Nisin E Is a Novel Nisin Variant Produced by Multiple Streptococcus equinus Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Isolation, Bacteriocin Activity Screening, and Speciation of Isolates
2.2. Strain Speciation and Genomic Comparison
2.3. Streptococcus Equinus Pangenome Analysis
2.4. Nisin Variant Cross-Immunity Assay
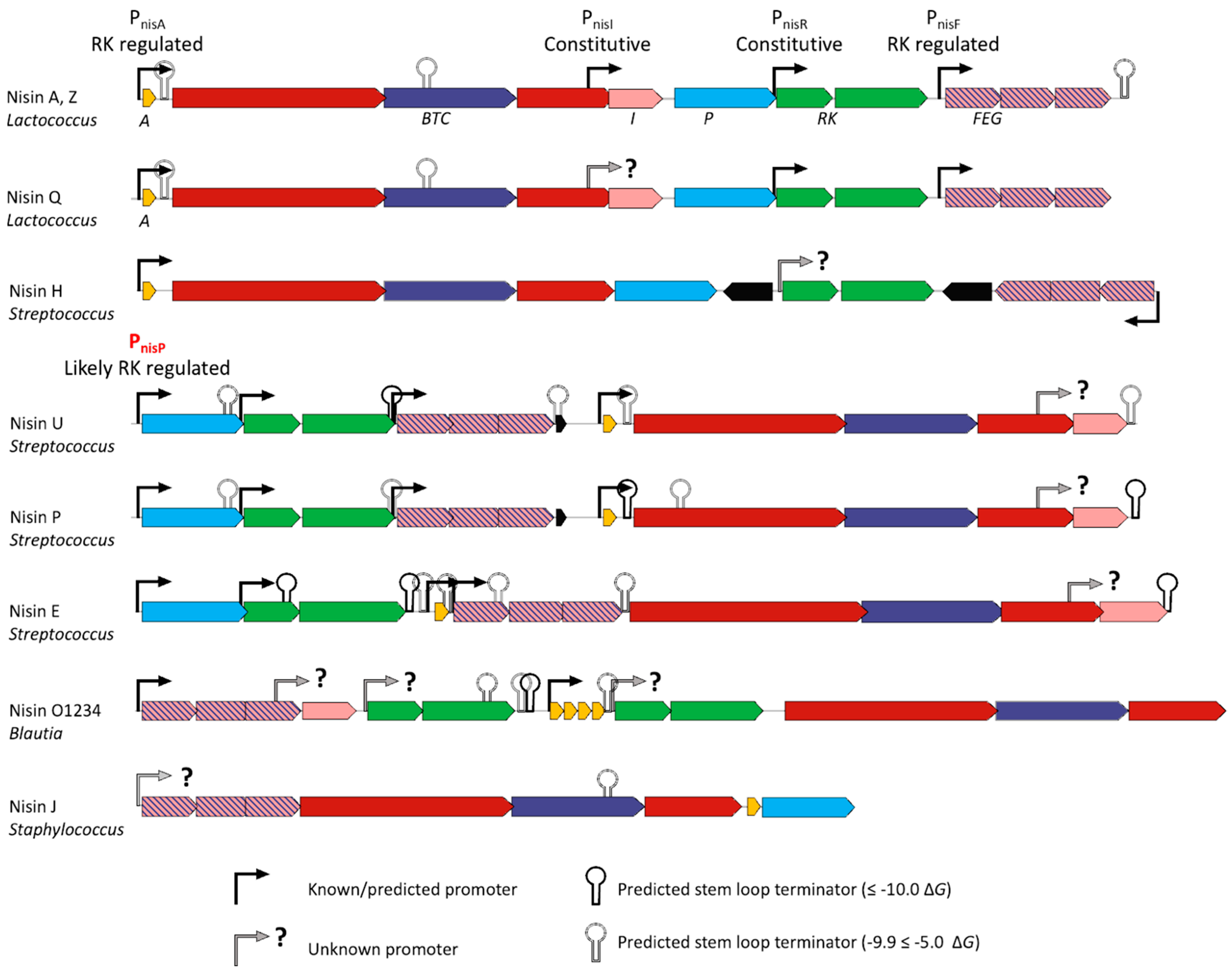
2.5. Promoter Prediction and Transcription Start Site Mapping
3. Results
3.1. Isolation of Two Bacteriocin-Producing Streptococcus equinus Strains
3.2. Nisin E Is a Novel Variant Unique to Streptococcus equinus
3.3. A Predicted Streptococcus-Specific Promoter for Expression of nisP
3.4. Spectrum of Inhibition of Nisin E Producers and Cross-Immunity to Other Nisin Producers
3.5. Nisin E Immunity Genes Are Spread throughout the Streptococcus equinus Pangenome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Interagency Coordination Group on Antimicrobial Resistance (IACG). No Time to Wait: Securing the Future from Drug-Resistant Infections; World Health Organization: Geneva, Switzerland, 2019. Available online: https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections (accessed on 29 December 2022).
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Hegarty, J.W.; Guinane, C.M.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocin production: A relatively unharnessed probiotic trait? F1000Research 2016, 5, 2587. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar]
- Xie, L.; van der Donk, W.A. Post-translational modifications during lantibiotic biosynthesis. Current Opin. Chem. Biol. 2004, 8, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.A. The inhibiting effect of streptococcus lactis on lactobacillus bulgaricus. J. Bacteriol. 1928, 16, 321–325. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS); Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Dusemund, B.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gundert-Remy, U.; et al. Safety of nisin (E 234) as a food additive in the light of new toxicological data and the proposed extension of use. EFSA J. 2017, 15, e05063. [Google Scholar]
- Gross, E.; Morell, J.L. Structure of nisin. J. Am. Chem. Soc. 1971, 93, 4634–4635. [Google Scholar] [CrossRef]
- Shin, J.; Gwak, J.; Kamarajan, P.; Fenno, J.; Rickard, A.; Kapila, Y. Biomedical applications of nisin. J. Appl. Microbiol. 2015, 120, 1449–1465. [Google Scholar] [CrossRef]
- Zhao, X.; Kuipers, O.P. Synthesis of silver-nisin nanoparticles with low cytotoxicity as antimicrobials against biofilm-forming pathogens. Colloids Surf. B: Biointerfaces 2021, 206, 111965. [Google Scholar] [CrossRef] [PubMed]
- Gut, I.M.; Blanke, S.R.; van der Donk, W.A. Mechanism of Inhibition of Bacillus anthracis Spore Outgrowth by the Lantibiotic Nisin. ACS Chem. Biol. 2011, 6, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Cotter, P.D.; Ross, R.P.; Hill, C. Bioengineering of the model lantibiotic nisin. Bioengineered 2015, 6, 187–192. [Google Scholar] [CrossRef]
- Field, D.; Begley, M.; O’Connor, P.M.; Daly, K.M.; Hugenholtz, F.; Cotter, P.D.; Hill, C.; Ross, R. Bioengineered Nisin A Derivatives with Enhanced Activity against Both Gram Positive and Gram Negative Pathogens. PLoS ONE 2012, 7, e46884. [Google Scholar] [CrossRef]
- Healy, B.; Field, D.; O’Connor, P.M.; Hill, C.; Cotter, P.D.; Ross, R.P. Intensive Mutagenesis of the Nisin Hinge Leads to the Rational Design of Enhanced Derivatives. PLoS ONE 2013, 8, e79563. [Google Scholar] [CrossRef] [PubMed]
- Mulders, J.W.M.; Boerrigter, I.J.; Rollema, H.S.; Siezen, R.J.; de Vos, W.M. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 1991, 201, 581–584. [Google Scholar] [CrossRef] [PubMed]
- De Kwaadsteniet, M.; Ten Doeschate, K.; Dicks, L.M.T. Characterization of the structural gene encoding nisin F, a new lantibiotic produced by a Lactococcus lactis subsp. lactis isolate from freshwater catfish (Clarias gariepinus). Appl. Environ. Microbiol. 2008, 74, 547–549. [Google Scholar] [CrossRef]
- Fukao, M.; Obita, T.; Yoneyama, F.; Kohda, D.; Zendo, T.; Nakayama, J.; Sonomoto, K. Complete Covalent Structure of Nisin Q, New Natural Nisin Variant, Containing Post-Translationally Modified Amino Acids. Biosci. Biotechnol. Biochem. 2008, 72, 1750–1755. [Google Scholar] [CrossRef]
- Zendo, T.; Ohashi, C.; Maeno, S.; Piao, X.; Salminen, S.; Sonomoto, K.; Endo, A. Kunkecin A, a New Nisin Variant Bacteriocin Produced by the Fructophilic Lactic Acid Bacterium, Apilactobacillus kunkeei FF30-6 Isolated From Honey Bees. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; O’Connor, P.M.; Rea, M.C.; O’Sullivan, O.; Walsh, C.J.; Healy, B.; Mathur, H.; Field, D.; Hill, C.; Ross, R.P. Nisin J, a Novel Natural Nisin Variant, Is Produced by Staphylococcus capitis Sourced from the Human Skin Microbiota. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef]
- Hatziioanou, D.; Gherghisan-Filip, C.; Saalbach, G.; Horn, N.; Wegmann, U.; Duncan, S.H.; Flint, H.J.; Mayer, M.J.; Narbad, A. Discovery of a novel lantibiotic nisin O from Blautia obeum A2-162, isolated from the human gastrointestinal tract. Microbiology 2017, 163, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, R.E.; Klesse, N.A.; Jack, R.W.; Tagg, J.R. Molecular and Genetic Characterization of a Novel Nisin Variant Produced by Streptococcus uberis. Appl. Environ. Microbiol. 2006, 72, 1148–1156. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, P.M.; O’Shea, E.F.; Guinane, C.M.; O’Sullivan, O.; Cotter, P.D.; Ross, R.P.; Hill, C. Nisin H Is a New Nisin Variant Produced by the Gut-Derived Strain Streptococcus hyointestinalis DPC6484. Appl. Environ. Microbiol. 2015, 81, 3953–3960. [Google Scholar] [CrossRef]
- Aldarhami, A.; Felek, A.; Sharma, V.; Upton, M. Purification and characterization of nisin P produced by a strain of Streptococcus gallolyticus. J. Med. Microbiol. 2020, 69, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, E.; O’Connor, P.M.; Saalbach, G.; Walsh, C.J.; Hegarty, J.W.; Guinane, C.M.; Mayer, M.J.; Narbad, A.; Cotter, P.D. First evidence of production of the lantibiotic nisin P. Sci. Rep. 2020, 10, 3738. [Google Scholar] [CrossRef]
- Lawrence, G.W.; Garcia-Gutierrez, E.; Walsh, C.J.; O’Connor, P.M.; Begley, M.; Cotter, P.D.; Guinane, C.M. Nisin G is a novel nisin variant produced by a gut-derived Streptococcus salivarius. BioRxiv 2022. Preprint. [Google Scholar] [CrossRef]
- Kleerebezem, M. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 2004, 25, 1405–1414. [Google Scholar] [CrossRef]
- de Ruyter, P.G.; Kuipers, O.P.; Beerthuyzen, M.M.; van Alen-Boerrigter, I.; de Vos, W.M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 1996, 178, 3434–3439. [Google Scholar] [CrossRef]
- Khosa, S.; AlKhatib, Z.; Smits, S.H. NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol. Chem. 2013, 394, 1543–1549. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, I.; O’Connor, P.M.; Hill, C.; Stanton, C.; Ross, R.P. Actinomyces produces defensin-like bacteriocins (actifensins) with a highly degenerate structure and broad antimicrobial activity. J. Bacteriol. 2020, 202, e00529-19. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr Protoc Bioinform. 2014, 48, 1.25.1–1.25.33. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Naville, M.; Ghuillot-Gaudeffroy, A.; Marchais, A.; Gautheret, D. ARNold: A web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 2011, 8, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Di Bonaventura, G.; Gherardi, G. An Overview on Streptococcus bovis/Streptococcus equinus Complex Isolates: Identification to the Species/Subspecies Level and Antibiotic Resistance. Int. J. Mol. Sci. 2019, 20, 480. [Google Scholar] [CrossRef]
- Mantovani, H.C.; Hu, H.; Worobo, R.W.; Russell, J.B. Bovicin HC5, a bacteriocin from Streptococcus bovis HC5. Microbiology 2002, 148, 3347–3352. [Google Scholar] [CrossRef]
- Xiao, H.; Chen, X.; Chen, M.; Tang, S.; Zhao, X.; Huan, L. Bovicin HJ50, a novel lantibiotic produced by Streptococcus bovis HJ50. Microbiology 2004, 150, 103–108. [Google Scholar] [CrossRef]
- Whitford, M.F.; McPherson, M.A.; Forster, R.J.; Teather, R.M. Identification of bacteriocin-like inhibitors from rumen Streptococcus spp. and isolation and characterization of bovicin 255. Appl. Environ. Microbiol. 2001, 67, 569–574. [Google Scholar] [CrossRef]
- Georgalaki, M.D.; Van den Berghe, E.; Kritikos, D.; Devreese, B.; Van Beeumen, J.; Kalantzopoulos, G.; De Vuyst, L.; Tsakalidou, E. Macedocin, a Food-Grade Lantibiotic Produced by Streptococcus macedonicus ACA-DC 198. Appl. Environ. Microbiol. 2002, 68, 5891–5903. [Google Scholar]
- Georgalaki, M.; Papadimitriou, K.; Anastasiou, R.; Pot, B.; Van Driessche, G.; Devreese, B.; Tsakalidou, E. Macedovicin, the second food-grade lantibiotic produced by Streptococcus macedonicus ACA-DC 198. Food Microbiol. 2013, 33, 124–130. [Google Scholar] [CrossRef]
- Aymeric, L.; Donnadieu, F.; Mulet, C.; du Merle, L.; Nigro, G.; Saffarian, A.; Bérard, M.; Poyart, C.; Robine, S.; Regnault, B.; et al. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc. Natl. Acad. Sci. USA 2017, 115, 201715112. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.; O’Connor, P.M.; Altermann, E.; Day, L.; Hill, C.; Stanton, C.; Ross, R.P. Extensive bacteriocin gene shuffling in the Streptococcus bovis/Streptococcus equinus complex reveals gallocin D with activity against vancomycin resistant enterococci. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Harrington, A.; Proutière, A.; Mull, R.W.; du Merle, L.; Dramsi, S.; Tal-Gan, Y. Secretion, Maturation, and Activity of a Quorum Sensing Peptide (GSP) Inducing Bacteriocin Transcription in Streptococcus gallolyticus. Mbio 2021, 12, e03189-20. [Google Scholar] [CrossRef]
- Chandrapati, S.; O’Sullivan, D.J. Nisin independent induction of the nisA promoter in Lactococcus lactis during growth in lactose or galactose. FEMS Microbiol. Lett. 1999, 170, 191–198. [Google Scholar]
- Chandrapati, S.; O’Sullivan, D.J. Characterization of the promoter regions involved in galactose- and nisin-mediated induction of the nisA gene in Lactococcus lactis ATCC 11454: Characterization of the nisA promoter. Mol. Microbiol. 2002, 46, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Montalban-Lopez, M.; Kuipers, O.P. Increasing the Antimicrobial Activity of Nisin-Based Lantibiotics against Gram-Negative Pathogens. Appl. Environ. Microbiol. 2018, 84, e00052-18. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; O’Sullivan, D.J. Identification of a nisI Promoter within the nisABCTIP Operon That May Enable Establishment of Nisin Immunity Prior to Induction of the Operon via Signal Transduction. J. Bacteriol. 2006, 188, 8496–8503. [Google Scholar] [CrossRef]
- Cheigh, C.-I.; Park, H.; Choi, H.-J.; Pyun, Y.-R. Enhanced nisin production by increasing genes involved in nisin Z biosynthesis in Lactococcus lactis subsp. lactis A164. Biotechnol. Lett. 2005, 27, 155–160. [Google Scholar] [CrossRef]
- Furtmann, F.; Porta, N.; Hoang, D.T.; Reiners, J.; Schumacher, J.; Gottstein, J.; Gohlke, H.; Smits, S.H. Characterization of the nucleotide-binding domain NsrF from the BceAB-type ABC-transporter NsrFP from the human pathogen Streptococcus agalactiae. Sci. Rep. 2020, 10, 15208. [Google Scholar]
- Simões, P.M.; Lemriss, H.; Dumont, Y.; Lemriss, S.; Rasigade, J.-P.; Assant-Trouillet, S.; Ibrahimi, A.; El Kabbaj, S.; Butin, M.; Laurent, F. Single-Molecule Sequencing (PacBio) of the Staphylococcus capitis NRCS-A Clone Reveals the Basis of Multidrug Resistance and Adaptation to the Neonatal Intensive Care Unit Environment. Front. Microbiol. 2016, 7, 1991. [Google Scholar] [CrossRef]
- Wels, M.; Siezen, R.; van Hijum, S.; Kelly, W.J.; Bachmann, H. Comparative Genome Analysis of Lactococcus lactis Indicates Niche Adaptation and Resolves Genotype/Phenotype Disparity. Front. Microbiol. 2019, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Van Gijtenbeek, L.A.; Eckhardt, T.H.; Herrera-Dominguez, L.; Brockmann, E.; Jensen, K.; Geppel, A.; Nielsen, K.F.; Vindeloev, J.; Neves, A.R.; Oregaard, G. Gene-Trait Matching and Prevalence of Nisin Tolerance Systems in Lactococus lactis. Front. Bioeng. Biotechnol. 2021, 9, 622835. [Google Scholar] [PubMed]
- Mierau, I.; Kleerebezem, M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2005, 68, 705–717. [Google Scholar] [CrossRef] [PubMed]






| Organism | Strain | Nisin Variant | Temp. | O2 | Medium |
|---|---|---|---|---|---|
| Lactococcus lactis ssp. lactis | ATCC11454 | A | 30 | Aerobic | M17, 0.5% glucose |
| Lactococcus lactis | NZ9800 pCI372-nisA | A | 30 | Aerobic | M17, 0.5% glucose, 10 µg·mL−1 Chloramphenicol |
| Lactococcus lactis | NZ9800 pCI372-nisZ | Z | 30 | Aerobic | M17, 0.5% glucose, 10 µg·mL−1 Chloramphenicol |
| Lactococcus lactis | NZ9800 pCI372-nisF | F | 30 | Aerobic | M17, 0.5% glucose, 10 µg·mL−1 Chloramphenicol |
| Lactococcus lactis | NZ9800 pCI372-nisQ | Q | 30 | Aerobic | M17, 0.5% glucose, 10 µg·mL−1 Chloramphenicol |
| Staphylococcus capitis | APC2923 | J | 37 | Aerobic | BHI |
| Streptococcus uberis | 42 | U | 37 | Aerobic | BHI |
| Streptococcus equinus | APC4007 | E | 37 | Aerobic | BHI |
| Streptococcus equinus | APC4008 | E | 37 | Aerobic | BHI |
| Streptococcus hyointestinalis | DPC6484 | H | 37 | Anaerobic | BHI |
| Streptococcus agalactiae | DPC7040 | P | 37 | Aerobic | BHI |
| Organism | Strain | Temp | O2 | Media | Inhibition | |
|---|---|---|---|---|---|---|
| 4007 | 4008 | |||||
| Bacillus cereus | NCIMB700577 | 37 | Aerobic | BHI | − | − |
| Bacillus subtilis | S249 | 37 | Aerobic | BHI | − | − |
| Bacillus thuringiensis | DPC6341 | 37 | Aerobic | BHI | − | − |
| Bacillus firmis | DPC6349 | 37 | Aerobic | BHI | +++ | +++ |
| Clostridioides difficile | DPC6534 | 37 | Anaerobic | RCM | + | + |
| Clostridioides sporogenes | LMG10143 | 37 | Anaerobic | RCM | + | + |
| Enterococcus faecium | NCDO0942 | 37 | Aerobic | BHI | − | − |
| Enterococcus faecium (VRE) | APC1026 | 37 | Aerobic | BHI | − | − |
| Enterococcus faecium (VRE) | APC1032 | 37 | Aerobic | BHI | − | − |
| Enterococcus faecium (VRE) | APC1033 | 37 | Aerobic | BHI | − | − |
| Enterococcus faecium (VRE) | APC1039 | 37 | Aerobic | BHI | − | − |
| Enterococcus faecium (VRE) | APC1044 | 37 | Aerobic | BHI | − | − |
| Enterococcus faecium (VRE) | APC1055 | 37 | Aerobic | BHI | − | − |
| Lactococcus lactis | HP | 30 | Aerobic | GM17 | + | + |
| Lactococcus lactis * | ATCC11454 | 30 | Aerobic | GM17 | − | − |
| Lactobacillus delbrueckii ssp. bulgaricus | LMG6901 | 37 | Anaerobic | MRS | +++ | +++ |
| Lactobacillus delbrueckii ssp. lactis | DPC5387 | 37 | Anaerobic | MRS | +++ | +++ |
| Lactobacillus helveticus | DPC5358 | 37 | Anaerobic | MRS | + | + |
| Ligilactobacillus salivarius | DPC6502 | 37 | Anaerobic | MRS | + | + |
| Listeria innocua | DPC1768 | 37 | Aerobic | BHI | − | − |
| Listeria monocytogenes | DPC3572 | 37 | Aerobic | BHI | − | − |
| Listeria monocytogenes | L028 | 37 | Aerobic | BHI | − | − |
| Staphylococcus aureus | 32679 | 37 | Aerobic | BHI | − | − |
| Staphylococcus aureus | C5M | 37 | Aerobic | BHI | − | − |
| Staphylococcus aureus | 47.9 | 37 | Aerobic | BHI | − | − |
| Staphylococcus aureus | DPC5243 | 37 | Aerobic | BHI | − | − |
| Staphylococcus aureus | DPC7673 | 37 | Aerobic | BHI | − | − |
| Staphylococcus aureus | R693 | 37 | Aerobic | BHI | − | − |
| Staphylococcus aureus (MRSA) | DPC5646 | 37 | Aerobic | BHI | − | − |
| Staphylococcus epidermidis | DSM3095 | 37 | Aerobic | BHI | − | − |
| Staphylococcus intermedius | DSM20373 | 37 | Aerobic | BHI | + | + |
| Streptococcus agalactiae | 35 | 37 | Aerobic | BHI | − | − |
| Streptococcus agalactiae | 119 | 37 | Aerobic | BHI | − | − |
| Streptococcus agalactiae | APC1055 | 37 | Aerobic | BHI | − | − |
| Streptococcus agalactiae | ATCC13813 | 37 | Aerobic | BHI | − | − |
| Streptococcus pneumoniae | APC3850 | 37 | Aerobic | BHI | − | − |
| Streptococcus pneumoniae | APC3857 | 37 | Aerobic | BHI | − | − |
| Streptococcus pyogenes | DPC6992 | 37 | Aerobic | BHI | − | − |
| Streptococcus uberis | ATCC5344 | 37 | Aerobic | BHI | − | − |
| Streptococcus uberis | LL383 | 37 | Aerobic | BHI | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugrue, I.; Hill, D.; O’Connor, P.M.; Day, L.; Stanton, C.; Hill, C.; Ross, R.P. Nisin E Is a Novel Nisin Variant Produced by Multiple Streptococcus equinus Strains. Microorganisms 2023, 11, 427. https://doi.org/10.3390/microorganisms11020427
Sugrue I, Hill D, O’Connor PM, Day L, Stanton C, Hill C, Ross RP. Nisin E Is a Novel Nisin Variant Produced by Multiple Streptococcus equinus Strains. Microorganisms. 2023; 11(2):427. https://doi.org/10.3390/microorganisms11020427
Chicago/Turabian StyleSugrue, Ivan, Daragh Hill, Paula M. O’Connor, Li Day, Catherine Stanton, Colin Hill, and R. Paul Ross. 2023. "Nisin E Is a Novel Nisin Variant Produced by Multiple Streptococcus equinus Strains" Microorganisms 11, no. 2: 427. https://doi.org/10.3390/microorganisms11020427
APA StyleSugrue, I., Hill, D., O’Connor, P. M., Day, L., Stanton, C., Hill, C., & Ross, R. P. (2023). Nisin E Is a Novel Nisin Variant Produced by Multiple Streptococcus equinus Strains. Microorganisms, 11(2), 427. https://doi.org/10.3390/microorganisms11020427






