C-di-AMP Is a Second Messenger in Corynebacterium glutamicum That Regulates Expression of a Cell Wall-Related Peptidase via a Riboswitch
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Culture Conditions
2.2. Construction of Plasmids and Recombinant Strains
2.3. Recombinant Protein Production and Purification
2.4. Diadenylate Cyclase and Phosphodiesterase Assays
2.5. Detection of Adenylpurines by HPLC
2.6. Real-Time Quantitative PCR
2.7. Fluorescence Reporter Assays
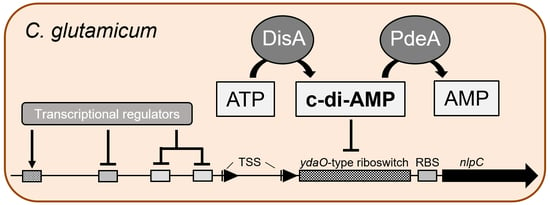
3. Results
3.1. DisA Is a Diadenylate Cyclase in C. glutamicum
3.2. PdeA Is the Sole c-di-AMP Phosphodiesterase in C. glutamicum
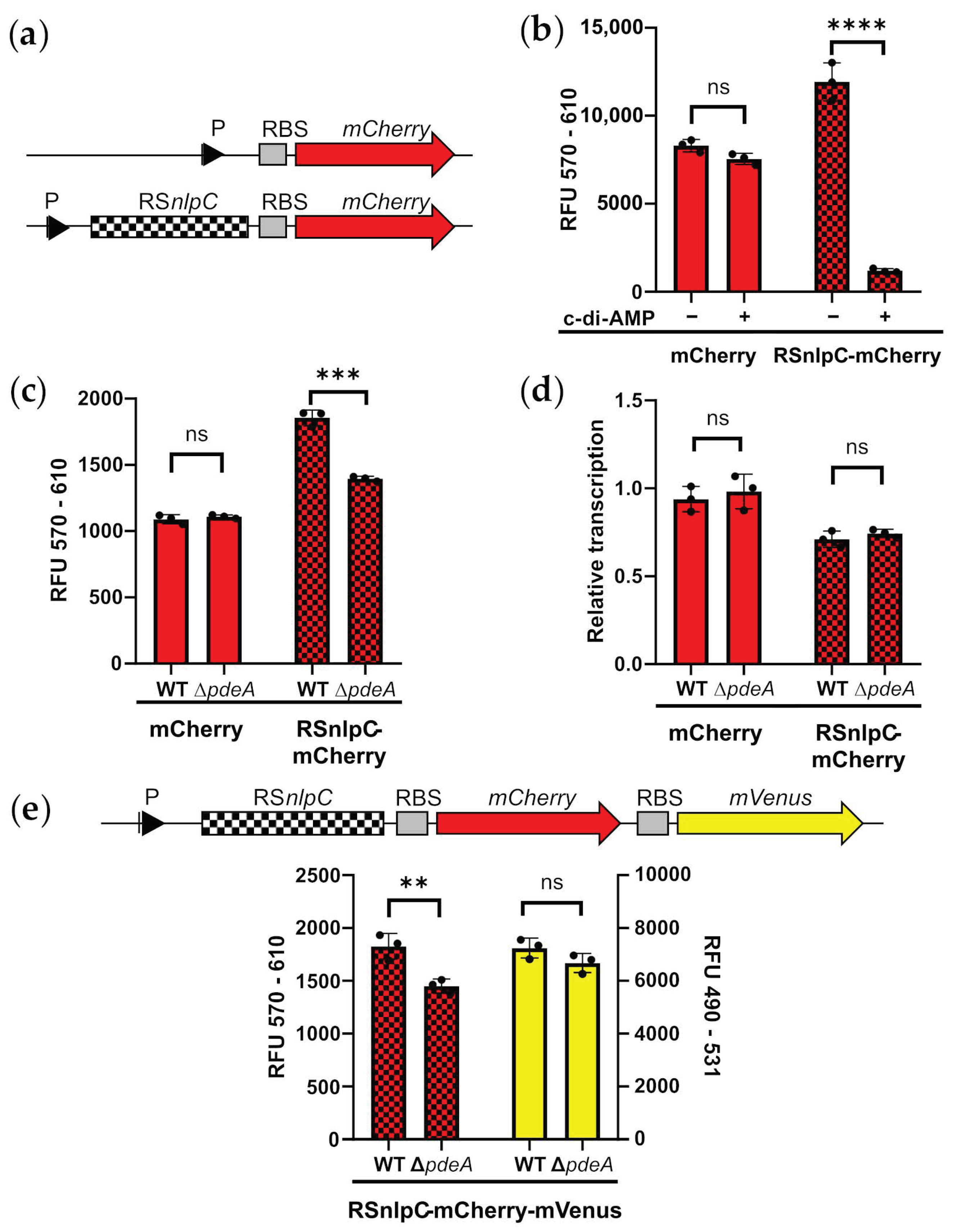
3.3. C-di-AMP Regulates nlpC via a c-di-AMP Riboswitch
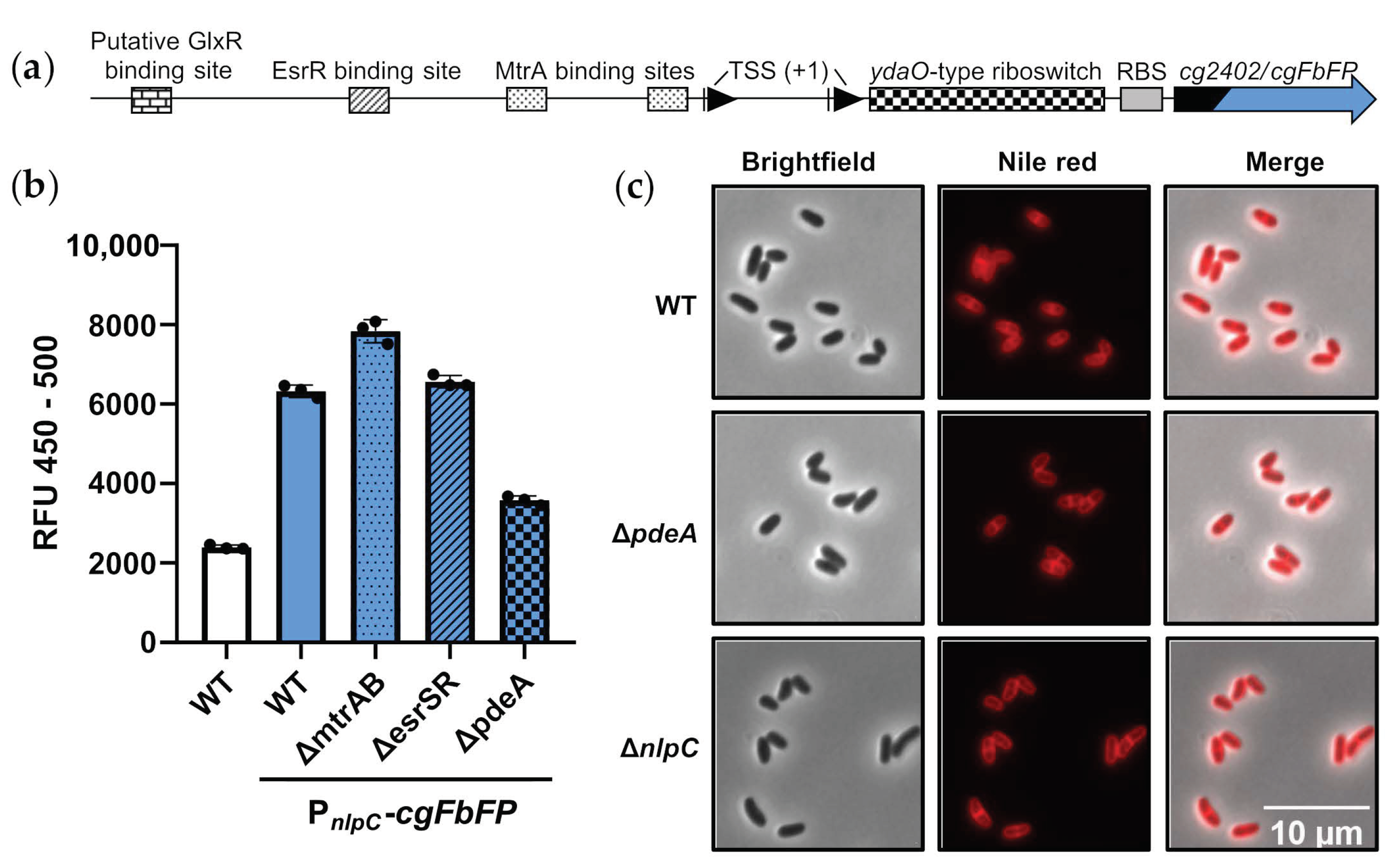
3.4. The Gene nlpC Is Strongly Regulated but Neglectable for Maintenance of Cell Morphology
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Gomelsky, M. cAMP, c-di-GMP, c-di-AMP and now cGMP: Bacteria use them all! Mol. Microbiol. 2011, 79, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Mehne, F.M.P.; Gunka, K.; Eilers, H.; Herzberg, C.; Kaever, V.; Stülke, J. Cyclic Di-AMP Homeostasis in Bacillus subtilis both lack and high level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem. 2013, 288, 2004–2017. [Google Scholar] [CrossRef]
- Whiteley, A.T.; Pollock, A.J.; Portnoy, D.A. The PAMP c-di-AMP Is Essential for Listeria monocytogenes Growth in Rich but Not Minimal Media due to a Toxic Increase in (p)ppGpp. Cell Host Microbe 2015, 17, 788–798. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, J.; Herzberg, C.; Kaever, V.; Gunka, K.; Hoffmann, T.; Weiß, M.; Gibhardt, J.; Thürmer, A.; Hertel, D.; Daniel, R.; et al. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci. Signal. 2017, 10, eaal3011. [Google Scholar] [CrossRef]
- Stülke, J.; Krüger, L. cyclic di-AMP signaling in bacteria. Annu. Rev. Microbiol. 2020, 74, 159–179. [Google Scholar] [CrossRef]
- Oppenheimer-Shaanan, Y.; Wexselblatt, E.; Katzhendler, J.; Yavin, E.; Ben-Yehuda, S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011, 12, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Campeotto, I.; Jeganathan, T.; Roelofs, K.G.; Lee, V.T.; Grundling, A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 9084–9089. [Google Scholar] [CrossRef]
- Gándara, C.; Alonso, J.C. DisA and c-di-AMP act at the intersection between DNA-damage response and stress homeostasis in exponentially growing Bacillus subtilis cells. DNA Repair (Amst). 2015, 27, 1–8. [Google Scholar] [CrossRef]
- Gibhardt, J.; Hoffmann, G.; Turdiev, A.; Wang, M.; Lee, V.T.; Commichau, F.M. c-di-AMP assists osmoadaptation by regulating the Listeria monocytogenes potassium transporters KimA and KtrCD. J. Biol. Chem. 2019, 294, 16020–16033. [Google Scholar] [CrossRef] [PubMed]
- Tosi, T.; Hoshiga, F.; Millership, C.; Singh, R.; Eldrid, C.; Patin, D.; Mengin-Lecreulx, D.; Thalassinos, K.; Freemont, P.; Gründling, A. Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM. PLoS Pathog. 2019, 15, e1007537. [Google Scholar] [CrossRef]
- Woodward, J.J.; Iavarone, A.T.; Portnoy, D.A. c-di-AMP Secreted by Intracellular Listeria monocytogenes Activates a Host Type I Interferon Response. Science 2010, 328, 1703–1705. [Google Scholar] [CrossRef]
- Barker, J.R.; Koestler, B.J.; Carpenter, V.K.; Burdette, D.L.; Waters, C.M.; Vance, R.E.; Valdivia, R.H. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. MBio 2013, 4, e00018-13. [Google Scholar] [CrossRef] [PubMed]
- McFarland, A.P.; Luo, S.; Ahmed-Qadri, F.; Zuck, M.; Thayer, E.F.; Goo, Y.A.; Hybiske, K.; Tong, L.; Woodward, J.J. Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-κB Activation and Shapes a Proinflammatory Antibacterial State. Immunity 2017, 46, 433–445. [Google Scholar] [CrossRef]
- Witte, G.; Hartung, S.; Büttner, K.; Hopfner, K.P. Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates. Mol. Cell 2008, 30, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Gándara, C.; de Lucena, D.K.C.; Torres, R.; Serrano, E.; Altenburger, S.; Graumann, P.L.; Alonso, J.C. Activity and in vivo dynamics of Bacillus subtilis DisA are affected by RadA/Sms and by Holliday junction-processing proteins. DNA Repair (Amst) 2017, 55, 17–30. [Google Scholar] [CrossRef]
- Commichau, F.M.; Heidemann, J.L.; Ficner, R.; Stülke, J. Making and breaking of an essential poison: The cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in bacteria. J. Bacteriol. 2018, 201, e00462-18. [Google Scholar] [CrossRef]
- Mehne, F.M.P.; Schröder-Tittmann, K.; Eijlander, R.T.; Herzberg, C.; Hewitt, L.; Kaever, V.; Lewis, R.J.; Kuipers, O.P.; Tittmann, K.; Stülke, J. Control of the Diadenylate Cyclase CdaS in Bacillus subtilis. J. Biol. Chem. 2014, 289, 21098–21107. [Google Scholar] [CrossRef]
- Rosenberg, J.; Dickmanns, A.; Neumann, P.; Gunka, K.; Arens, J.; Kaever, V.; Stülke, J.; Ficner, R.; Commichau, F.M. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J. Biol. Chem. 2015, 290, 6596–6606. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Gründling, A. Cyclic di-AMP: Another second messenger enters the fray. Nat. Rev. Microbiol. 2013, 11, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Rao, F.; See, R.Y.; Zhang, D.; Toh, D.C.; Ji, Q.; Liang, Z.X. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 2010, 285, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Bai, Y.; Zhang, Y.; Gabrielle, V.D.; Jin, L.; Bai, G. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol. Microbiol. 2014, 93, 65–79. [Google Scholar] [CrossRef]
- Ye, M.; Zhang, J.J.; Fang, X.; Lawlis, G.B.; Troxell, B.; Zhou, Y.; Gomelsky, M.; Lou, Y.; Yang, X.F. DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect. Immun. 2014, 82, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Drexler, D.J.; Müller, M.; Rojas-Cordova, C.A.; Bandera, A.M.; Witte, G. Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima. Structure 2017, 25, 1887–1897. [Google Scholar] [CrossRef] [PubMed]
- Latoscha, A.; Drexler, D.J.; Al-Bassam, M.M.; Bandera, A.M.; Kaever, V.; Findlay, K.C.; Witte, G.; Tschowri, N. c-di-AMP hydrolysis by the phosphodiesterase AtaC promotes differentiation of multicellular bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 7392–7400. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.T.; Nhiep, N.T.H.; Vu, T.N.M.; Huynh, T.N.; Zhu, Y.; Huynh, A.L.D.; Chakrabortti, A.; Marcellin, E.; Lo, R.; Howard, C.B.; et al. Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLoS Genet. 2018, 14, e1007574. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.W.; Sudarsan, N.; Furukawa, K.; Weinberg, Z.; Wang, J.X.; Breaker, R.R. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 2013, 9, 834–839. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Ma, H.; Yin, W.; Zhu, L.; Li, X.; Lim, H.M.; Chou, S.-H.; He, J. A c-di-AMP riboswitch controlling kdpFABC operon transcription regulates the potassium transporter system in Bacillus thuringiensis. Commun. Biol. 2019, 2, 182. [Google Scholar] [CrossRef]
- St-Onge, R.J.; Haiser, H.J.; Yousef, M.R.; Sherwood, E.; Tschowri, N.; Al-Bassam, M.; Elliot, M.A. Nucleotide Second Messenger-Mediated Regulation of a Muralytic Enzyme in Streptomyces. Mol. Microbiol. 2015, 96, 779–795. [Google Scholar] [CrossRef]
- Bai, Y.; Yang, J.; Zhou, X.; Ding, X.; Eisele, L.E.; Bai, G. Mycobacterium tuberculosis Rv3586 (DacA) Is a Diadenylate Cyclase That Converts ATP or ADP into c-di-AMP. PLoS ONE 2012, 7, e35206. [Google Scholar] [CrossRef]
- Manikandan, K.; Sabareesh, V.; Singh, N.; Saigal, K.; Mechold, U.; Sinha, K.M. Two-Step Synthesis and Hydrolysis of Cyclic di-AMP in Mycobacterium tuberculosis. PLoS ONE 2014, 9, e86096. [Google Scholar] [CrossRef]
- Zhang, L.; He, Z.G. Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein a, to negatively affect cyclic Di-AMP synthesis activity in Mycobacterium smegmatis. J. Biol. Chem. 2013, 288, 22426–22436. [Google Scholar] [CrossRef]
- Becker, J.; Wittmann, C. Advanced biotechnology: Metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew. Chemie Int. Ed. 2015, 54, 3328–3350. [Google Scholar] [CrossRef] [PubMed]
- Wendisch, V.F.; Jorge, J.M.P.; Pérez-García, F.; Sgobba, E. Updates on industrial production of amino acids using Corynebacterium glutamicum. World J. Microbiol. Biotechnol. 2016, 32, 105. [Google Scholar] [CrossRef] [PubMed]
- Schröder, J.; Tauch, A. Transcriptional regulation of gene expression in Corynebacterium glutamicum: The role of global, master and local regulators in the modular and hierarchical gene regulatory network. FEMS Microbiol. Rev. 2010, 34, 685–737. [Google Scholar] [CrossRef]
- Zahoor, A.; Lindner, S.N.; Wendisch, V.F. Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products. Comput. Struct. Biotechnol. J. 2012, 3, e201210004. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, K.; Inui, M. Regulons of global transcription factors in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2016, 100, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Blombach, B.; Gauttam, R.; Eikmanns, B.J. The RamA regulon: Complex regulatory interactions in relation to central metabolism in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2018, 102, 5901–5910. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, V.; Aravind, L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003, 4, R11. [Google Scholar] [CrossRef]
- Brocker, M.; Bott, M. Evidence for activator and repressor functions of the response regulator MtrA from Corynebacterium glutamicum. FEMS Microbiol. Lett. 2006, 264, 205–212. [Google Scholar] [CrossRef]
- Tsuge, Y.; Ogino, H.; Teramoto, H.; Inui, M.; Yukawa, H. Deletion of cgR_1596 and cgR_2070, encoding NlpC/P60 proteins, causes a defect in cell separation in Corynebacterium glutamicum R. J. Bacteriol. 2008, 190, 8204–8214. [Google Scholar] [CrossRef]
- Kohl, T.A.; Baumbach, J.; Jungwirth, B.; Pühler, A.; Tauch, A. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: In silico and in vitro detection of DNA binding sites of a global transcription regulator. J. Biotechnol. 2008, 135, 340–350. [Google Scholar] [CrossRef]
- Kleine, B.; Chattopadhyay, A.; Polen, T.; Pinto, D.; Mascher, T.; Bott, M.; Brocker, M.; Freudl, R. The three-component system EsrISR regulates a cell envelope stress response in Corynebacterium glutamicum. Mol. Microbiol. 2017, 106, 719–741. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Eikmanns, B.J.; Metzger, M.; Reinscheid, D.; Kircher, M.; Sahm, H. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl. Microbiol. Biotechnol. 1991, 34, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Möker, N.; Brocker, M.; Schaffer, S.; Krämer, R.; Morbach, S.; Bott, M. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 2004, 54, 420–438. [Google Scholar] [CrossRef]
- Kočan, M.; Schaffer, S.; Ishige, T.; Sorger-Herrmann, U.; Wendisch, V.F.; Bott, M. Two-Component Systems of Corynebacterium glutamicum: Deletion Analysis and Involvement of the PhoS-PhoR System in the Phosphate Starvation Response. J. Bacteriol. 2006, 188, 724–732. [Google Scholar] [CrossRef]
- Chang, A.C.; Cohen, S.N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978, 134, 1141–1156. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Cremer, J.; Eggeling, L.; Sahm, H. Cloning the dapA dapB cluster of the lysine-secreting bacterium Corynebacterium glutamicum. Mol. Gen. Genet. 1990, 220, 478–480. [Google Scholar] [CrossRef]
- Jiang, Y.; Qian, F.; Yang, J.; Liu, Y.; Dong, F.; Xu, C.; Sun, B.; Chen, B.; Xu, X.; Li, Y.; et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017, 8, 15179. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Ngouoto-Nkili, C.-E.; Burkovski, A. Construction and application of new Corynebacterium glutamicum vectors. Biotechnol. Tech. 1999, 13, 437–441. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Van der Rest, M.E.; Lange, C.; Molenaar, D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 1999, 52, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Tauch, A.; Kirchner, O.; Löffler, B.; Götker, S.; Pühler, A.; Kalinowski, J. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 2002, 45, 362–367. [Google Scholar] [CrossRef]
- Katayama, M.; Matsuda, Y.; Shimokawa, K.; Tanabe, S.; Kaneko, S.; Hara, I.; Sato, H. Simultaneous determination of six adenyl purines in human plasma by high-performance liquid chromatography with fluorescence derivatization. J. Chromatogr. B Biomed. Sci. Appl. 2001, 760, 159–163. [Google Scholar] [CrossRef]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef]
- Ponchel, F.; Toomes, C.; Bransfield, K.; Leong, F.T.; Douglas, S.H.; Field, S.L.; Bell, S.M.; Combaret, V.; Puisieux, A.; Mighell, A.J.; et al. Real-time PCR based on SYBR-Green I fluorescence: An alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003, 3, 18. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Drepper, T.; Eggert, T.; Circolone, F.; Heck, A.; Krauß, U.; Guterl, J.-K.; Wendorff, M.; Losi, A.; Gärtner, W.; Jaeger, K.-E. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 2007, 25, 443–445. [Google Scholar] [CrossRef] [PubMed]
- Diver, W.P.; Sargentini, N.J.; Smith, K.C. A Mutation (radA100) in Escherichia coli That Selectively Sensitizes Cells Grown in Rich Medium to X- or U.V.-radiation, or Methyl Methanesulphonate. Int. J. Radiat. Biol. Relat. Stud. Physics, Chem. Med. 1982, 42, 339–346. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Berg, D.E.; Stauffer, G.V. Mutational analysis of the Escherichia coli serB promoter region reveals transcriptional linkage to a downstream gene. Gene 1992, 120, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer-Sancar, K.; Mentz, A.; Rückert, C.; Kalinowski, J. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genomics 2013, 14, 888. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yang, J.; Eisele, L.E.; Underwood, A.J.; Koestler, B.J.; Waters, C.M.; Metzger, D.W.; Bai, G. Two DHH Subfamily 1 Proteins in Streptococcus pneumoniae Possess Cyclic Di-AMP Phosphodiesterase Activity and Affect Bacterial Growth and Virulence. J. Bacteriol. 2013, 195, 5123–5132. [Google Scholar] [CrossRef]
- Tang, Q.; Luo, Y.; Zheng, C.; Yin, K.; Ali, M.K.; Li, X.; He, J. Functional analysis of a c-di-AMP-specific phosphodiesterase MsPDE from Mycobacterium smegmatis. Int. J. Biol. Sci. 2015, 11, 813–824. [Google Scholar] [CrossRef]
- Bowman, L.; Zeden, M.S.; Schuster, C.F.; Kaever, V.; Gründling, A. New Insights into the Cyclic Di-adenosine Monophosphate (c-di-AMP) Degradation Pathway and the Requirement of the Cyclic Dinucleotide for Acid Stress Resistance in Staphylococcus aureus. J. Biol. Chem. 2016, 291, 26970–26986. [Google Scholar] [CrossRef]
- Jochmann, N.; Kurze, A.K.; Czaja, L.F.; Brinkrolf, K.; Brune, I.; Hüser, A.T.; Hansmeier, N.; Pühler, A.; Borovok, I.; Tauch, A. Genetic makeup of the Corynebacterium glutamicum LexA regulon deduced from comparative transcriptomics and in vitro DNA band shift assays. Microbiology 2009, 155, 1459–1477. [Google Scholar] [CrossRef]
- St-Onge, R.J.; Elliot, M.A. Regulation of a muralytic enzyme-encoding gene by two non-coding RNAs. RNA Biol. 2017, 14, 1592–1605. [Google Scholar] [CrossRef]
- Jones, C.P.; Ferré-D’Amaré, A.R. Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA. EMBO J. 2014, 33, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Patel, D.J. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry–related pockets. Nat. Chem. Biol. 2014, 10, 780–786. [Google Scholar] [CrossRef]
- Hett, E.C.; Chao, M.C.; Deng, L.L.; Rubin, E.J. A Mycobacterial Enzyme Essential for Cell Division Synergizes with Resuscitation-Promoting Factor. PLoS Pathog. 2008, 4, e1000001. [Google Scholar] [CrossRef]
- Zhou, X.; Rodriguez-Rivera, F.P.; Lim, H.C.; Bell, J.C.; Bernhardt, T.G.; Bertozzi, C.R.; Theriot, J.A. Sequential assembly of the septal cell envelope prior to V snapping in Corynebacterium glutamicum. Nat. Chem. Biol. 2019, 15, 221–231. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, T.H.; Kim, Y.; Lee, H.S. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J. Bacteriol. 2004, 186, 3453–3460. [Google Scholar] [CrossRef]
- Kohl, T.A.; Tauch, A. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: Detection of the corynebacterial core regulon and integration into the transcriptional regulatory network model. J. Biotechnol. 2009, 143, 239–246. [Google Scholar] [CrossRef]
- Brocker, M.; Mack, C.; Bott, M. Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. J. Bacteriol. 2011, 193, 1237–1249. [Google Scholar] [CrossRef]
- Weixler, D.; Goldbeck, O.; Seibold, G.M.; Eikmanns, B.J. Towards improved resistance of Corynebacterium glutamicum against nisin. bioRxiv 2021, 49, 0–1. [Google Scholar] [CrossRef]
- Goldbeck, O.; Seibold, G.M. Construction of pOGOduet—An inducible, bicistronic vector for synthesis of recombinant proteins in Corynebacterium glutamicum. Plasmid 2018, 95, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Reich, S.J. Cyclic Diadenosine Monophosphate in Corynebacterium glutamicum: Exploration of an Essential Second Messenger Molecule. PhD Thesis, Ulm University, Ulm, Germany, 2020. [Google Scholar] [CrossRef]


| Strain or Plasmid | Relevant Characteristics | Source or Reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F-thi-1 endA1 hsdR17 (r– m-) supE44 ΔlacU169 (φ80lacZΔM15) recA1 gyrA96 relA1 F- λ-ilvG rfb-50 rph-1 | [45] |
| Escherichia coli BL21 | ompT, hsdSB (rB-, mB-), dcm, gal (DE3) | [46] |
| Corynebacterium glutamicum ATCC 13032 | Wild type | American Type Culture Collection |
| C. glutamicum ΔpdeA | ATCC 13032 with in-frame deletion of the cyclic-diadenosine monophosphate (c-di-AMP) phosphodiesterase (PDE) gene pdeA (cg2174) | This work |
| C. glutamicum ΔnlpC | ATCC 13032 with in-frame deletion of the cell wall peptidase gene nlpC (cg2402) | This work |
| C. glutamicum ΔmtrAB | ATCC 13032 with in-frame deletion of the two-component system (TCS) mtrAB | [47] |
| C. glutamicum ΔesrSR | ATCC 13032 with in-frame deletion of the TCS esrSR (cgtSR7) | [48] |
| Plasmids | ||
| pACYC184 | Cmr, Tcr, Orip15A | [49] |
| pACYC_disA-strep | Tcr, replacement of Cmr region with araC-PBAD-disA-strep region from pBAD33_disA-strep | This work |
| pACYC_disA’-strep | As above, in-frame deletion of 414 bp in disA by PstI digestion and subsequent religation | This work |
| pBAD33 | Cmr, Orip15A, araC, PBAD | [50] |
| pBAD33_disA-strep | Expression plasmid carrying the cg2951/disA gene from C. glutamicum with a C-terminal strep tag | This work |
| pJC1 | Shuttle vector, Kmr, Orip15A, OripHM1519 | [51] |
| pJC1_Pcg2402-cgFbFP | Promoter reporter comprising 594 bp upstream of the cg2402/nlpC gene and including its first 30 bp fused to the reporter gene cgFbFP | This work |
| pJYS3 | cpf1, Kmr, OripBL1, OripSC101 | [52] |
| pJYS3_dpdeA | CRISPR/Cpf1-mediated deletion of of the c-di-AMP PDE gene pdeA (cg2174) | This work |
| pJYS3_dnlpC | CRISPR/Cpf1-mediated deletion of of the cell wall peptidase gene nlpC (cg2402) | This work |
| pXMJ19 | Shuttle expression vector, Ptac, lacIq, Cmr, OripMB1, OripBL1 | [53] |
| pXMJ19_mCherry | Expression plasmid carrying mCherry under control of the IPTG inducible Ptac promoter | This work |
| pXMJ19_RSnlpC-mCherry | Expression plasmid carrying mCherry under control of the IPTG inducible Ptac promoter and additional control of the riboswitch from the 5′ UTR of nlpC | This work |
| pXMJ19_RSnlpC-mCherry-mVenus | As above, with additional control of the putative riboswitch sequence upstream of cg2402 according to Nelson et al., 2013 | This work |
| pXMJ19_pdeA-strep | Cmr, IPTG inducible expression of the c-di-AMP PDE gene pdeA (cg2174) | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reich, S.J.; Goldbeck, O.; Lkhaasuren, T.; Weixler, D.; Weiß, T.; Eikmanns, B.J. C-di-AMP Is a Second Messenger in Corynebacterium glutamicum That Regulates Expression of a Cell Wall-Related Peptidase via a Riboswitch. Microorganisms 2023, 11, 296. https://doi.org/10.3390/microorganisms11020296
Reich SJ, Goldbeck O, Lkhaasuren T, Weixler D, Weiß T, Eikmanns BJ. C-di-AMP Is a Second Messenger in Corynebacterium glutamicum That Regulates Expression of a Cell Wall-Related Peptidase via a Riboswitch. Microorganisms. 2023; 11(2):296. https://doi.org/10.3390/microorganisms11020296
Chicago/Turabian StyleReich, Sebastian J., Oliver Goldbeck, Tsenguunmaa Lkhaasuren, Dominik Weixler, Tamara Weiß, and Bernhard J. Eikmanns. 2023. "C-di-AMP Is a Second Messenger in Corynebacterium glutamicum That Regulates Expression of a Cell Wall-Related Peptidase via a Riboswitch" Microorganisms 11, no. 2: 296. https://doi.org/10.3390/microorganisms11020296
APA StyleReich, S. J., Goldbeck, O., Lkhaasuren, T., Weixler, D., Weiß, T., & Eikmanns, B. J. (2023). C-di-AMP Is a Second Messenger in Corynebacterium glutamicum That Regulates Expression of a Cell Wall-Related Peptidase via a Riboswitch. Microorganisms, 11(2), 296. https://doi.org/10.3390/microorganisms11020296