Characterisation of Soil Bacterial Communities That Exhibit Chemotaxis to Root Exudates from Phosphorus-Limited Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Collection of Root Exudates from P-Sufficient Control and P-Deficient Plants
2.3. Capture of Soil Microbes That Exhibit Chemotaxis to Root Exudates
2.4. Preparing the Source Community
2.5. Preparing the Control and Exudate-Baited Trap Solutions
2.6. Setting the Traps
2.7. Enumeration of Chemotactic Microbial Cells by Flow Cytometry
2.8. Identifying the Composition and Relative Abundance of Microbial Populations
2.8.1. DNA Extraction, PCR, and Sequencing
2.8.2. Bioinformatic Processing
2.8.3. Statistical Analyses
3. Results
3.1. Absolute Abundances of Captured Cells
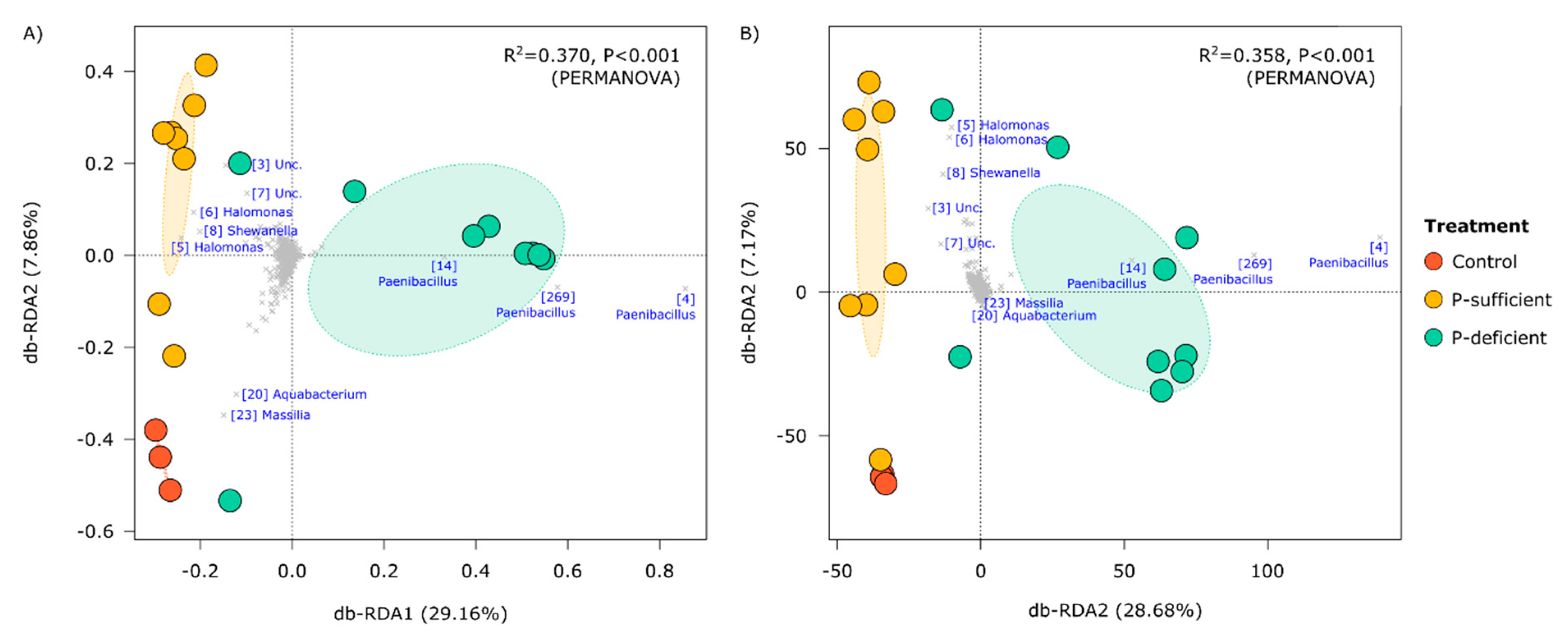
3.2. Identities of Captured Cells
3.3. Alpha Diversity of Captured Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef] [PubMed]
- Neset, T.S.; Bader, H.P.; Scheidegger, R.; Lohm, U. The flow of phosphorus in food production and consumption—Linköping, Sweden, 1870–2000. Sci. Total Environ. 2008, 396, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.A.; Bending, G.D.; White, P.J. Biological costs and benefits to plant–microbe interactions in the rhizosphere. J. Exp. Bot. 2005, 56, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Péret, B.; Clément, M.; Nussaume, L.; Desnos, T. Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci. 2011, 16, 442–450. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Brito, L.F.; López, M.G.; Straube, L.; Passaglia, L.M.; Wendisch, V.F. Inorganic phosphate solubilization by rhizosphere bacterium Paenibacillus sonchi: Gene expression and physiological functions. Front. Microbiol. 2020, 11, 588605. [Google Scholar] [CrossRef]
- Kawasaki, A.; Dennis, P.G.; Forstner, C.; Raghavendra, A.K.H.; Mathesius, U.; Richardson, A.E.; Delhaize, E.; Gilliham, M.; Watt, M.; Ryan, P.R. Manipulating exudate composition from root apices shapes the microbiome throughout the root system. J. Plant Physiol. 2021, 187, 2279–2295. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Carvalhais, L.C.; Dennis, P.G.; Badri, D.V.; Kidd, B.N.; Vivanco, J.M.; Schenk, P.M. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol. Plant-Microbe Interact. 2015, 28, 1049–1058. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Carvalhais, L.C.; Percy, C.D.; Prakash Verma, J.; Schenk, P.M.; Singh, B.K. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytol. 2021, 229, 2873–2885. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, S.; Zhalnina, K.; Kosina, S.; Northen, T.R.; Sasse, J. The core metabolome and root exudation dynamics of three phylogenetically distinct plant species. Nat. Commun. 2023, 14, 1649. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.; de Vries, F.T. Plant root exudation under drought: Implications for ecosystem functioning. New Phytol. 2020, 225, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Carvalhais, L.C.; Dennis, P.G.; Fan, B.; Fedoseyenko, D.; Kierul, K.; Becker, A.; von Wiren, N.; Borriss, R. Linking plant nutritional status to plant-microbe interactions. PLoS ONE 2013, 8, e68555. [Google Scholar] [CrossRef] [PubMed]
- Shakir, M.A.; Bano, A.; Arshad, M. Short Communication Rhizosphere bacteria containing ACC-deaminase conferred drought tolerance in wheat grown under semi-arid climate. J. Soil Sci. Environ. 2012, 31, 108–112. [Google Scholar]
- Belimov, A.A.; Dodd, I.C.; Hontzeas, N.; Theobald, J.C.; Safronova, V.I.; Davies, W.J. Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol. 2009, 181, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Marasco, R.; Rolli, E.; Ettoumi, B.; Vigani, G.; Mapelli, F.; Borin, S.; Abou-Hadid, A.F.; El-Behairy, U.A.; Sorlini, C.; Cherif, A.; et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS ONE 2012, 7, e48479. [Google Scholar] [CrossRef]
- Lambers, H.; Mougel, C.; Jaillard, B.; Hinsinger, P. Plant-microbe-soil interactions in the rhizosphere: An evolutionary perspective. Plant Soil. 2009, 321, 83–115. [Google Scholar] [CrossRef]
- Dodd, I.C.; Ruiz-Lozano, J.M. Microbial enhancement of crop resource use efficiency. COBIOT 2012, 23, 236–242. [Google Scholar] [CrossRef]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Law, A.M.; Aitken, M.D. Continuous-flow capillary assay for measuring bacterial chemotaxis. Appl. Environ. Microbiol. 2005, 71, 3137–3143. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, D.; Liu, Y.; Li, S.; Shen, Q.; Zhang, R. Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil. 2014, 374, 689–700. [Google Scholar] [CrossRef]
- Dennis, P.G.; Seymour, J.; Kumbun, K.; Tyson, G.W. Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J. 2013, 7, 1661–1664. [Google Scholar] [CrossRef] [PubMed]
- Weigh, K.V.; Batista, B.D.; Dennis, P.G. A Bait-Trap Assay to Characterize Soil Microbes that Exhibit Chemotaxis to Root Exudates. In The Plant Microbiome: Methods and Protocols; Carvalhais, L.C., Dennis, P.G., Eds.; Springer: Berlin/Heidelberg, Germany, 2021; pp. 283–289. [Google Scholar]
- Lindsey, B.E., III; Rivero, L.; Calhoun, C.S.; Grotewold, E.; Brkljacic, J. Standardized method for high-throughput sterilization of Arabidopsis seeds. J. Vis. Exp. 2017, 128, 56587. [Google Scholar]
- Carvalhais, L.C.; Dennis, P.G.; Fedoseyenko, D.; Hajirezaei, M.R.; Borriss, R.; von Wirén, N. Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. JPNSS 2011, 174, 3–11. [Google Scholar] [CrossRef]
- Forstner, C.; Orton, T.G.; Skarshewski, A.; Wang, P.; Kopittke, P.M.; Dennis, P.G. Effects of graphene oxide and graphite on soil bacterial and fungal diversity. Sci. Total Environ. 2019, 671, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Isbell, R. The Australian Soil Classification; CSIRO Publishing: Clayton, Australia, 2016. [Google Scholar]
- Soil Survey Staff. Keys to Soil Taxonomy, 12th ed.; USDA-Natural Resources Conservation Service: Washington, DC, USA, 2014. [Google Scholar]
- Engelbrektson, A.; Kunin, V.; Wrighton, K.C.; Zvenigorodsky, N.; Chen, F.; Ochman, H.; Hugenholtz, P. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010, 4, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2021. [Google Scholar]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Li, M.; Wang, Y.; Xu, Z.; Li, L. Root exudates and chemotactic strains mediate bacterial community assembly in the rhizosphere soil of Casuarina equisetifolia L. Front. Plant Sci. 2022, 13, 988442. [Google Scholar] [CrossRef] [PubMed]
- Vora, S.M.; Joshi, P.; Belwalkar, M.; Archana, G. Root exudates influence chemotaxis and colonization of diverse plant growth promoting rhizobacteria in the pigeon pea–maize intercropping system. Rhizosphere 2021, 18, 100331. [Google Scholar] [CrossRef]
- Jiao, H.; Lyu, C.; Xu, W.; Chen, W.; Hu, Y.; Wang, Z. An improved chemotaxis assay for the rapid identification of rhizobacterial chemoattractants in root exudates. J. Vis. Exp. 2022, 181, e63249. [Google Scholar]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Baligar, V.C.; Fageria, N.K. Nutrient Use Efficiency in Plants: An Overview. In Nutrient Use Efficiency: From Basics to Advances; Rakshit, A., Singh, H.B., Sen, A., Eds.; Springer: New Delhi, India, 2015; pp. 1–14. [Google Scholar]
- Mohd Din, A.R.; Rosli, M.A.; Mohamad Azam, Z.; Othman, N.Z.; Sarmidi, M.R. Paenibacillus polymyxa role involved in phosphate solubilization and growth promotion of Zea mays under abiotic stress condition. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2020, 90, 63–71. [Google Scholar] [CrossRef]
- Haldar, S.; Sengupta, S. Plant-microbe cross-talk in the rhizosphere: Insight and biotechnological potential. Open Microbiol. J. 2015, 9, 1–7. [Google Scholar] [CrossRef]
- Hartmann, A.; Schmid, M.; Tuinen, D.V.; Berg, G. Plant-driven selection of microbes. Plant Soil. 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Li, M.; Song, Z.; Li, Z.; Qiao, R.; Zhang, P.; Ding, C.; Xie, J.; Chen, Y.; Guo, H. Populus root exudates are associated with rhizosphere microbial communities and symbiotic patterns. Front. Microbiol. 2022, 13, 1042944. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef]
- Khan, M.S.; Gao, J.; Chen, X.; Zhang, M.; Yang, F.; Du, Y.; Moe, T.S.; Munir, I.; Xue, J.; Zhang, X. Isolation and characterization of plant growth-promoting endophytic bacteria Paenibacillus polymyxa SK1 from Lilium lancifolium. Biomed Res. Int. 2020, 2020, 8650957. [Google Scholar] [CrossRef]
- Cherchali, A.; Boukhelata, N.; Kaci, Y.; Abrous-Belbachir, O.; Djebbar, R. Isolation and identification of a phosphate-solubilizing Paenibacillus polymyxa strain GOL 0202 from durum wheat (Triticum durum Desf.) rhizosphere and its effect on some seedlings morphophysiological parameters. Biocatal. Agric. Biotechnol. 2019, 19, 101087. [Google Scholar] [CrossRef]
- Das, S.N.; Dutta, S.; Kondreddy, A.; Chilukoti, N.; Pullabhotla, S.V.; Vadlamudi, S.; Podil, A.R. Plant growth-promoting chitinolytic Paenibacillus elgii responds positively to tobacco root exudates. J. Plant Growth Regul. 2010, 29, 409–418. [Google Scholar] [CrossRef]
- Xu, S.; Kim, B.S. Evaluation of Paenibacillus polymyxa strain SC09-21 for biocontrol of Phytophthora blight and growth stimulation in pepper plants. Trop. Plant Pathol. 2016, 41, 162–168. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, Y.; Li, B.; Shan, C.; Ibrahim, M.; Jabeen, A.; Xie, G.; Sun, G. Phosphate solubilization of Paenibacillus polymyxa and Paenibacillus macerans from mycorrhizal and non-mycorrhizal cucumber plants. Afr. J. Microbiol. Res. 2012, 6, 4567–4573. [Google Scholar]
- Xie, J.; Shi, H.; Du, Z.; Wang, T.; Liu, X.; Chen, S. Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 2016, 6, 21329. [Google Scholar] [CrossRef]
- Sukweenadhi, J.; Kim, Y.J.; Choi, E.S.; Koh, S.C.; Lee, S.W.; Kim, Y.J.; Yang, D.C. Paenibacillus yonginensis DCY84T induces changes in Arabidopsis thaliana gene expression against aluminum, drought, and salt stress. Microbiol. Res. 2015, 172, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Ravnskov, S.; Xie, G.; Larsen, J. Biocontrol of Pythium damping-off in cucumber by arbuscular mycorrhiza-associated bacteria from the genus Paenibacillus. Biocontrol 2007, 52, 863–875. [Google Scholar] [CrossRef]
- Pantigoso, H.A.; Yuan, J.; He, Y.; Guo, Q.; Vollmer, C.; Vivanco, J.M. Role of root exudates on assimilation of phosphorus in young and old Arabidopsis thaliana plants. PLoS ONE 2020, 15, e0234216. [Google Scholar] [CrossRef]
- Ling, N.; Raza, W.; Ma, J.; Huang, Q.; Shen, Q. Identification and role of organic acids in watermelon root exudates for recruiting Paenibacillus polymyxa SQR-21 in the rhizosphere. Eur. J. Soil Biol. 2011, 47, 374–379. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weigh, K.V.; Batista, B.D.; Hoang, H.; Dennis, P.G. Characterisation of Soil Bacterial Communities That Exhibit Chemotaxis to Root Exudates from Phosphorus-Limited Plants. Microorganisms 2023, 11, 2984. https://doi.org/10.3390/microorganisms11122984
Weigh KV, Batista BD, Hoang H, Dennis PG. Characterisation of Soil Bacterial Communities That Exhibit Chemotaxis to Root Exudates from Phosphorus-Limited Plants. Microorganisms. 2023; 11(12):2984. https://doi.org/10.3390/microorganisms11122984
Chicago/Turabian StyleWeigh, Katherine V., Bruna D. Batista, Huong Hoang, and Paul G. Dennis. 2023. "Characterisation of Soil Bacterial Communities That Exhibit Chemotaxis to Root Exudates from Phosphorus-Limited Plants" Microorganisms 11, no. 12: 2984. https://doi.org/10.3390/microorganisms11122984
APA StyleWeigh, K. V., Batista, B. D., Hoang, H., & Dennis, P. G. (2023). Characterisation of Soil Bacterial Communities That Exhibit Chemotaxis to Root Exudates from Phosphorus-Limited Plants. Microorganisms, 11(12), 2984. https://doi.org/10.3390/microorganisms11122984





