Abstract
Bivalves are filter-feeding organisms and biomarkers of bacterial pollution. Our study aimed to analyze the occurrence and characteristics of extended-spectrum β-lactamase (ESBL)- and carbapenemase-producing Escherichia coli among bivalves. A total of 522 bivalve samples were collected along Portuguese shellfish production areas. Homogenized samples were screened for E. coli contamination on corresponding selective plates, allowing for concomitant growth of Klebsiella pneumoniae. E. coli growth was observed in 39% of the samples. Subsequent selective screening identified nine samples (4.4%) contaminated with ESBL producers, corresponding to E. coli (n = 7) and K. pneumoniae (n = 2), while a single carbapenemase-producing K. pneumoniae (0.5%) was identified. ESBLs were all CTX-M-types commonly identified in human isolates, i.e., CTX-M-32 (n = 4), CTX-M-15 (n = 4), and CTX-M-14 (n = 1). The carbapenemase producer harbored the blaGES-5 gene located on a ColE plasmid. Clonality was evaluated by multilocus sequence typing, identifying E. coli backgrounds as ST10, ST23, ST540, ST617, ST746, SLV206, and SLV2325, commonly identified among environmental and human strains. The K. pneumoniae isolates belonged to ST834, ST15, and DLV644. The occurrence of ESBL- and carbapenemase-producing Enterobacteriaceae in bivalves reveals how the marine environment constitutes a reservoir of critical bacterial pathogens, thus potentially representing a risk to human health.
1. Introduction
The emergence of extended-spectrum β-lactamase (ESBL)- and carbapenemase-producing Enterobacterales, and their subsequent spread, remains a major global threat. The transmission of such multidrug-resistant bacteria may occur not only from human to human, but also from animal sources to humans via the food chain [1]. In fact, high rates of Enterobacterales producing ESBLs or carbapenemases are seen frequently in food-producing animals, namely in pigs, cattle, and poultry [2]. Although studies reporting ESBL- and carbapenemase-producing Enterobacterales in seafood products are scarce, more frequent surveillance studies have recently been initiated [3,4,5,6,7,8,9,10,11].
Marine bivalves represent a significant proportion of the world’s seafood production sector with a global volume exceeding 15 million tons per year [12]. Due to its privileged geographical position, Portugal combines several characteristics favorable to the exploitation of bivalves: (1) 1793 km of coast under the influence of different currents; (2) a Mediterranean climate in the south and a temperate climate in the north; (3) average water temperatures ranging between 13 and 18 °C; and (4) the existence of several estuaries and lagoons along the coast [13]. The exploitation of bivalves arises from the north to the south of Portugal, with a significant impact on the national economy.
Bivalve species can be distributed in four major groups: clams, oysters, mussels, and cockles. These organisms feed by filtering phytoplankton and organic matter, a process that allows for the accumulation of various contaminants in these species, including bacteria of both aquatic and anthropogenic origin [14]. Bivalves are therefore good indicators of bacterial, as well as chemical, contamination in a given marine environment. In addition, since bivalves are often eaten raw or lightly cooked, they might constitute a risk for human health.
Bivalve production areas in Europe are assigned to different classes (A, B, or C) according to the content of fecal contamination. Class A areas meet the health standards, whereas class B and C areas require treatment to reduce microbiological contamination before marketing [15]. In Portugal, compliance in bivalve production areas is monitored weekly by the Instituto Português do Mar e Atmosfera (IPMA) and includes the screening for Escherichia coli contamination in bivalves collected in all production areas.
According to the European Centre for Disease Prevention and Control data of 2020, multidrug resistance among Enterobacterales remains a major problem in Portuguese hospitals [16]: (a) the mean proportion of ESBL-producing E. coli isolates causing human invasive infections was estimated at 14.4%; (b) the prevalence of ESBL producers in invasive Klebsiella pneumoniae was estimated at 47.6%; and (c) there has been a notorious, increasing trend in carbapenem resistance among K. pneumoniae, reaching 11.6% in 2020. Given this context, it is essential to better understand the sources and modes of dissemination of multidrug-resistant bacteria, especially those involved in human infections, using a global One Health approach, as recommended by the World Health Organization [17]. Following this approach, we have performed surveillance studies on antimicrobial resistance among multidrug-resistant Enterobacterales in different settings, i.e., among hospitalized patients and individuals at the time of hospital admission [18,19], among healthy community individuals (students) [20], among pigs [21], and among wild birds [22]. A single study conducted in 2019 evaluated the bacterial contamination in bivalves collected in Portuguese shellfish farms but did not report any ESBL producers among Enterobacteriaceae [23]. Therefore, the aim of the present study was to analyze the occurrence and characteristics of ESBL- and carbapenemase-producing E. coli and K. pneumoniae among bivalve samples collected in production areas from Portuguese estuaries and coastal waters, using the same prospective methodology as in our previous epidemiological studies.
2. Materials and Methods
2.1. Bivalve Samples
As part of the national monitoring program for shellfish production area management, 300 batch samples of bivalve mollusks were collected between December 2021 and September 2022 by IPMA from 19 different shellfish production areas along the Portuguese coast (12 estuaries and 7 coastal waters)—Figure 1 and Table 1. A total of 522 bivalve samples were collected.
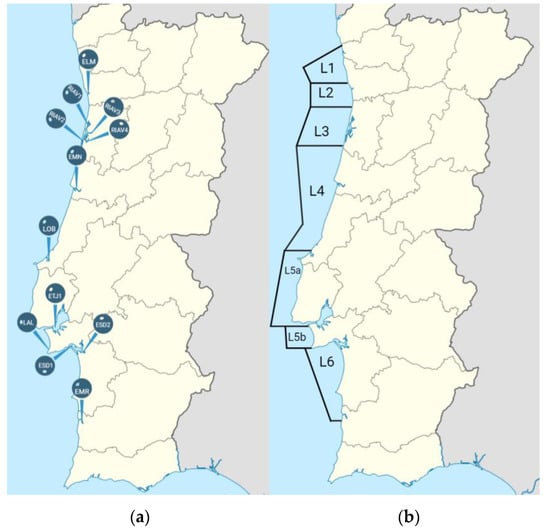
Figure 1.
Localization of the 19 shellfish production areas. (a) Estuaries, (b) Coastal waters. ELM—Estuário do Lima; RIAV1—Ria de Aveiro, Triângulo das Correntes-Moacha; RIAV2—Ria de Aveiro, Canal de Mira; RIAV3—Ria de Aveiro, Canal Principal-Espinheiro; RIAV4—Ria de Aveiro, Canal de Ílhavo; EMN—Estuário do Mondego; LOB—Lagoa de Óbidos; ETJ1—Estuário do Tejo—Jusante Ponte Vasco da Gama; LAL—Lagoa de Albufeira; ESD1—Estuário do Sado, Esteiro da Marateca; ESD2—Estuário do Sado, Canal de Alcácer; EMR—Estuário do Rio Mira; L1—Litoral de Viana; L2—Litoral de Matosinhos; L3—Litoral de Aveiro; L4—Litoral Figueira da Foz—Nazaré; L5a—Litoral Peniche—Cabo Raso; L5b—Litoral Cabo Raso—Cabo Espichel; L6—Litoral Setúbal—Sines.

Table 1.
ESBL and carbapenemase producers by bivalve production area.
Each batch sample comprised 10–20 individual bivalve specimens. Approximately 25 g of muscle and intervalvular liquid per bivalve sample were extracted into a sterile container using a scalpel, diluted in maximum recovery diluent (Oxoid, Heysham, Ireland), and homogenized for 60 s using a Stomacher 400 (Seward Laboratory System, London, UK). Homogenized samples were subsequently screened for E. coli contamination by plating onto Tryptone Bile X-glucuronide (TBX) selective plates (Oxoid), allowing for concomitant growth of K. pneumoniae. A total of 206 TBX plates showing presumptive E. coli growth were provided by IPMA for this study.
2.2. Bacterial Isolates
The bacterial growth of each TBX plate was resuspended in 3 mL of tryptic soy broth (TSB) (Becton, Dickinson & Co., Franklin Lakes, NJ, USA) for enrichment and incubated at 37 °C overnight. Afterwards, a volume of 25 μL of each culture was inoculated into two selective media: (i) CHROMagar ESBL (Frilabo, Maia, Portugal) for ESBL producers, and (ii) ChromID Carba Smart selective medium (bioMérieux, La Balme-les-Grottes, France) for carbapenem-resistant isolates. For quality control, the selective agar plates were also inoculated with the following control strains: ESBL-positive K. pneumoniae ATCC 700603, carbapenemase-positive K. pneumoniae ATCC BAA-1705, and ESBL- and carbapenemase-negative E. coli ATCC 25922.
The isolates selected for the different media were identified at the species level using the API20E system (bioMérieux).
2.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed on all isolates recovered from the two selective media using the disc diffusion method on Mueller–Hinton (MH) agar plates (Neogen, Lansing, Michigan) for ticarcillin (75 μg), amoxicillin/clavulanic acid (20–10 μg), cefotaxime (30 μg), ceftazidime (10 μg), temocillin (30 μg), cefoxitin (30 μg), ertapenem (10 μg), imipenem (10 μg), meropenem (10 μg), ceftazidime/avibactam (10–4 μg), aztreonam (30 μg), ciprofloxacin (5 μg), trimethoprim-sulfamethoxazole (SXT) (1.25–23.75 μg), tetracycline (30 μg), amikacin (30 μg), gentamicin (15 μg), and tobramycin (10 μg) (Bio-Rad Laboratories, Algés, Portugal), following EUCAST recommendations and breakpoint tables. Susceptibility to fosfomycin was evaluated by the disk diffusion method (50 μg) on MH agar plates supplemented with 25 μg/mL glucose-6-phosphate, according to EUCAST guidelines [24]. Strain E. coli ATCC 25922 was used for quality control. Multidrug resistance was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories [25].
2.4. Molecular Analysis
Identification of ESBL and carbapenemase genes was performed by PCR as previously reported [26,27]. All positive amplicons were sent out for sequencing (Eurofins Genomics, Ebersberg, Germany).
The clonal relationship of the ESBL- and carbapenemase-producing isolates was evaluated by multilocus sequence typing (MLST) [28], and sequence types (STs) were assigned using the MLST databases for E. coli (http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search; accessed on 6 December 2022) and K. pneumoniae (https://bigsdb.pasteur.fr/klebsiella/; accessed on 6 December 2022).
2.5. Conjugation Experiments and Plasmid Analysis
Mating-out assays were performed using the azide-resistant E. coli J53 as the recipient. E. coli J53 and the blaGES-5-positive donor were separately inoculated into 5 mL of tryptic soy broth (TSB (Frilabo)) and incubated at 37 °C for 5 h. The samples were subsequently mixed at a ratio of 1:4 (200 µL donor:800 µL recipient), centrifuged for 1 min at 3500 rpm, and 800 µL of the supernatant were discarded. The pellet was resuspended in the remaining 200 µL. This volume was deposited onto 22 µm filters onto a tryptic soy agar (TSA (Frilabo)) plate and incubated for 3 h. After 3 h, the filter was resuspended in 5 mL of 0.9% NaCl, and 100 µL of the mixture were plated onto TSA agar plates supplemented with cefoxitin (50 μg/mL) and azide (100 μg/mL). Susceptibility testing was performed for the E. coli transconjugants, and positivity for blaGES-5 was assessed by PCR.
The plasmid harboring the GES-5 carbapenemase-encoding gene was classified according to its incompatibility group using the PCR-based replicon typing (PBRT) method performed on DNA recovered from E. coli transconjugants, as described previously [29].
2.6. Statistical Analysis
Statistical analysis was performed using GNU R v 4.0.3 under RStudio 2022.07.1 + 554. Fisher’s independence test was used to identify variables associated with the E. coli contamination and carriage of ESBL and carbapenemase producers. All p-values below 0.05 were considered statistically significant.
3. Results
3.1. Population Description
Out of the 522 bivalve samples collected during a nine-month period, 206 (39%) grew E. coli on the TBX selective plates. Those 206 bivalve samples were recovered from the 19 production areas included in the study (Table 1). Of note, E. coli contamination rates were over 40% in 11 of the 19 production areas, all but one (L5b) being located in estuaries. In contrast, the production areas with lower contamination rates (<15%) were all situated in coastal waters (p < 0.001). However, the two coastal production areas closest to the Lisbon metropolitan area (L5a/b and L6) showed a significant amount (30–42%) of E. coli contamination.
E. coli isolates were detected in 18 (out of the 19) bivalve species (Table 2). Four species showed higher rates (>50%) of contamination: Venus casina (1/1; 100%), Scrobicularia plana (10/15; 67%), Dosinia exoleta (3/5; 60%), and Solen marginatus (29/57; 51%). Only the species Scrobicularia plana showed a statistically significant result (p = 0.03).

Table 2.
ESBL and carbapenemase producers by bivalve species.
3.2. Carriage of Multidrug-Resistant Enterobacterales
Out of the 206 samples recovered on the TBX plates, nine enterobacterial isolates grew on the CHROMagar ESBL agar, and one isolate grew on the ChromID Carba Smart selective medium.
The antimicrobial susceptibility testing of the nine former isolates showed non-susceptibility to ticarcillin, cefotaxime and temocillin (100%), ceftazidime and aztreonam (n = 8; 89%), tetracycline (n = 6; 66%), amoxicillin/clavulanic acid (n = 4; 44%), ciprofloxacin, SXT and tobramycin (n = 3; 33%), gentamicin (n = 2; 22%), and amikacin (n = 1; 11%) (Table 3). All isolates remained susceptible to cefoxitin, ertapenem, imipenem, meropenem, and fosfomycin. The single isolate growing on the selective medium for carbapenem resistance showed non-susceptibility to amoxicillin/clavulanic acid, ceftazidime, temocillin, cefoxitin, ertapenem, imipenem, meropenem, and tobramycin. None of the 10 isolates were resistant to the newly developed ceftazidime/avibactam combination. Half of the isolates presented resistance to at least two antibiotic classes other than β-lactams and were considered multidrug-resistant.

Table 3.
Characteristics of the ESBL- and carbapenemase-producing isolates.
Nine bivalve samples out of the 206 that grew on the TBX plates carried an ESBL producer (9/206; 4.4%), and one was contaminated by a carbapenemase producer (1/206; 0.5%)—Table 1. The 10 isolates belonged to two species: E. coli (n = 7; 70%) and K. pneumoniae (n = 3; 30%). All ESBLs were from the CTX-M-type, with CTX-M-32 and CTX-M-15 being the most frequently found (n = 4; 40% each), followed by CTX-M-14 (n = 1). The single carbapenemase producer (K. pneumoniae) harbored blaGES-5.
The ESBL-producing isolates were recovered from six production areas (Table 1), out of which two showed higher frequencies of ESBL producers (p < 0.01 in both cases): ELM (22%) and ESD2 (17%)—Figure 1. The single carbapenemase-producing isolate was found in EMN. All ESBL- and carbapenemase-producing isolates were harvested from estuaries, although this result was not statistically significant (p = 0.16).
The nine ESBL-producing Enterobacterales were isolated from five different bivalve species (Table 2): Venerupis corrugate (n = 2), Magallana gigas (n = 2), Magallana angulata (n = 2), Scrobicularia plana (n = 1), and Solen marginatus (n = 2), whereas the carbapenemase-producing isolate was recovered from Cerastoderma edule. Four (25%) ESBL-producing isolates were recovered from oysters (Magallana gigas and Magallana angulata), although this result was not statistically significant (p = 0.057).
The MLST analysis showed high heterogeneity among the seven ESBL-producing E. coli isolates that were distributed into seven different clones; although two isolates, belonging to ST617 (CTX-M-32) and ST10 (CTX-M-15), were classified into the same clonal complex (CC), CC10—Table 3. The two ESBL-producing (CTX-M-15) K. pneumoniae isolates belonged to ST834 and ST15, while the GES-5 carbapenemase producer was classified as a double-locus variant (DLV) of ST644. The mating-out assay followed by PBRT revealed that the blaGES-5 gene was located on a ColE plasmid.
4. Discussion
Here we present a comprehensive study on the occurrence and characteristics of ESBL- and carbapenemase-producing isolates among bivalve samples, during a nine-month period, from different shellfish production areas in Portuguese estuaries and coastal waters. Our study revealed that a high proportion (39%) of bivalves were contaminated with E. coli, and among these, 4% were contaminated with ESBL producers. In addition, one sample was contaminated with a carbapenemase-producing K. pneumoniae. Of note, all ESBL- and carbapenemase-producing isolates were harvested from estuaries, where effluent flow is less affected by tides and there is lower salinity compared to coastal waters.
The rates of ESBL-producing isolates in the present study were slightly higher than those reported among bivalves in other countries: (i) a study on antibiotic resistance of E. coli from marine bivalves collected in Norway identified ESBL-encoding genes in 1% of isolates (2/199) [5], and (ii) 1.6% of clams bought from retail markets in Tunisia were contaminated with ESBL-producing Enterobacterales [9]. In contrast, the proportion was much lower than the 15% (21/141) reported among E. coli isolates recovered from clams in Italy [6] and India, where 53% of fresh shellfish from retail markets exhibited ESBL-producing E. coli isolates [11].
All ESBLs found in the present study belonged exclusively to the CTX-M family, i.e., CTX-M-32, CTX-M-15, and CTX-M-14. These enzymes are commonly identified in human isolates worldwide, including Portugal [19,20]. Accordingly, we may speculate that the ESBL producers identified among bivalves might correspond to strains colonizing the human gut.
One out of the three K. pneumoniae identified in the present study displayed a carbapenemase gene, which is usually much less frequent among bivalve samples than are ESBLs. Sporadic carbapenem-producing Enterobacterial isolates were previously reported: In Germany, one VIM-1-producing E. coli was isolated from clams collected in Italy [10]. In Tunisia, two K. pneumoniae isolates (blaNDM-1 and blaOXA-48) were isolated in clams, and a KPC-3-producing E. coli was recovered in mussels purchased in retail markets [8,9]. In Myanmar, two NDM-1 producers (one E. coli and one K. pneumoniae) were isolated from a clam and a prawn, respectively [30], and in Canada, three and two clam samples imported from Vietnam contained E. cloacae harboring blaIMI-1 and blaNDM-1, respectively [31]. We identified in the present study the first enterobacterial isolate producing a carbapenemase of GES-type (GES-5) recovered from bivalve samples. This carbapenemase has been previously detected in Portugal among humans [18], gulls [22], the aquatic environment [32], and now bivalves.
The seven ESBL-producing E. coli strains belonged to distinct genetic backgrounds (seven STs), which suggests multiple sources of E. coli contamination in the harvesting production areas. Most of these STs have been previously found among humans and/or animals and/or in the environment, namely in Portugal. In fact, ST10 was the predominant clone among E. coli recovered from healthy students in Lisbon [20], was frequently isolated in Portuguese hospitals [33], and was found among healthy and sick cats in the country [34]. Moreover, ST10 was the predominant lineage among E. coli present in irrigation water and vegetables from 16 household farms in Portugal [35]. Concerning bivalves, one E. coli ST10 producing VIM-1 was isolated from a clam harvested in the Mediterranean Sea (Italy) [10]. ST617, which also belongs to CC10, has been formerly reported in Portugal in a CTX-M-55 E. coli strain colonizing a healthy dog [36] and from two CTX-M-15 E. coli strains isolated from the effluent from a wastewater treatment plant [37]. In addition, ST617 has been previously described in a CTX-M-14 E. coli isolate from clams sold in Tunisian markets [9].
In the present study, one E. coli isolate belonged to a single-locus variant (SLV) of ST206. This ST has been reported only once in Portugal, corresponding to a single CTX-M-15 isolate recently recovered from a pigeon fecal sample in Lisbon [38]. ST206 is mainly associated with animals, being the main clone among chickens in Nigeria [39] and frequently found among food-chain animals in China [40,41]. Concerning E. coli ST23, it has also been previously reported in Portugal among hospital outpatients [33]. This ST was frequently found among human isolates, namely causing urinary tract infections (UTIs) in nursing homes in the Netherlands [42] and also causing mastitis in cattle in France [43].
One ESBL-producing K. pneumoniae strain found in the present study belonged to ST15. ST15 was the main (32%) clonal type among 509 K. pneumoniae isolates collected nationwide in Portugal from 16 hospitals and environmental settings between 1980 and 2019 [44]. ST15 includes clinical, animal, and environment isolates, being an international lineage with a wide geographic distribution [45]. Overall, the clinically relevant bacteria identified in our study seem to qualitatively mirror the human panorama, not only in terms of β-lactamases circulating in this specific environment, but also in terms of clonal backgrounds. These results suggest a very likely human fecal contamination, possibly through sewage effluents that are discharged in estuaries where bivalve production areas are located.
In conclusion, this study constitutes the first report of enterobacterial ESBL and carbapenemase producers among bivalves in Portuguese coastal waters. Although a relatively low occurrence of ESBL and carbapenemase producers was detected, our study demonstrates that bivalves collected for human consumption may act as a potential reservoir of multidrug-resistant bacterial pathogens that eventually may be transmitted through the food chain. Therefore, in the future, the compliance of bivalve production areas routinely monitored by IPMA should also include screening for ESBL- and carbapenemase-producing Enterobacterales contamination in bivalves collected in the different production areas.
Author Contributions
Conceptualization, M.A.-d.-S., A.M. and L.P.; Investigation, S.F., T.G., B.R., C.E. and R.O.; Supervising, M.A.-d.-S. and L.P.; writing—original draft preparation, M.A.-d.-S.; writing—review and editing, A.M. and L.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially financed by the Portuguese Red Cross, by the University of Fribourg, and by the Swiss National Science Foundation (projects FNS-31003A_163432 and FNS-407240_177381).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are all available in the main text.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pérez-Rodríguez, F.; Mercanoglu Taban, B. A state-of-art review on multi-drug resistant pathogens in foods of animal origin: Risk factors and mitigation strategies. Front. Microbiol. 2019, 10, 2091. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.L.E.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as commensal and pathogenic bacteria among food-producing animals: Health implications of extended spectrum β-lactamase (ESBL) production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Morrison, B.J.; Rubin, J.E. Carbapenemase producing bacteria in the food supply escaping detection. PLoS ONE 2015, 10, e0126717. [Google Scholar] [CrossRef]
- Håkonsholm, F.; Hetland, M.A.K.; Svanevik, C.S.; Sundsfjord, A.; Lunestad, B.T.; Marathe, N.P. Antibiotic sensitivity screening of Klebsiella spp. and Raoultella spp. isolated from marine bivalve molluscs reveal presence of CTX-M-producing K. pneumoniae. Microorganisms 2020, 8, 1909. [Google Scholar] [CrossRef]
- Grevskott, D.H.; Svanevik, C.S.; Sunde, M.; Wester, A.L.; Lunestad, B.T. Marine bivalve mollusks as possible indicators of multidrug-resistant Escherichia coli and other species of the Enterobacteriaceae family. Front. Microbiol. 2017, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Vignaroli, C.; Di Sante, L.; Leoni, F.; Chierichetti, S.; Ottaviani, D.; Citterio, B.; Biavasco, F. Multidrug-resistant and epidemic clones of Escherichia coli from natural beds of Venus clam. Food Microbiol. 2016, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mani, Y.; Mansour, W.; Lupo, A.; Saras, E.; Bouallègue, O.; Madec, J.Y.; Haenni, M. Spread of blaCTX-M-15-producing Enterobacteriaceae and OXA-23-producing Acinetobacter baumannii sequence type 2 in Tunisian seafood. Antimicrob. Agents Chemother. 2018, 62, e00727-18. [Google Scholar] [CrossRef]
- Mani, Y.; Mansour, W.; Mammeri, H.; Denamur, E.; Saras, E.; Boujâafar, N.; Bouallègue, O.; Madec, J.Y.; Haenni, M. KPC-3-producing ST167 Escherichia coli from mussels bought at a retail market in Tunisia. J. Antimicrob. Chemother. 2017, 72, 2403–2404. [Google Scholar] [CrossRef]
- Sola, M.; Mani, Y.; Saras, E.; Drapeau, A.; Grami, R.; Aouni, M.; Madec, J.Y.; Haenni, M.; Mansour, W. Prevalence and characterization of Extended-Spectrum β-Lactamase- and carbapenemase-producing Enterobacterales from Tunisian seafood. Microorganisms 2022, 10, 1364. [Google Scholar] [CrossRef]
- Roschanski, N.; Guenther, S.; Vu, T.T.T.; Fischer, J.; Semmler, T.; Huehn, S.; Alter, T.; Roesler, U. VIM-1 carbapenemase-producing Escherichia coli isolated from retail seafood, Germany 2016. Euro Surveill. 2017, 22, 17–00032. [Google Scholar] [CrossRef]
- Singh, A.S.; Nayak, B.B.; Kumar, S.H. High prevalence of multiple antibiotic-resistant, Escherichia coli in fresh seafood sold in retail markets of Mumbai, India. Vet Sci. 2020, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Wijsman, J.W.M.; Troost, K. Global production of marine bivalves. Trends and challenges. In Goods and Services of Marine Bivalves, 1st ed.; Smaal, A.C., Ferreira, J.G., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; pp. 7–26. [Google Scholar]
- Oliveira, J.; Castilho, F.; Cunha, A.; Pereira, M.J. Bivalve harvesting and production in Portugal: An overview. J. Shellfish. Res. 2013, 32, 911–924. [Google Scholar]
- Rehnstam-Holm, A.S.; Hernroth, B. Shellfish and public health: A Swedish perspective. Ambio 2005, 34, 139–144. [Google Scholar] [CrossRef] [PubMed]
- EUR-Lex. Official Journal of the European Union. Commission Implementing Regulation (EU) 2019/627 of 15 March 2019. Available online: file:///Volumes/GoogleDrive/O%20meu%20disco/Marta/Publicac%CC%A7oes%20MAS/Artigos/Bivalves/CELEX_32019R0627_EN_TXT.pdf (accessed on 12 December 2022).
- European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe—Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2020. 2021. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/AER-EARS-Net-2020.pdf (accessed on 6 December 2022).
- Diallo, O.O.; Baron, S.A.; Abat, C.; Colson, P.; Chaudet, H.; Rolain, J.M. Antibiotic resistance surveillance systems: A review. J. Glob. Antimicrob. Resist. 2020, 23, 430–438. [Google Scholar] [CrossRef]
- Aires-de-Sousa, M.; Ortiz de la Rosa, J.M.; Gonçalves, M.L.; Pereira, A.L.; Nordmann, P.; Poirel, L. Epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital, Portugal. Emerg. Infect. Dis. 2019, 25, 1632–1638. [Google Scholar] [CrossRef] [PubMed]
- Aires-de-Sousa, M.; Lopes, E.; Gonçalves, M.L.; Pereira, A.L.; Machado Costa, A.E.; de Lencastre, H.; Poirel, L. Intestinal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae at admission in a Portuguese hospital. Eur J Clin. Microbiol. Infect. Dis. 2020, 39, 783–790. [Google Scholar] [CrossRef]
- Fournier, C.; Aires de Sousa, M.; Fuster Escriva, B.; Sales, L.; Nordmann, P.; Poirel, L. Epidemiology of extended-spectrum β-lactamase-producing Enterobacteriaceae among healthcare students, at the Portuguese Red Cross Health School of Lisbon, Portugal. J. Glob. Antimicrob. Resist. 2020, 22, 733–737. [Google Scholar] [CrossRef]
- Fournier, C.; Aires-de-Sousa, M.; Nordmann, P.; Poirel, L. Occurrence of CTX-M-15- and MCR-1-producing Enterobacterales in pigs in Portugal: Evidence of direct links with antibiotic selective pressure. Int. J. Antimicrob. Agents. 2020, 55, 105802. [Google Scholar] [CrossRef]
- Aires-de-Sousa, M.; Fournier, C.; Lopes, E.; de Lencastre, H.; Nordmann, P.; Poirel, L. High colonization rate and heterogeneity of ESBL- and carbapenemase-producing Enterobacteriaceae isolated from gull feces in Lisbon, Portugal. Microorganisms 2020, 8, 1487. [Google Scholar] [CrossRef]
- Salgueiro, V.; Reis, L.; Jo, M.; Manageiro, V.; Caniça, M. Assessing the bacterial community composition of bivalve mollusks collected in aquaculture farms and respective susceptibility to antibiotics. Antibiotics 2021, 10, 1135. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 12 November 2022).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Lartigue, M.F.; Zinsius, C.; Wenger, A.; Bille, J.; Poirel, L.; Nordmann, P. Extended-spectrum beta-lactamases of the CTX-M type now in Switzerland. Antimicrob. Agents Chemother. 2007, 51, 2855–2860. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.A.; Brisse, S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005, 43, 4178–4182. [Google Scholar] [CrossRef]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Sugawara, Y.; Hagiya, H.; Akeda, Y.; Aye, M.M.; Myo Win, H.P.; Sakamoto, N.; Shanmugakani, R.K.; Takeuchi, D.; Nishi, I.; Ueda, A.; et al. Dissemination of carbapenemase-producing Enterobacteriaceae harbouring blaNDM or blaIMI in local market foods of Yangon, Myanmar. Sci. Rep. 2019, 9, 14455. [Google Scholar] [CrossRef] [PubMed]
- Janecko, N.; Martz, S.L.; Avery, B.P.; Daignault, D.; Desruisseau, A.; Boyd, D.; Irwin, R.J.; Mulvey, M.R.; Reid-Smith, R.J. Carbapenem-resistant Enterobacter spp. in retail seafood imported from Southeast Asia to Canada. Emerg. Infect. Dis. 2016, 22, 1675–1677. [Google Scholar] [CrossRef]
- Manageiro, V.; Ferreira, E.; Caniça, M.; Manaia, C.M. GES-5 among the β-lactamases detected in ubiquitous bacteria isolated from aquatic environment samples. FEMS Microbiol. Lett. 2014, 351, 64–69. [Google Scholar] [CrossRef]
- Rodrigues, C.; Machado, E.; Pires, J.; Ramos, H.; Novais, Â.; Peixe, L. Increase of widespread A, B1 and D Escherichia coli clones producing a high diversity of CTX-M-types in a Portuguese hospital. Future Microbiol. 2015, 10, 1125–1131. [Google Scholar] [CrossRef]
- Carvalho, I.; Safia Chenouf, N.; Cunha, R.; Martins, C.; Pimenta, P.; Pereira, A.R.; Martínez-Álvarez, S.; Ramos, S.; Silva, V.; Igrejas, G.; et al. Antimicrobial resistance genes and diversity of clones among ESBL- and acquired AmpC-producing Escherichia coli isolated from fecal samples of healthy and sick cats in Portugal. Antibiotics 2021, 10, 262. [Google Scholar] [CrossRef]
- Araújo, S.; Silva, I.A.T.; Tacão, M.; Patinha, C.; Alves, A.; Henriques, I. Characterization of antibiotic resistant and pathogenic Escherichia coli in irrigation water and vegetables in household farms. Int. J. Food Microbiol. 2017, 257, 192–200. [Google Scholar] [CrossRef]
- Carvalho, I.; Cunha, R.; Martins, C.; Martínez-Álvarez, S.; Safia Chenouf, N.; Pimenta, P.; Pereira, A.R.; Ramos, S.; Sadi, M.; Martins, Â.; et al. Antimicrobial resistance genes and diversity of clones among faecal ESBL-producing Escherichia coli isolated from healthy and sick dogs living in Portugal. Antibiotics 2021, 10, 1013. [Google Scholar] [CrossRef] [PubMed]
- Tavares, R.D.S.; Tacão, M.; Figueiredo, A.S.; Duarte, A.S.; Esposito, F.; Lincopan, N.; Manaia, C.M.; Henriques, I. Genotypic and phenotypic traits of blaCTX-M-carrying Escherichia coli strains from an UV-C-treated wastewater effluent. Water Res. 2020, 184, 116079. [Google Scholar] [CrossRef] [PubMed]
- Freire, S.; Grilo, T.; Poirel, L.; Aires-de-Sousa, M. Urban pigeons (Columba livia) as a source of broad-spectrum β-lactamase-producing Escherichia coli in Lisbon, Portugal. Antibiotics 2022, 11, 1368. [Google Scholar] [CrossRef] [PubMed]
- Ayeni, F.A.; Falgenhauer, J.; Schmiedel, J.; Schwengers, O.; Chakraborty, T.; Falgenhauer, L. Detection of blaCTX-M-27-encoding Escherichia coli ST206 in Nigerian poultry stocks. J. Antimicrob. Chemother. 2020, 75, 3070–3072. [Google Scholar] [CrossRef]
- Li, F.; Cheng, P.; Li, X.; Liu, R.; Liu, H.; Zhang, X. Molecular epidemiology and colistin-resistant mechanism of mcr-positive and mcr-negative Escherichia coli isolated from animal in Sichuan province, China. Front Microbiol. 2022, 13, 818548. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qin, L.; Hao, Z. High prevalence and diversity characteristics of blaNDM, mcr, and blaESBLs harboring multidrug-resistant Escherichia coli from chicken, pig, and cattle in China. Front. Cell Infect. Microbiol. 2022, 11, 755545. [Google Scholar] [CrossRef]
- Hidad, S.; van der Putten, B.; van Houdt, R.; Schneeberger, C.; Kuil, S.D. Recurrent E. coli urinary tract infections in nursing homes: Insight in sequence types and antibiotic resistance patterns. Antibiotics 2022, 11, 1638. [Google Scholar] [CrossRef]
- Dahmen, S.; Métayer, V.; Gay, E.; Madec, J.Y.; Haenni, M. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Vet. Microbiol. 2013, 162, 793–799. [Google Scholar] [CrossRef]
- Spadar, A.; Phelan, J.; Elias, R.; Modesto, A.; Caneiras, C.; Marques, C.; Lito, L.; Pinto, M.; Cavaco-Silva, P.; Ferreira, H.; et al. Genomic epidemiological analysis of Klebsiella pneumoniae from portuguese hospitals reveals insights into circulating antimicrobial resistance. Sci. Rep. 2022, 12, 13791. [Google Scholar] [CrossRef]
- Rocha, J.; Henriques, I.; Gomila, M.; Manaia, C.M. Common and distinctive genomic features of Klebsiella pneumoniae thriving in the natural environment or in clinical settings. Sci. Rep. 2022, 12, 10441. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).