Activation of MyD88-Dependent TLR Signaling Modulates Immune Response of the Mouse Heart during Pasteurella multocida Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. P. multocida Challenge Experiments
2.3. Histopathological Examination
2.4. RNA Extraction and Transcriptome Sequencing
2.5. Bioinformatics Analysis of Sequence Data
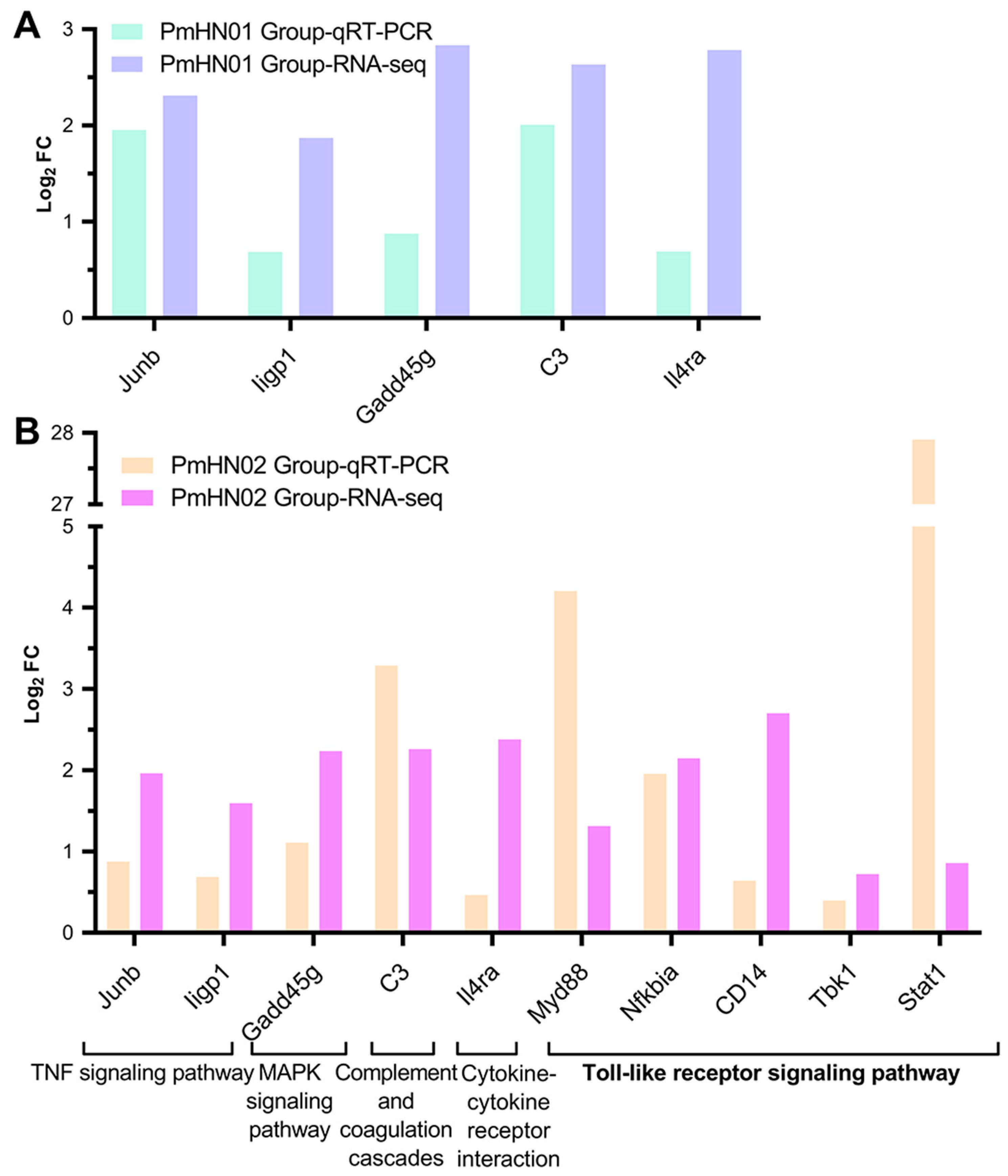
2.6. Quantitative Real-Time PCR
2.7. Immunohistochemistry Staining
2.8. Statistical Analysis
3. Results
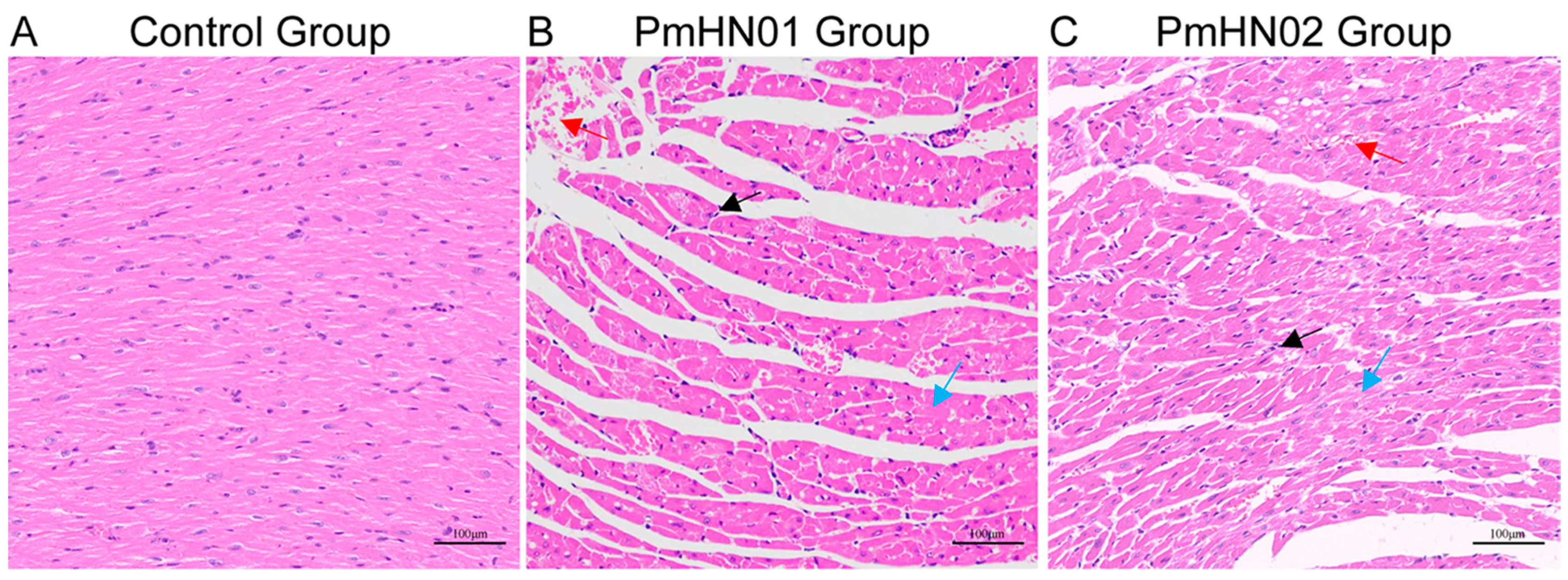
3.1. Pm HN01 and Pm HN02 Cause Different Pathological Changes in the Heart
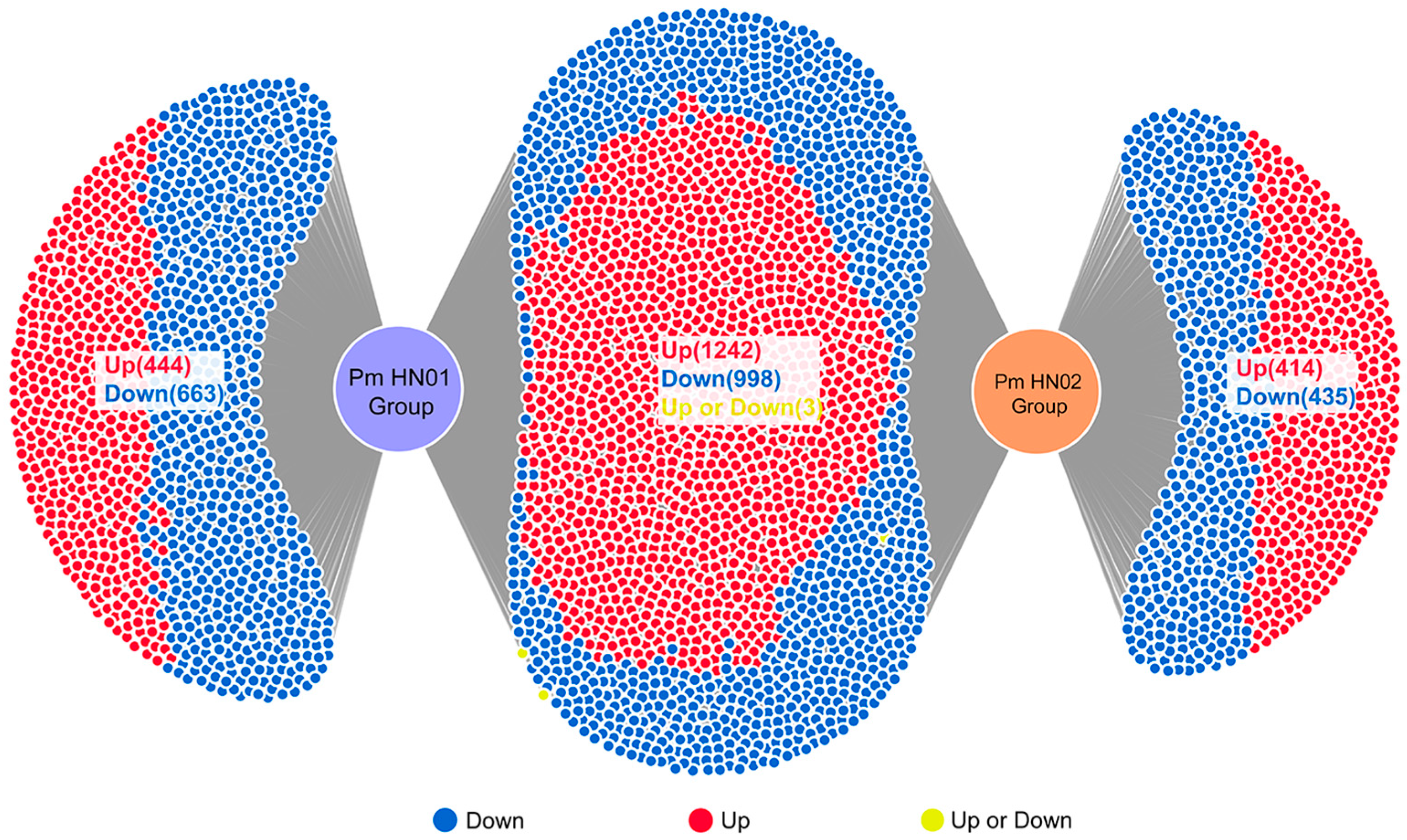
3.2. Pm HN01 and Pm HN02 Induce Unique and Common DEGs
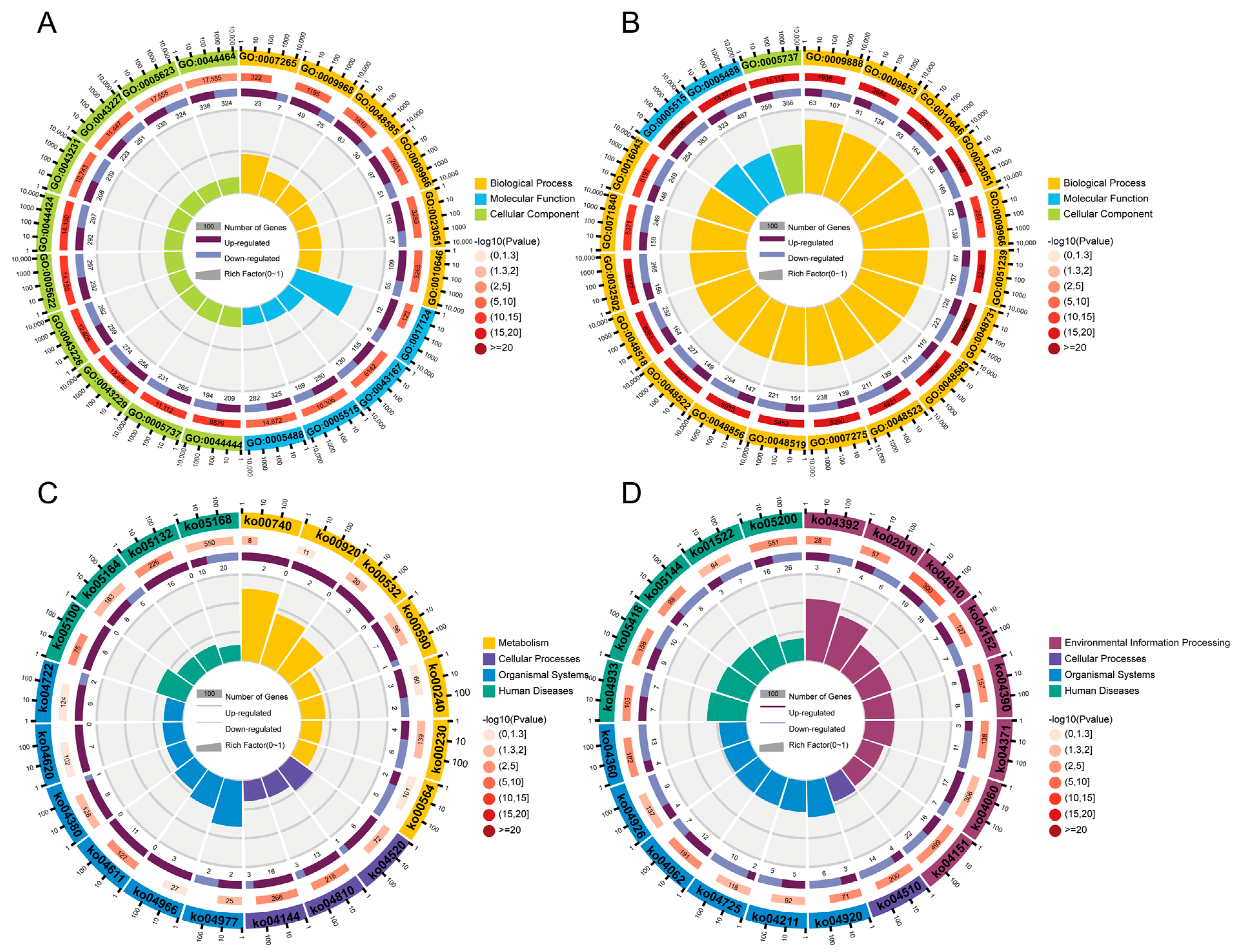
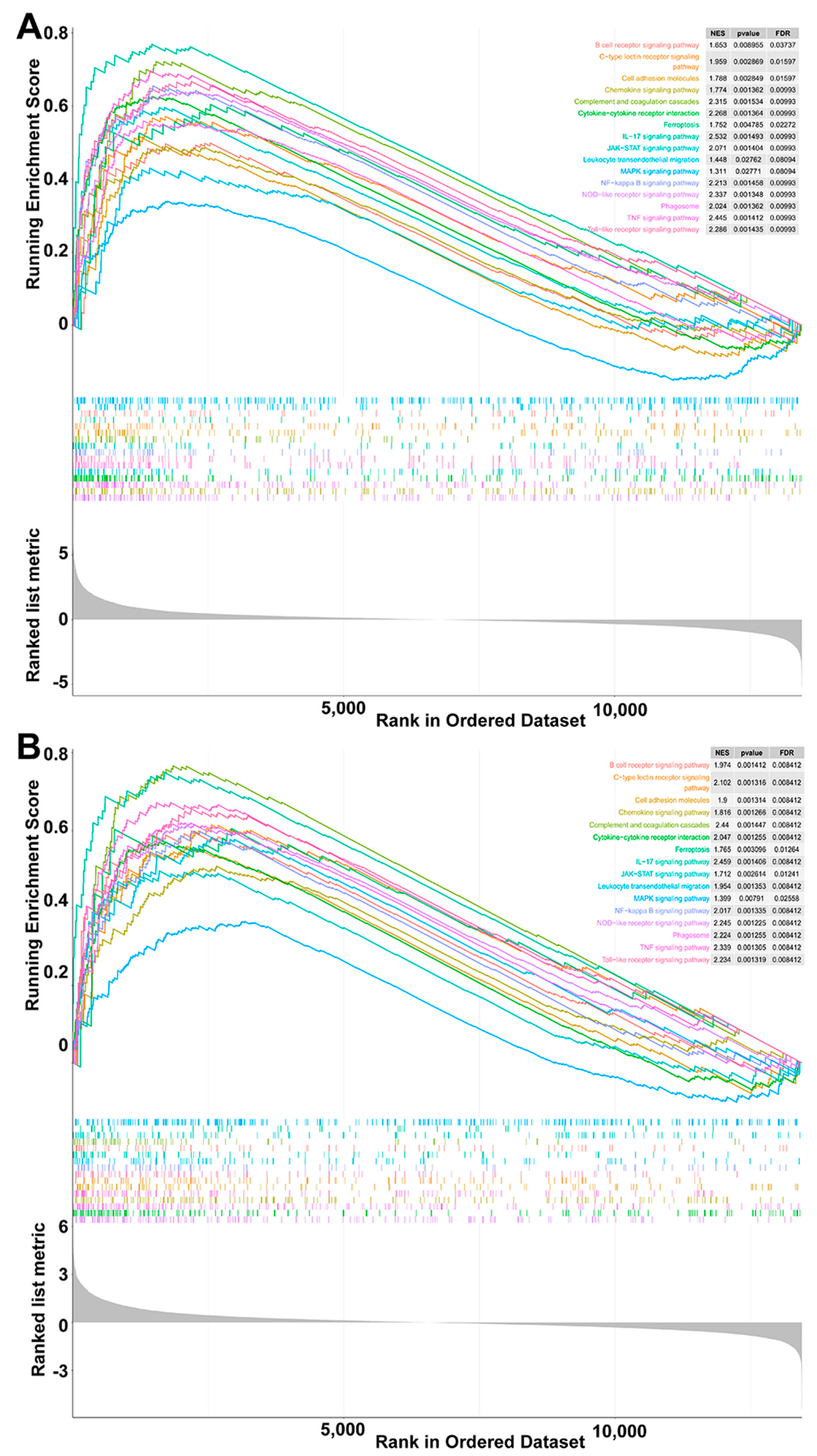
3.3. Common DEGs Regulated by Pm HN01 and Pm HN02 Are Mainly Related to Innate Immunity
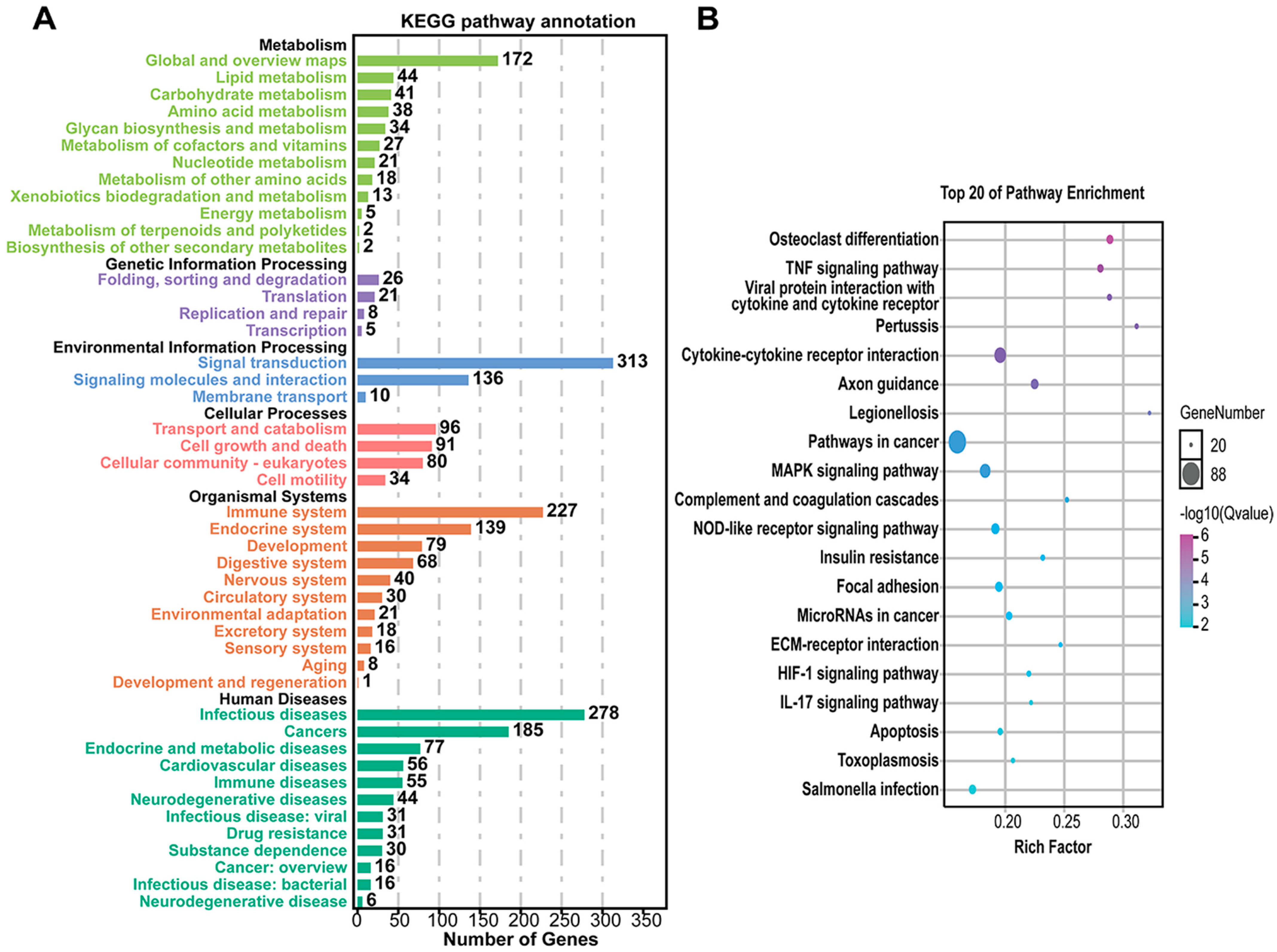
3.4. Unique DEGs Caused by Pm HN01 and Pm HN02 Are Functionally Distinct
3.5. Toll-like Receptor Signaling Pathway Significantly Activated by Pm HN02
3.6. Pm HN02 Induced the Activation of Myd88 at Transcription and Protein Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Shi, L.; Zhang, Y.; Wu, L.; Xun, W.; Liu, Q.; Cao, T.; Hou, G.; Zhou, H. Moderate Coconut Oil Supplement Ameliorates Growth Performance and Ruminal Fermentation in Hainan Black Goat Kids. Front. Vet. Sci. 2020, 7, 622259. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, X.; Zhou, R.; Chen, H.; Wilson, B.A.; Wu, B. Pasteurella multocida: Genotypes and Genomics. Microbiol. Mol. Biol. Rev. 2019, 83, e00014-19. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Liang, W.; Wang, F.; Xu, Z.; Xie, Z.; Lian, Z.; Hua, L.; Zhou, R.; Chen, H.; Wu, B. Genetic and Phylogenetic Characteristics of Pasteurella multocida Isolates From Different Host Species. Front. Microbiol. 2018, 9, 1408. [Google Scholar] [CrossRef]
- Gharib Mombeni, E.; Gharibi, D.; Ghorbanpoor, M.; Jabbari, A.R.; Cid, D. Toxigenic and nontoxigenic Pasteurella multocida genotypes, based on capsular, LPS, and virulence profile typing, associated with pneumonic pasteurellosis in Iran. Vet. Microbiol. 2021, 257, 109077. [Google Scholar] [CrossRef] [PubMed]
- Petruzzi, B.; Briggs, R.E.; Tatum, F.M.; Swords, W.E.; de Castro, C.; Molinaro, A.; Inzana, T.J. Capsular Polysaccharide Interferes with Biofilm Formation by Pasteurella multocida Serogroup A. mBio 2017, 8, e01843-17. [Google Scholar] [CrossRef]
- Deangelis, P.L.; White, C.L. Identification and molecular cloning of a heparosan synthase from Pasteurella multocida type D. J. Biol. Chem. 2002, 277, 7209–7213. [Google Scholar] [CrossRef]
- Williams, A.; Gedeon, K.S.; Vaidyanathan, D.; Yu, Y.; Collins, C.H.; Dordick, J.S.; Linhardt, R.J.; Koffas, M.A.G. Metabolic engineering of Bacillus megaterium for heparosan biosynthesis using Pasteurella multocida heparosan synthase, PmHS2. Microb. Cell Fact. 2019, 18, 132. [Google Scholar] [CrossRef]
- An, Q.; Chen, S.; Zhang, L.; Zhang, Z.; Cheng, Y.; Wu, H.; Liu, A.; Chen, Z.; Li, B.; Chen, J.; et al. The mRNA and miRNA profiles of goat bronchial epithelial cells stimulated by Pasteurella multocida strains of serotype A and D. PeerJ 2022, 10, e13047. [Google Scholar] [CrossRef]
- He, F.; Yin, Z.; Wu, C.; Xia, Y.; Wu, M.; Li, P.; Zhang, H.; Yin, Y.; Li, N.; Zhu, G.; et al. l-Serine Lowers the Inflammatory Responses during Pasteurella multocida Infection. Infect. Immun. 2019, 87, e00677-19. [Google Scholar] [CrossRef]
- Li, Y.; Xie, M.; Zhou, J.; Lin, H.; Xiao, T.; Wu, L.; Ding, H.; Fang, B. Increased Antimicrobial Activity of Colistin in Combination With Gamithromycin Against Pasteurella multocida in a Neutropenic Murine Lung Infection Model. Front. Microbiol. 2020, 11, 511356. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, X.; Zhang, C.; Yong, K.; Clifton, A.C.; Ding, H.; Liu, Y. Pharmacokinetics and Pharmacodynamics of Gamithromycin Treatment of Pasteurella multocida in a Murine Lung Infection Model. Front. Pharmacol. 2019, 10, 1090. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, I.W.; Harper, M.; Boyce, J.D.; Adler, B. Pasteurella multocida: Diseases and pathogenesis. Curr. Top. Microbiol. Immunol. 2012, 361, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Weise, M.; Vettel, C.; Spiger, K.; Gilsbach, R.; Hein, L.; Lorenz, K.; Wieland, T.; Aktories, K.; Orth, J.H.C. A systemic Pasteurella multocida toxin aggravates cardiac hypertrophy and fibrosis in mice. Cell. Microbiol. 2015, 17, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Shivachandra, S.B.; Kumar, A.A.; Gautam, R.; Saxena, M.K.; Chaudhuri, P.; Srivastava, S.K. Detection of multiple strains of Pasteurella multocida in fowl cholera outbreaks by polymerase chain reaction-based typing. Avian Pathol. 2005, 34, 456–462. [Google Scholar] [CrossRef]
- Hasan, J.; Hug, M. Pasteurella Multocida; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Pors, S.E.; Hansen, M.S.; Bisgaard, M.; Jensen, H.E. Occurrence and associated lesions of Pasteurella multocida in porcine bronchopneumonia. Vet. Microbiol. 2011, 150, 160–166. [Google Scholar] [CrossRef]
- Panna, S.; Nazir, K.H.; Rahman, M.; Ahmed, S.; Saroare, M.; Chakma, S.; Kamal, T.; Majumder, U. Isolation and molecular detection of Pasteurella multocida Type A from naturally infected chickens, and their histopathological evaluation in artificially infected chickens in Bangladesh. J. Adv. Vet. Anim. Res. 2015, 2, 338. [Google Scholar] [CrossRef]
- Chung, E.L.T.; Abdullah, F.F.J.; Marza, A.D.; Saleh, W.M.M.; Ibrahim, H.H.; Abba, Y.; Zamri-Saad, M.; Haron, A.W.; Saharee, A.A.; Lila, M.A.M.; et al. Clinico-pathology and hemato-biochemistry responses in buffaloes infected with Pasteurella multocida type B:2 immunogen outer membrane protein. Microb. Pathog. 2017, 102, 89–101. [Google Scholar] [CrossRef]
- Ren, W.; Liu, S.; Chen, S.; Zhang, F.; Li, N.; Yin, J.; Peng, Y.; Wu, L.; Liu, G.; Yin, Y.; et al. Dietary L-glutamine supplementation increases Pasteurella multocida burden and the expression of its major virulence factors in mice. Amino Acids 2013, 45, 947–955. [Google Scholar] [CrossRef]
- Brown, A.O.; Mann, B.; Gao, G.; Hankins, J.S.; Humann, J.; Giardina, J.; Faverio, P.; Restrepo, M.I.; Halade, G.V.; Mortensen, E.M.; et al. Streptococcus pneumoniae translocates into the myocardium and forms unique microlesions that disrupt cardiac function. PLoS Pathog. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Kharb, S.; Charan, S. Mouse model of haemorrhagic septicaemia: Dissemination and multiplication of Pasteurella multocida B:2 in vital organs after intranasal and subcutaneous challenge in mice. Vet. Res. Commun. 2013, 37, 59–63. [Google Scholar] [CrossRef]
- Miao, W.; Han, Y.; Yang, Y.; Hao, Z.; An, N.; Chen, J.; Zhang, Z.; Gao, X.; Storey, K.B.; Chang, H.; et al. Dynamic Changes in Colonic Structure and Protein Expression Suggest Regulatory Mechanisms of Colonic Barrier Function in Torpor-Arousal Cycles of the Daurian Ground Squirrel. Int. J. Mol. Sci. 2022, 23, 9026. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Dong, S.; Ge, Y.; Fonseca, J.P.; Robinson, Z.T.; Mysore, K.S.; Mehta, P. DiVenn: An Interactive and Integrated Web-Based Visualization Tool for Comparing Gene Lists. Front. Genet. 2019, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, Z.; Ran, J.; Peng, L.; Wu, X.; Ye, C.; Dong, C.; Peng, Y.; Fang, R. The Critical Role of Potassium Efflux and Nek7 in Pasteurella multocida-Induced NLRP3 Inflammasome Activation. Front. Microbiol. 2022, 13, 849482. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Sun, M.; Lin, Z.; Li, M.; Gehring, R.; Zeng, Z. Pharmacokinetics and Pharmacodynamics of Tildipirosin Against Pasteurella multocida in a Murine Lung Infection Model. Front. Microbiol. 2018, 9, 1038. [Google Scholar] [CrossRef]
- Priya, G.B.; Nagaleekar, V.K.; Milton, A.A.P.; Saminathan, M.; Kumar, A.; Sahoo, A.R.; Wani, S.A.; Kumar, A.; Gupta, S.K.; Sahoo, A.P.; et al. Genome wide host gene expression analysis in mice experimentally infected with Pasteurella multocida. PLoS ONE 2017, 12, e0179420. [Google Scholar] [CrossRef]
- Wu, C.; Qin, X.; Li, P.; Pan, T.; Ren, W.; Li, N.; Peng, Y. Transcriptomic Analysis on Responses of Murine Lungs to Pasteurella multocida Infection. Front. Cell. Infect. Microbiol. 2017, 7, 251. [Google Scholar] [CrossRef]
- Esfahani, N.S.; Wu, Q.; Kumar, N.; Ganesan, L.P.; Lafuse, W.P.; Rajaram, M.V.S. Aging influences the cardiac macrophage phenotype and function during steady state and during inflammation. Aging Cell 2021, 20, e13438. [Google Scholar] [CrossRef]
- Mouton, A.J.; Li, X.; Hall, M.E.; Hall, J.E. Obesity, Hypertension, and Cardiac Dysfunction: Novel Roles of Immunometabolism in Macrophage Activation and Inflammation. Circ. Res. 2020, 126, 789–806. [Google Scholar] [CrossRef]
- Zhang, T.; Ding, C.; Chen, H.; Zhao, J.; Chen, Z.; Chen, B.; Mao, K.; Hao, Y.; Roulis, M.; Xu, H.; et al. m(6)A mRNA modification maintains colonic epithelial cell homeostasis via NF-κB-mediated antiapoptotic pathway. Sci. Adv. 2022, 8, eabl5723. [Google Scholar] [CrossRef]
- Trembinski, D.J.; Bink, D.I.; Theodorou, K.; Sommer, J.; Fischer, A.; van Bergen, A.; Kuo, C.-C.; Costa, I.G.; Schürmann, C.; Leisegang, M.S.; et al. Aging-regulated anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction. Nat. Commun. 2020, 11, 2039. [Google Scholar] [CrossRef]
- Xu, M.; Liu, P.P.; Li, H. Innate Immune Signaling and Its Role in Metabolic and Cardiovascular Diseases. Physiol. Rev. 2019, 99, 893–948. [Google Scholar] [CrossRef] [PubMed]
- Radakovics, K.; Battin, C.; Leitner, J.; Geiselhart, S.; Paster, W.; Stöckl, J.; Hoffmann-Sommergruber, K.; Steinberger, P. A Highly Sensitive Cell-Based TLR Reporter Platform for the Specific Detection of Bacterial TLR Ligands. Front. Immunol. 2021, 12, 817604. [Google Scholar] [CrossRef] [PubMed]
- Dickson, K.; Lehmann, C. Inflammatory Response to Different Toxins in Experimental Sepsis Models. Int. J. Mol. Sci. 2019, 20, 4341. [Google Scholar] [CrossRef]
- Saur, I.M.L.; Panstruga, R.; Schulze-Lefert, P. NOD-like receptor-mediated plant immunity: From structure to cell death. Nat. Rev. Immunol. 2021, 21, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Griffin, M.E.; Espinosa, J.; Becker, J.L.; Luo, J.-D.; Carroll, T.S.; Jha, J.K.; Fanger, G.R.; Hang, H.C. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 2021, 373, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, J.; Xu, X.; Wang, H.; Qiao, Y.; Chu, W.C.; Xu, S.; Chai, L.; Cottier, F.; Pavelka, N.; et al. Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat. Microbiol. 2019, 4, 766–773. [Google Scholar] [CrossRef]
- Mathé, J.; Benhammadi, M.; Kobayashi, K.S.; Brochu, S.; Perreault, C. Regulation of MHC Class I Expression in Lung Epithelial Cells during Inflammation. J. Immunol. 2022, 208, 1021–1033. [Google Scholar] [CrossRef]
- Bekassy, Z.; Lopatko Fagerström, I.; Bader, M.; Karpman, D. Crosstalk between the renin-angiotensin, complement and kallikrein-kinin systems in inflammation. Nat. Rev. Immunol. 2021, 22, 411–428. [Google Scholar] [CrossRef]
- Hjorth, M.; Febbraio, M.A. IL-1β delivers a sweet deal. Nat. Immunol. 2017, 18, 247–248. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.E.; Petri, W.A., Jr. TLR2 as a Therapeutic Target in Bacterial Infection. Trends Mol. Med. 2020, 26, 715–717. [Google Scholar] [CrossRef]
- Jang, J.C.; Li, J.; Gambini, L.; Batugedara, H.M.; Sati, S.; Lazar, M.A.; Fan, L.; Pellecchia, M.; Nair, M.G. Human resistin protects against endotoxic shock by blocking LPS-TLR4 interaction. Proc. Natl. Acad. Sci. USA 2017, 114, E10399–E10408. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Du, H.; Lei, G.; Liu, Y.; Feng, S.; Ye, C.; Li, N.; Peng, Y. NLRP3 inflammasome plays an important role in caspase-1 activation and IL-1β secretion in macrophages infected with Pasteurella multocida. Vet. Microbiol. 2019, 231, 207–213. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Qin, X.; Xu, N.; Li, P.; Wu, X.; Duan, L.; Du, Y.; Fang, R.; Hardwidge, P.R.; Li, N.; et al. Pasteurella multocida Pm0442 Affects Virulence Gene Expression and Targets TLR2 to Induce Inflammatory Responses. Front. Microbiol. 2020, 11, 1972. [Google Scholar] [CrossRef] [PubMed]
- Tiku, V.; Tan, M.-W. Host immunity and cellular responses to bacterial outer membrane vesicles. Trends Immunol. 2021, 42, 1024–1036. [Google Scholar] [CrossRef]
- Li, N.; Zhou, H.; Wu, H.; Wu, Q.; Duan, M.; Deng, W.; Tang, Q. STING-IRF3 contributes to lipopolysaccharide-induced cardiac dysfunction, inflammation, apoptosis and pyroptosis by activating NLRP3. Redox Biol. 2019, 24, 101215. [Google Scholar] [CrossRef]
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Núñez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016, 530, 354–357. [Google Scholar] [CrossRef]
- Keestra, A.M.; van Putten, J.P.M. Unique properties of the chicken TLR4/MD-2 complex: Selective lipopolysaccharide activation of the MyD88-dependent pathway. J. Immunol. 2008, 181, 4354–4362. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, D.; Sahr, A.; Wölfle, S.J.; Heeg, K.; Kubatzky, K.F. Regulation of Toll-like receptor 4-mediated immune responses through Pasteurella multocida toxin-induced G protein signalling. Cell Commun. Signal. 2012, 10, 22. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Q.; Jiang, J.; Li, X.; Zhai, Z.; Wang, X.; Li, C.; Chen, Q.; Man, C.; Du, L.; Wang, F.; et al. Activation of MyD88-Dependent TLR Signaling Modulates Immune Response of the Mouse Heart during Pasteurella multocida Infection. Microorganisms 2023, 11, 400. https://doi.org/10.3390/microorganisms11020400
Fu Q, Jiang J, Li X, Zhai Z, Wang X, Li C, Chen Q, Man C, Du L, Wang F, et al. Activation of MyD88-Dependent TLR Signaling Modulates Immune Response of the Mouse Heart during Pasteurella multocida Infection. Microorganisms. 2023; 11(2):400. https://doi.org/10.3390/microorganisms11020400
Chicago/Turabian StyleFu, Qiaoyu, Junming Jiang, Xubo Li, Zhe Zhai, Xuemei Wang, Chongrui Li, Qiaoling Chen, Churiga Man, Li Du, Fengyang Wang, and et al. 2023. "Activation of MyD88-Dependent TLR Signaling Modulates Immune Response of the Mouse Heart during Pasteurella multocida Infection" Microorganisms 11, no. 2: 400. https://doi.org/10.3390/microorganisms11020400
APA StyleFu, Q., Jiang, J., Li, X., Zhai, Z., Wang, X., Li, C., Chen, Q., Man, C., Du, L., Wang, F., & Chen, S. (2023). Activation of MyD88-Dependent TLR Signaling Modulates Immune Response of the Mouse Heart during Pasteurella multocida Infection. Microorganisms, 11(2), 400. https://doi.org/10.3390/microorganisms11020400








