Morphological and Phylogenetic Characterisation of Prorocentrum spinulentum, sp. nov. (Prorocentrales, Dinophyceae), a Small Spiny Species from the North Atlantic
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Cell Isolation, Cultivation
2.2. Microscopy
2.3. DNA Sequencing
2.4. Molecular Phylogeny
2.5. Terminology
3. Results
3.1. Formal Description
3.2. Detailed Description
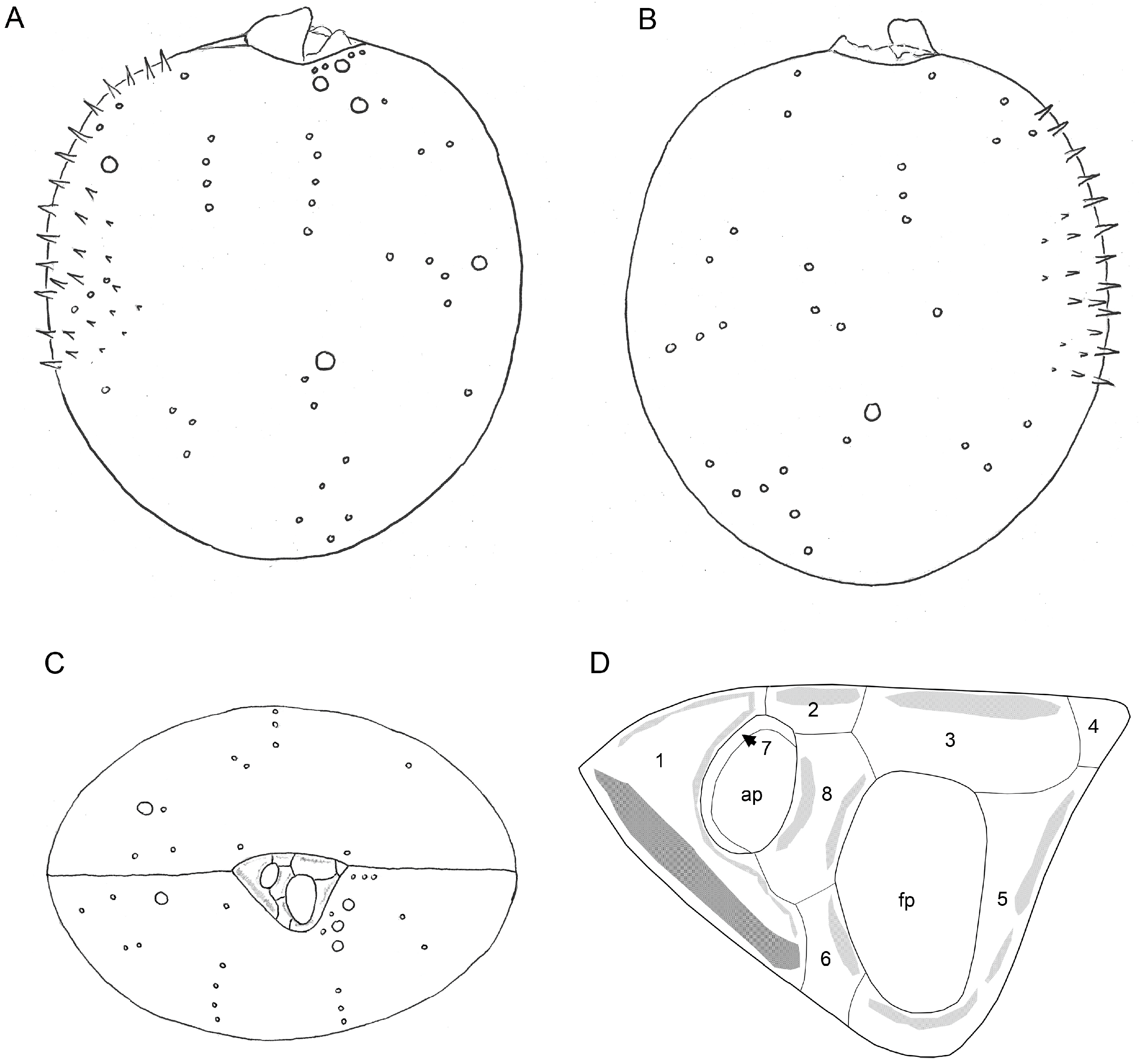
3.2.1. Light Microscopy
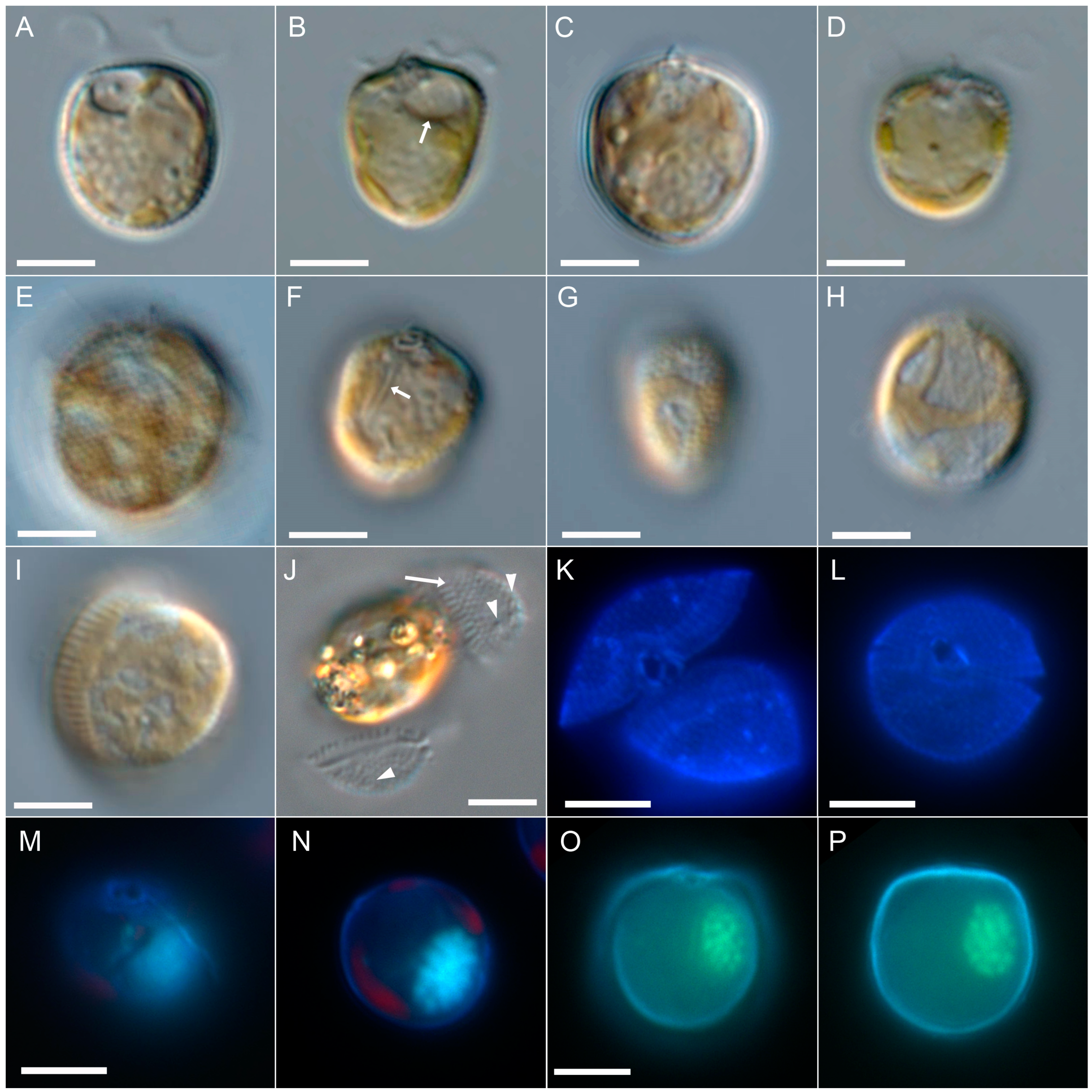
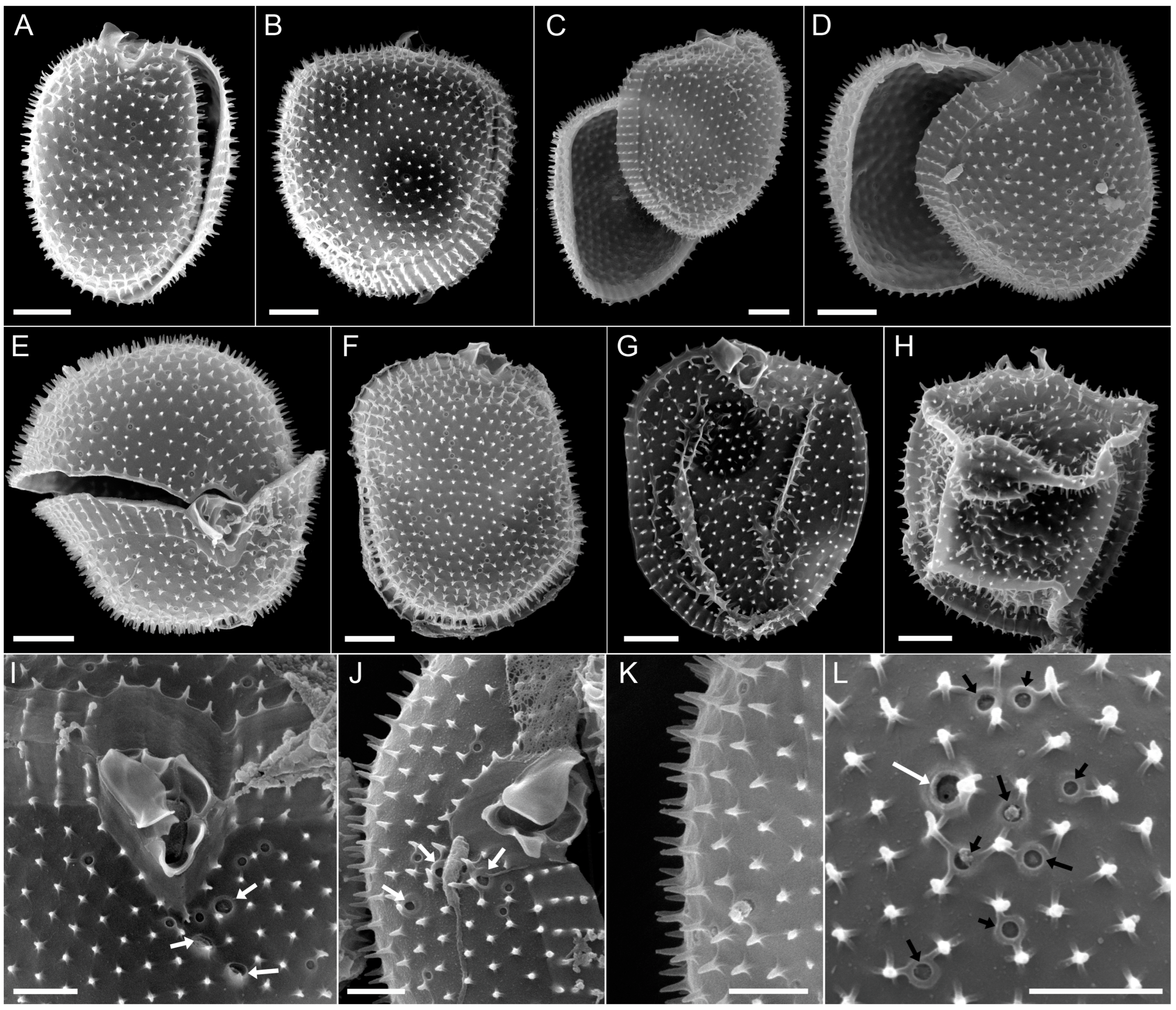
3.2.2. Scanning Electron Microscopy
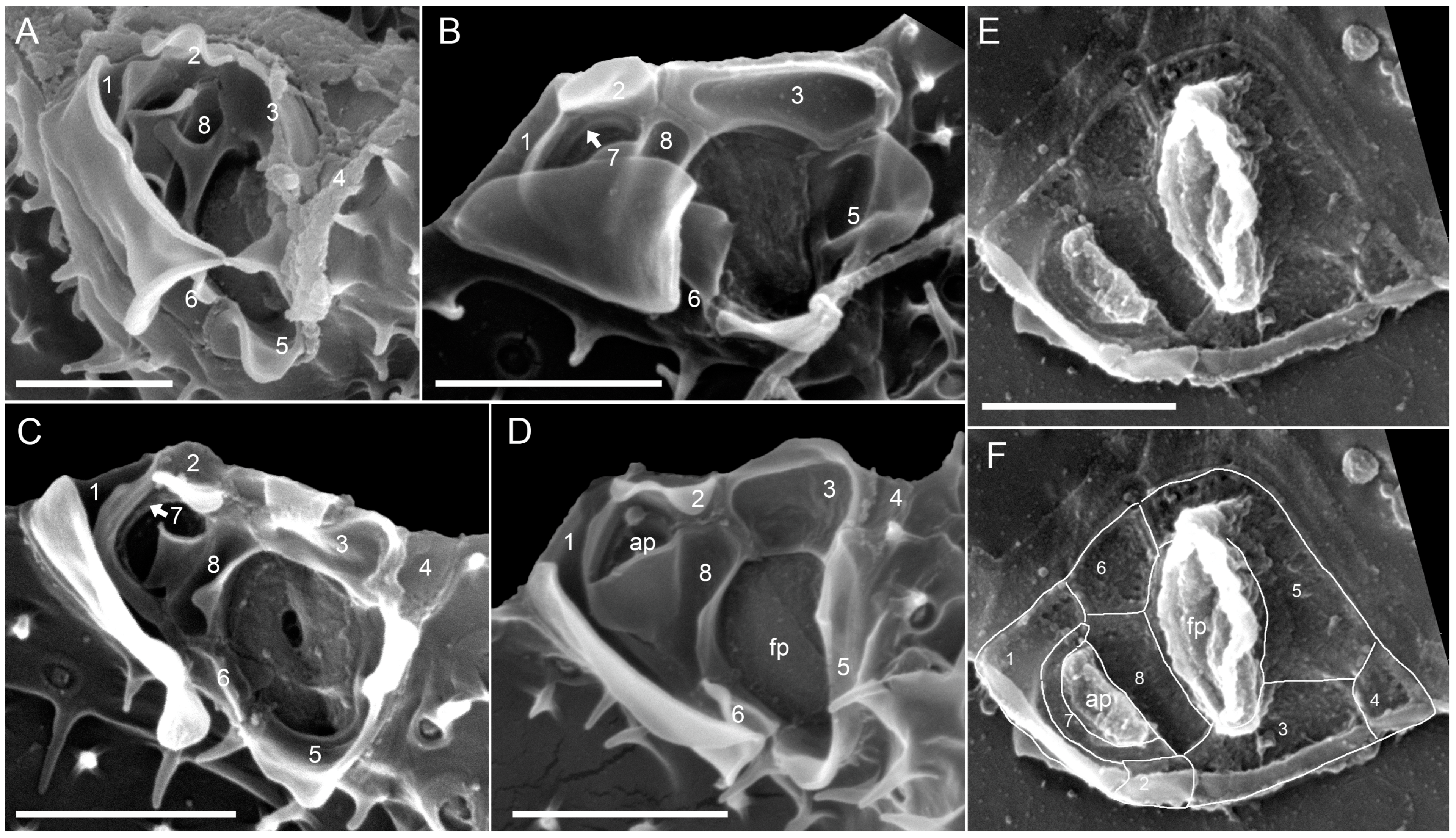
3.3. Molecular Phylogeny
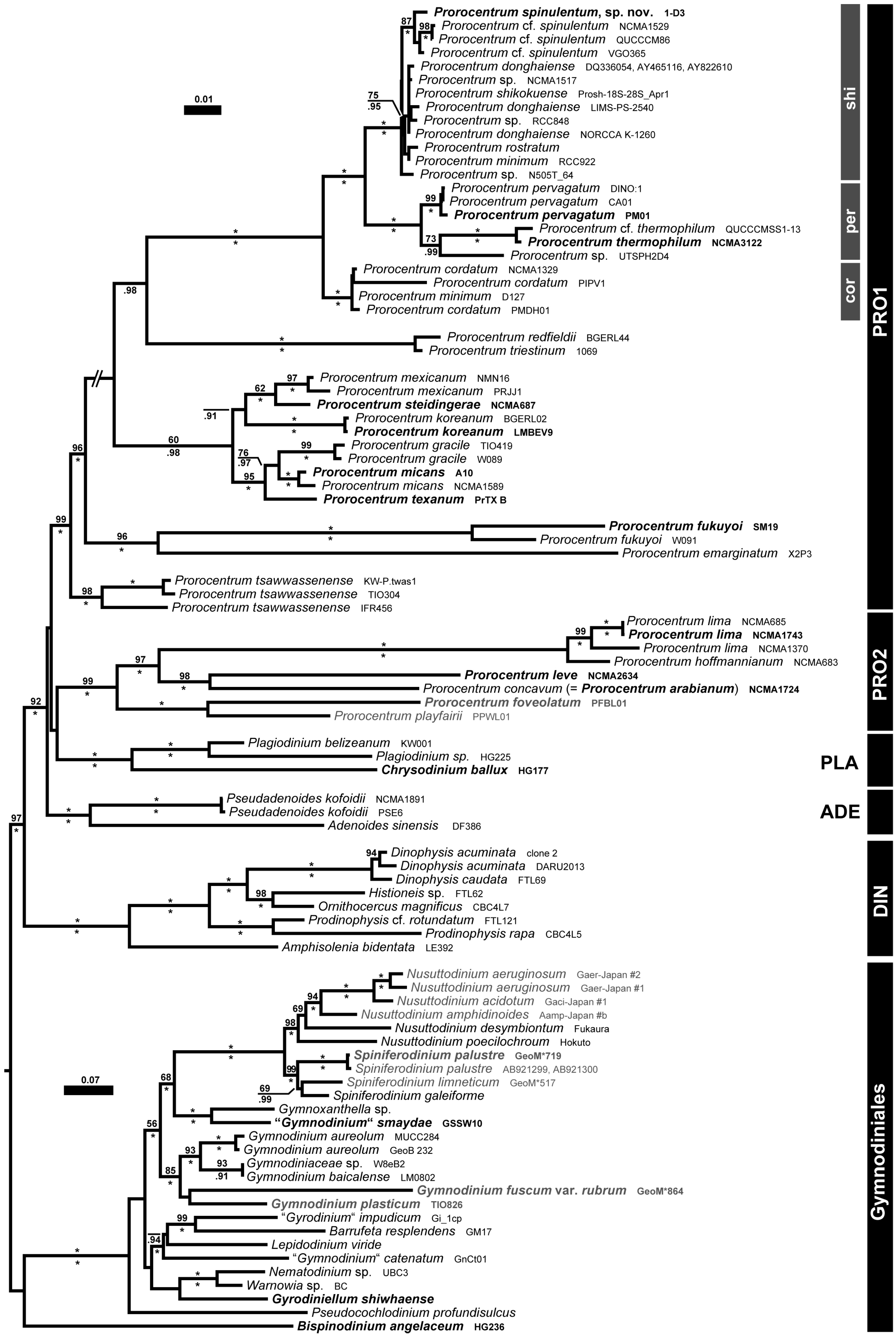
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dodge, J.D. The Prorocentrales (Dinophyceae). II Revision of the taxonomy within the genus Prorocentrum. Bot. J. Linn. Soc. 1975, 71, 103–125. [Google Scholar] [CrossRef]
- Hoppenrath, M.; Chomérat, N.; Horiguchi, T.; Schweikert, M.; Nagahama, Y.; Murray, S. Taxonomy and phylogeny of the benthic Prorocentrum species (Dinophyceae)—A proposal and review. Harmful Algae 2013, 27, 1–28. [Google Scholar] [CrossRef]
- Tillmann, U.; Wietkamp, S.; Gottschling, M.; Hoppenrath, M. Prorocentrum pervagatum sp. nov. (Prorocentrales, Dinophyceae): A new, small, planktonic species with a global distribution. Phycol. Res. 2023, 71, 56–71. [Google Scholar] [CrossRef]
- Shin, H.H.; Li, Z.; Mertens, K.N.; Seo, M.H.; Gu, H.; Lim, W.A.; Yoon, Y.H.; Soh, H.Y.; Matsuoka, K. Prorocentrum shikokuense Hada and P. donghaiense Lu are junior synonyms of P. obtusidens Schiller, but not of P. dentatum Stein (Prorocentrales, Dinophyceae). Harmful Algae 2019, 89, 101686. [Google Scholar] [CrossRef]
- Gómez, F.; Zhang, H.; Roselli, L.; Lin, S. Detection of Prorocentrum shikokuense in the Mediterranean Sea and evidence that P. dentatum, P. obtusidens and P. shikokuense are three different species (Prorocentrales, Dinophyceae). Acta Protozool. 2022, 60, 47–59. [Google Scholar] [CrossRef]
- Puigserver, M.; Zingone, A. Prorocentrum nux sp. nov. (Dinophyceae), a small planktonic dinoflagellate from the Mediterranean Sea, and discussion of P. nanum and P. pusillum. Phycologia 2002, 41, 29–38. [Google Scholar] [CrossRef]
- Krakhmalny, A.; Terenko, G.V. Prorocentrum ponticus Krachmalny & Terenko sp. nov., a new species of Dinophyta from the Black Sea. Int. J. Algae 2004, 6, 35–42. [Google Scholar]
- Larsson, M.E.; Bramucci, A.R.; Collins, S.; Hallegraeff, G.; Kahlke, T.; Raina, J.B.; Seymour, J.R.; Doblin, M.A. Mucospheres produced by a mixotrophic protist impact ocean carbon cycling. Nat. Commun. 2022, 13, 1301. [Google Scholar] [CrossRef] [PubMed]
- Gómez, F.; Gourvil, P.; Li, T.; Huang, Y.; Zhang, H.; Courcot, L.; Artigas, L.F.; Onis, E.S.; Gutiérrez-Rodriguez, A.; Lin, S. Molecular phylogeny of the spiny-surfaced species of the dinoflagellate Prorocentrum with the description of P. thermophilum sp. nov. and P. criophilum sp. nov (Prorocentrales, Dinophyceae). J. Phycol. in press. [CrossRef]
- Keller, M.D.; Selvin, R.C.; Claus, W.; Guillard, R.R.L. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 1987, 23, 633–638. [Google Scholar] [CrossRef]
- Fritz, L.; Triemer, R.E. A rapid simple technique utilizing Calcofluor white M2R for the visualization of dinoflagellate thecal plates. J. Phycol. 1985, 21, 662–664. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Adachi, M.; Sako, Y.; Ishida, Y. Analysis of Alexandrium (Dinophyceae) species using sequences of the 5.8S ribosomal DNA and internal transcribed spacer regions. J. Phycol. 1996, 32, 424–432. [Google Scholar] [CrossRef]
- Scholin, C.A.; Herzog, M.; Sogin, M.; Anderson, D.M. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. J. Phycol. 1994, 30, 999–1011. [Google Scholar] [CrossRef]
- Tillmann, U.; Wietkamp, S.; Krock, B.; Tillmann, A.; Voss, D.; Gu, H. Amphidomataceae (Dinophyceae) in the western Greenland area, including the description of Azadinium perforatum sp. nov. Phycologia 2020, 59, 63–88. [Google Scholar] [CrossRef]
- Tillmann, U.; Trefault, N.; Krock, B.; Parada-Pozo, G.; De la Iglesia, R.; Vásquez, M. Identification of Azadinium poporum (Dinophyceae) in the Southeast Pacific: Morphology, molecular phylogeny, and azaspiracid profile characterization. J. Plankton Res. 2017, 39, 350–367. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic logical alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tillmann, U.; Beran, A.; Gottschling, M.; Wietkamp, S.; Hoppenrath, M. Clarifying confusion–Prorocentrum triestinum J.Schiller and Prorocentrum redfieldii Bursa (Prorocentrales, Dinophyceae) are two different species. Eur. J. Phycol. 2022, 57, 207–226. [Google Scholar] [CrossRef]
- Tillmann, U.; Hoppenrath, M.; Gottschling, M. Reliable determination of Prorocentrum micans Ehrenb. (Prorocentrales, Dinophyceae) based on newly collected material from the type locality. Eur. J. Phycol. 2019, 54, 417–431. [Google Scholar] [CrossRef]
- Takano, Y.; Matsuoka, K. A comparative study between Prorocentrum shikokuense and P. donghaiense (Prorocentrales, Dinophyceae) based on morphology and DNA sequences. Plankton Benthos Res. 2011, 6, 179–186. [Google Scholar] [CrossRef]
- Percopo, I.; Siano, R.; Cerino, F.; Sarno, D.; Zingone, A. Phytoplankton diversity during the spring bloom in the northwestern Mediterranean Sea. Bot. Mar. 2011, 54, 243–276. [Google Scholar] [CrossRef]
- Ostenfeld, C.H. Phytoplankton fra det Kaspiske Hav. Vidensk. Medd. Dra Dan. Nat. Foren. 1902, 1901, 129–139. [Google Scholar]
- Pertola, S.; Faust, M.A.; Kuosa, H.; Hällfors, G. Morphology of Prorocentrum minimum (Dinophyceae) in the Baltic Sea and in Chesapeake Bay: Comparison of cell shapes and thecal ornamentation. Bot. Mar. 2003, 46, 477–486. [Google Scholar] [CrossRef]
- Monti, M.; Stoecker, D.K.; Cataletto, B.; Talarico, L. Morphology of the flagellar pore complex in Prorocentrum minimum (Dinophyceae) from the Adriatic and Baltic Seas. Bot. Mar. 2010, 53, 357–365. [Google Scholar] [CrossRef]
- Pei, L.; Hu, W.; Wang, P.; Kang, J.; Mohamed, H.F.; Wang, C.; Liu, L.; Luo, Z. Morphologic and phylogenic characterization of two bloom-forming planktonic Prorocentrum (Dinophyceae) species and their potential distribution in the China Sea. Algal Res. 2022, 66, 102788. [Google Scholar] [CrossRef]
- Lohmann, H. Untersuchungen zur Feststellung des vollständigen Gehaltes des Meeres an Plankton. Wiss. Meeresunters. 1908, 10, 129–370. [Google Scholar]
- Faust, M.A.; Larsen, J.; Moestrup, O. Leaflet no 184. Potentially toxic phytoplankton 3. Genus Prorocentrum (Dinophyceae). In ICES Identification Leaflets for Plankton; Lindley, J.A., Ed.; International Council for the Exploration of the Sea: Copenhagen, Denmark, 1999; Volume 184, pp. 1–24. [Google Scholar]
- Larsen, J.; Nguyen-Ngoc, L. Potentially toxic microalgae of Vietnamese waters. Opera Bot. 2004, 140, 5–216. [Google Scholar]
- Hoppenrath, M. A revised checklist of planktonic diatoms and dinoflagellates from Helgoland (North Sea, German Bight). Helgol. Mar. Res. 2004, 58, 243–251. [Google Scholar] [CrossRef]
- Schiller, J. Dinoflagellatae (Peridineae) in monographischer Behandlung. In Dr. L. Rabenhorst’s Kryptogamen-Flora von Deutschland, Österreich und der Schweiz; Kolkwitz, R., Ed.; Akademische Verlagsgesellschaft: Leipzig, Germany, 1933; Volume 10, pp. 1–615. [Google Scholar]
- Schiller, J. Über neue Prorocentrum- und Exuviella-Arten aus der Adria. Arch. Protistenkd. 1918, 38, 250–262. [Google Scholar]
- Schiller, J. Die planktische Vegetationen des adriatischen Meeres. C. Dinoflagellata I Teil. Adiniferidae, Dinophysidaceae. Arch. Protistenkd. 1928, 61, 46–91. [Google Scholar]
- Bursa, A.S. The genus Prorocentrum Ehrenberg. Morphodynamics, protoplasmic structure, and taxonomy. Can. J. Bot. 1959, 37, 1–31. [Google Scholar] [CrossRef]
- Dodge, J.D.; Bibby, B.T. The Prorocentrales (Dinophyceae) I. A comparative account of fine structure in the genera Prorocentrum and Exuviaella. Bot. J. Linn. Soc. 1973, 67, 175–187. [Google Scholar] [CrossRef]
- Berdieva, M.; Kalinina, V.; Lomert, E.; Knyazev, N.; Skarlato, S. Life cycle stages and evidence for sexual reproduction in the marine dinoflagellate Prorocentrum minimum (Dinophyceae, Prorocentrales). J. Phycol. 2020, 56, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Wulff, A. Über das Kleinplankton der Barentssee. Wissenschaftliche Meeresuntersuchungen, neue Folge, Abteilung Helgoland 1916, 13, 95–125. [Google Scholar]
- Honsell, G.; Talarico, L. The importance of flagellar arrangement and insertion in the interpretation of the theca of Prorocentrum (Dinophyceae). Bot. Mar. 1985, 28, 15–21. [Google Scholar] [CrossRef]
- Loeblich, A.R., III. Dinoflagellate evolution: Speculation and evidence. J. Protozool. 1976, 23, 13–28. [Google Scholar] [CrossRef]
- Taylor, F.J.R. On dinoflagellate evolution. BioSystems 1980, 13, 65–108. [Google Scholar] [CrossRef] [PubMed]
- Kretschmann, J.; Žerdoner Čalasan, A.; Kusber, W.-H.; Gottschling, M. Still curling after all these years: Glenodinium apiculatum Ehrenb. (Peridiniales, Dinophyceae) repeatedly found at its type locality in Berlin (Germany). Syst. Biodivers. 2018, 16, 200–209. [Google Scholar] [CrossRef]
- Tillmann, U.; Bantle, A.; Krock, B.; Elbrächter, M.; Gottschling, M. Recommendations for epitypification of dinophytes exemplified by Lingulodinium polyedra and molecular phylogenetics of the Gonyaulacales based on curated rRNA sequence data. Harmful Algae 2021, 104, 101956. [Google Scholar] [CrossRef]
| Spines | Pore Size | ||
|---|---|---|---|
| Length (µm) | Density (µm−2) | Large (µm) | Small (µm) |
| Mean ± SD Min–Max | mean ± SD Min–Max | Mean ± SD Min–Max | Mean ± SD Min–Max |
| 0.36 ± 0.03 0.30–0.43 n = 18 | 4.6 ± 0.2 4.2–5.0 n = 12 | 0.23 ± 0.02 0.20–0.28 n = 13 | 0.14 ± 0.01 0.11–0.16 n = 25 |
| Periflagellar Area | Size Accessory Pore | Size Flagellar Pore | Size Wing on Platelet 1 | ||||
|---|---|---|---|---|---|---|---|
| Depth (µm) | Width (µm) | Length (µm) | Width (µm) | Length (µm) | Width (µm) | Length | Height |
| Mean ± SD Min–Max | Mean ± SD Min–Max | Mean ± SD Min–Max | Mean ± SD Min–Max | Large | Small | Mean ± SD Min–Max | Mean ± SD Min–Max |
| 2.15 ± 0.17 1.78–2.37 n = 15 | 1.48 ± 0.20 1.14–1.83 n = 15 | 0.59 ± 0.03 0.54–0.63 n = 10 | 0.38 ± 0.03 0.33–0.44 n = 10 | 0.94 ± 0.11 0.74–1.12 n = 13 | 0.60 ± 0.04 0.50–0.67 n = 12 | 1.24 ± 0.13 0.93–1.49 n = 25 | 1.09 ± 0.17 0.83–1.44 n = 20 |
| trait | P. spinulentum | P. pervagatum | P. thermophilum | P. cordatum 1 | P. balticum | P. sphaeroideum2 | P. nux | P. nanum | P. ovum | P. pusillum3 | P. ponticum | P. pomoideum4 | P. cordiforme5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| shape in outline | round | round | almost round | obcordate emarginate heart-shaped | round, ovate | round | irregularily ovate | irregular, almost angular | ovate, anterior end square 6 | ovate | orbicular to broadly elliptic | apple-shaped (often with posterior constriction) | heart-shaped |
| length [µm] | 9.0–12.8 | 12–16 | 12–17 | 22–24 18 | 9–12 | 13 | 6.3–9 | 10–14 | 14 | 8–10 | 10.4–15.1 | 9–20 | 6–12 |
| depth [µm] | 8.5–11.9 | 12–16 | 10–15 | 18–20 15 | 9–12 | 13 | 5.3–10 | 10–14 | 10 | 6.5–7.5 | 9.6-11.5 | 11–16 | 5–9 |
| compression | slightly compressed | young cells slightly compressed; round | no | compressed strongly compressed | no | not reported | no | strongly compressed | scarcely compressed | strongly compressed | slightly or strongly compressed | flattened | not reported |
| nucleus shape; position | ellipsoid; median, ventral | spherical to ellipsoid; posterior | more or less rotund; posterior | not reported not reported 7 | not reported 8 | not reported | round; posterior | not reported | not reported | not reported | shape not reported; posterior position | subspherical; posterior | not reported |
| apical spine | absent | 1.3–1.7 µm, on platelet 6 | absent | absent small tooth with minute wing | absent | a solid spine without wing | absent | short spine 9 | absent | absent | absent | one main apical tooth without a wing | short simple spine 10 |
| projections | high and wide wing on platelet 1 | flat wings | spine-like prolongation | absent absent | not reported | not reported | absent | two irregular bumps bordering the flagellar pore | absent | absent | small projections | two more and smaller “spines” | additional smaller spine opposite to the main spine |
| surface | spines | knobs | knobs | with fine pores 11 densly perforated with subtle pores 12 | not reported | not reported | smooth | not reported | not reported | with subtle pores 13 | knobs | spines or smooth 14 | smooth |
| spine/knob density [µm−2] | 4.2–5.0 | 7.0-10.5 | 5.0–7.0 | not reported not reported | not reported | not reported | not applicable | not reported | not reported | not reported | 9 15 | not reported | not applicable |
| periflagellar plateletts | 8 | 8 | not determined | not determined not determined 17 | not determined | not determined | 7 | not determined | not determined | not determined | not determined | not determined | not determined |
| pores | present | present | present | not reported not reported | not reported | present | present | present | present | present | one line of fine pores positioned in periphery of each thecal plate | numerous small pores | absent 16 |
| pore size | large (0.23 µm), small (0.14 µm) | large (0.3 µm), small (0.16 µm) | large (0.3 µm), small (0.15 µm) | not reported not reported | not reported | large | large, small | large | large | subtle | fine pores | small | absent |
| pore number | ca. 30 per plate | ca. 20–30 per plate | ca. 10–15 per plate | not reported not reported | not reported | few | few | scarce | few | not specified or drawn | not specified | numerous | absent |
| pore position | scattered over plates | scattered on plate margins | scattered on plate margins | not reported not reported | not reported | all over theca | adjacent to plate margins | scattered all over theca | scatted all over theca | not specified | arranged in one line in periphery of each thecal plate | scattered all over theca | absent |
| type locality | Celtic Sea | Labrador Sea | Gulf of Mexixo, Sarasota, Florida | Caspian Sea Golfe de Lion, France | Baltic Sea (Kiel, Germany) | Adriatic Sea | Tyrrhenian Sea, off Naples | Adriatic Sea | Adriatic Sea | Adriatic Sea | Black Sea | English Channel (Tamar estuary) | English Channel (Plymouth) |
| distribution | Celtic Sea | Antarctica, Labrador Sea, North Sea | Gulf of Mexico, New Zealand, Arabian Gulf | Caspian Sea Mediteranean | Baltic Sea (Kiel) | Mediterranen | Tyrrhenia Sea, English chanel | Adriatic Sea, North Sea, Atlantic | Adriatic Sea | Adriatic Sea | Black Sea | English Channel (Tamar estuary) | English Channel (Plymouth) |
| references | this study | Tillmann et al. 2022 [3] | Gomez et al. 2022 [9] | Ostenfeld 1902 [23] Schiller 1933 [31] | Lohman 1908 [27] | Schiller 1918 [32] | Puigserver & Zingone 2002 [6] | Schiller 1918 [32] | Schiller 1918 [32] | Schiller 1928 [33] | Krakhmalny & Terenko 2004 [7] | Bursa 1959 [34] | Bursa 1959 [34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tillmann, U.; Gottschling, M.; Wietkamp, S.; Hoppenrath, M. Morphological and Phylogenetic Characterisation of Prorocentrum spinulentum, sp. nov. (Prorocentrales, Dinophyceae), a Small Spiny Species from the North Atlantic. Microorganisms 2023, 11, 271. https://doi.org/10.3390/microorganisms11020271
Tillmann U, Gottschling M, Wietkamp S, Hoppenrath M. Morphological and Phylogenetic Characterisation of Prorocentrum spinulentum, sp. nov. (Prorocentrales, Dinophyceae), a Small Spiny Species from the North Atlantic. Microorganisms. 2023; 11(2):271. https://doi.org/10.3390/microorganisms11020271
Chicago/Turabian StyleTillmann, Urban, Marc Gottschling, Stephan Wietkamp, and Mona Hoppenrath. 2023. "Morphological and Phylogenetic Characterisation of Prorocentrum spinulentum, sp. nov. (Prorocentrales, Dinophyceae), a Small Spiny Species from the North Atlantic" Microorganisms 11, no. 2: 271. https://doi.org/10.3390/microorganisms11020271
APA StyleTillmann, U., Gottschling, M., Wietkamp, S., & Hoppenrath, M. (2023). Morphological and Phylogenetic Characterisation of Prorocentrum spinulentum, sp. nov. (Prorocentrales, Dinophyceae), a Small Spiny Species from the North Atlantic. Microorganisms, 11(2), 271. https://doi.org/10.3390/microorganisms11020271







