0.237 | −0.363 * | 0.225 | −0.584 ** | −0.160 | −0.225 | −0.035 | −0.244 | 1 | ||||||
| allS2 | 0.217 | −0.095 | 0.282 | −0.024 | 0.298 | 0.115 | 0.227 | 0.429 ** | 0.010 | 0.402 ** | −0.096 | 0.314 * | 0.402 ** | 0.065 | 0.493 ** | 0.188 | −0.116 | 0.429 ** | −0.154 | 0.116 | 0.492 ** | 0.568 ** | −0.376 * | 1 | |||||
| iutA | 0.454 ** | 0.469 ** | 0.191 | 0.314 * | 0.120 | 0.155 | −0.172 | 0.201 | 0.373 * | 0.895 ** | 0.250 | 0.577 ** | 1.000 ** | 0.120 | 0.009 | 0.075 | −0.213 | 0.201 | −0.283 | 0.213 | 0.736 ** | 0.158 | −0.015 | 0.402 ** | 1 | ||||
| ybtA | 0.493 ** | −0.081 | 0.159 | −0.138 | 0.193 | 0.350 * | 0.076 | 0.132 | 0.120 | 0.201 | −0.283 | 0.227 | 0.094 | −0.220 | 0.424 ** | −0.083 | 0.193 | −0.193 | −0.268 | −0.193 | −0.011 | 0.429 ** | 0.260 | 0.138 | 0.094 | 1 | |||
| rmpA1 | 0.310 * | −0.131 | 0.170 | −0.179 | 0.053 | 0.007 | 0.499 ** | 0.493 ** | 0.206 | 0.463 ** | −0.288 | 0.210 | 0.328 * | −0.348* | 0.431 ** | 0.034 | −0.127 | 0.357 * | −0.169 | −0.121 | 0.278 | 0.799 ** | −0.434 ** | 0.729 ** | 0.328 * | 0.220 | 1 | ||
| kfu | −0.203 | −0.324 * | 0.180 | −0.051 | 0.119 | 0.029 | 0.028 | 0.296 | 0.120 | 0.042 | 0.125 | 0.162 | 0.042 | 0.140 | 0.185 | 0.140 | 0.129 | 0.814 ** | 0.121 | 0.249 | 0.079 | 0.184 | −0.602 ** | 0.468 ** | 0.042 | −0.119 | 0.382 * | 1 | |
| k2A | 0.464 ** | 0.281 | 0.191 | 0.116 | 0.202 | 0.181 | −0.064 | −0.202 | 0.288 | 0.370 * | 0.092 | 0.213 | 0.370 * | 0.044 | −0.138 | 0.127 | −0.079 | −0.202 | −0.105 | 0.079 | −0.181 | −0.160 | 0.225 | −0.116 | 0.370 * | 0.390 * | −0.127 | −0.249 | 1 |
| Antimicrobial Agent | AK | SXT | TGC | ATM | AMP | AML | AMC | C | DO | TE | LEV | NOR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | |||||||||||||
| Colorectal Cancer | PKS +ve (n = 3) | - | - | - | - | 2 | 3 | - | - | - | - | - | - |
| PKS-ve (n = 12) | 1 | 3 | 2 | 1 | 10 | 12 | 1 | 5 | 2 | 3 | 2 | - | |
| Healthy | PKS +ve (n = 7) | 1 | - | - | - | 4 | 7 | - | - | - | - | - | - |
| PKS-ve (n = 18) | - | 1 | - | 1 | 17 | 18 | - | 1 | 1 | 1 | 1 | 1 | |
| Total | 2 | 4 | 2 | 2 | 33 | 40 | 1 | 6 | 3 | 4 | 3 | 1 | |
| Resistance Gene | Colorectal Cancer | Healthy | ||
|---|---|---|---|---|
| PKS +ve (n = 3) | PKS –ve (n = 12) | PKS +ve (n = 7) | PKS -ve (n = 18) | |
| blaKPC | 0 | 0 | 0 | 0 |
| blaNDM-1 | 0 | 0 | 0 | 0 |
| blaOXA-48 | 0 | 1 | 0 | 2 |
| blaIMP | 0 | 0 | 0 | 0 |
| blaVIM | 0 | 0 | 0 | 0 |
| Biofilm Forming Capability | Colorectal Cancer | Healthy | ||
|---|---|---|---|---|
| PKS +ve (n = 3) | PKS –ve (n = 12) | PKS +ve (n = 7) | PKS -ve (n = 18) | |
| Non-Biofilm | 0 | 4 (33%) | 2 (29%) | 6 (33%) |
| Weak | 0 | 1 (8%) | 1 (14%) | 2 (11%) |
| Moderate | 1 (33%) | 3 (25%) | 1 (14%) | 1 (6%) |
| Strong | 2 (67%) | 4 (33%) | 3 (43%) | 9 (50%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, C.P.; Iyadorai, T.; Sears, C.; Roslani, A.C.; Vadivelu, J.; Samudi, C. Presence of Polyketide Synthase (PKS) Gene and Counterpart Virulence Determinants in Klebsiella pneumoniae Strains Enhances Colorectal Cancer Progression In-Vitro. Microorganisms 2023, 11, 443. https://doi.org/10.3390/microorganisms11020443
Kaur CP, Iyadorai T, Sears C, Roslani AC, Vadivelu J, Samudi C. Presence of Polyketide Synthase (PKS) Gene and Counterpart Virulence Determinants in Klebsiella pneumoniae Strains Enhances Colorectal Cancer Progression In-Vitro. Microorganisms. 2023; 11(2):443. https://doi.org/10.3390/microorganisms11020443
Chicago/Turabian StyleKaur, Christina Parvinder, Thevambiga Iyadorai, Cynthia Sears, April Camilla Roslani, Jamuna Vadivelu, and Chandramathi Samudi. 2023. "Presence of Polyketide Synthase (PKS) Gene and Counterpart Virulence Determinants in Klebsiella pneumoniae Strains Enhances Colorectal Cancer Progression In-Vitro" Microorganisms 11, no. 2: 443. https://doi.org/10.3390/microorganisms11020443
APA StyleKaur, C. P., Iyadorai, T., Sears, C., Roslani, A. C., Vadivelu, J., & Samudi, C. (2023). Presence of Polyketide Synthase (PKS) Gene and Counterpart Virulence Determinants in Klebsiella pneumoniae Strains Enhances Colorectal Cancer Progression In-Vitro. Microorganisms, 11(2), 443. https://doi.org/10.3390/microorganisms11020443







