Exploring and Engineering Novel Strong Promoters for High-Level Protein Expression in Bacillus subtilis DB104 through Transcriptome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions, and Transformation
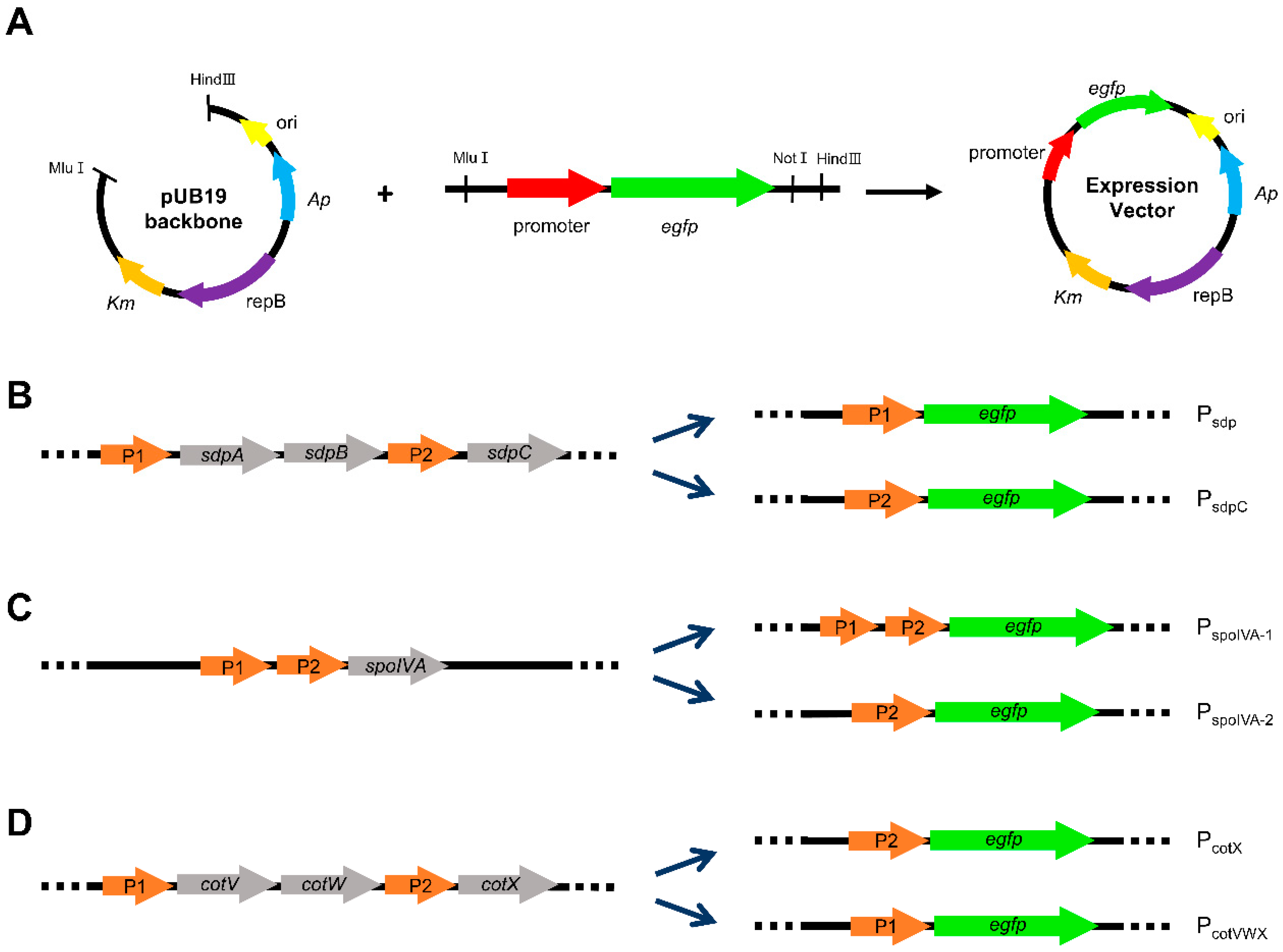
2.2. Analysis of Transcriptome Data and Construction of Expression Vectors
2.3. Engineering of Expression Cassettes
2.4. Measurement of Fluorescence Intensity of Enhanced Green Fluorescent Protein (eGFP)
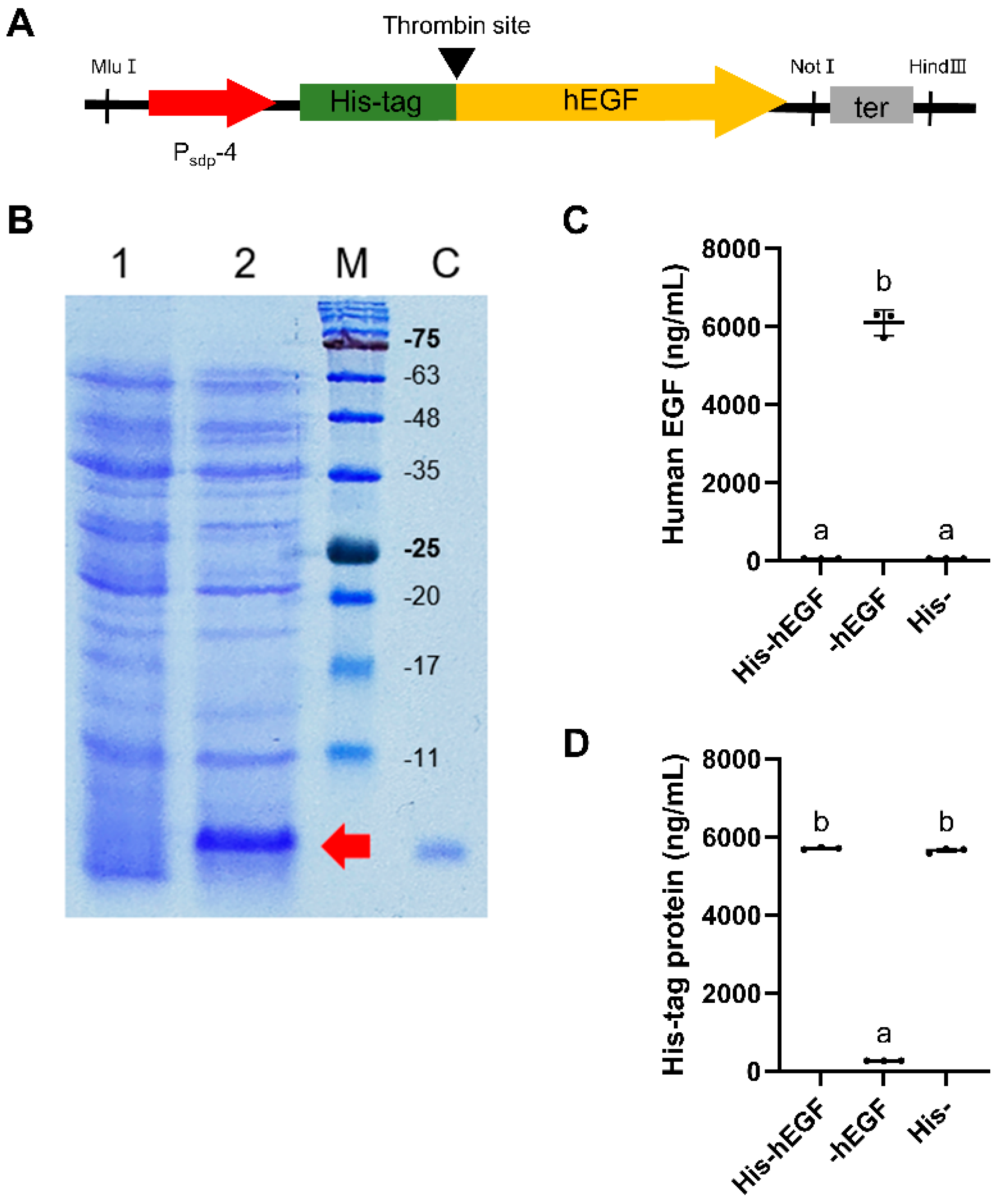
2.5. Human Epidermal Growth Factor (hEGF) Expression Using Developed Expression Cassette
2.6. Purification of His-Tagged Human Epidermal Growth Factor (hEGF) and Thrombin Treatment
2.7. Enzyme-Linked Immunosorbent Assay (ELISA)
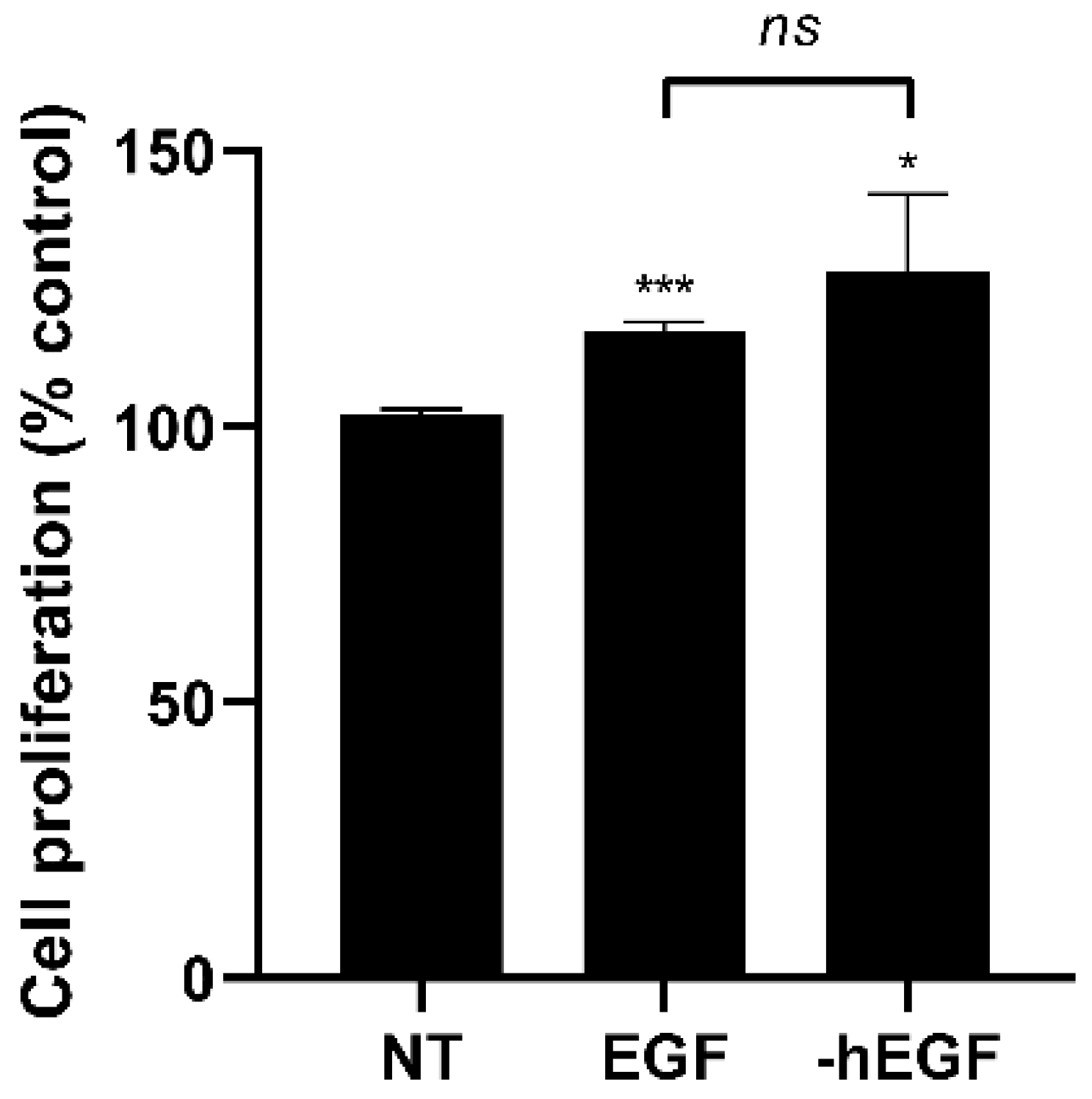
2.8. Cell Proliferation
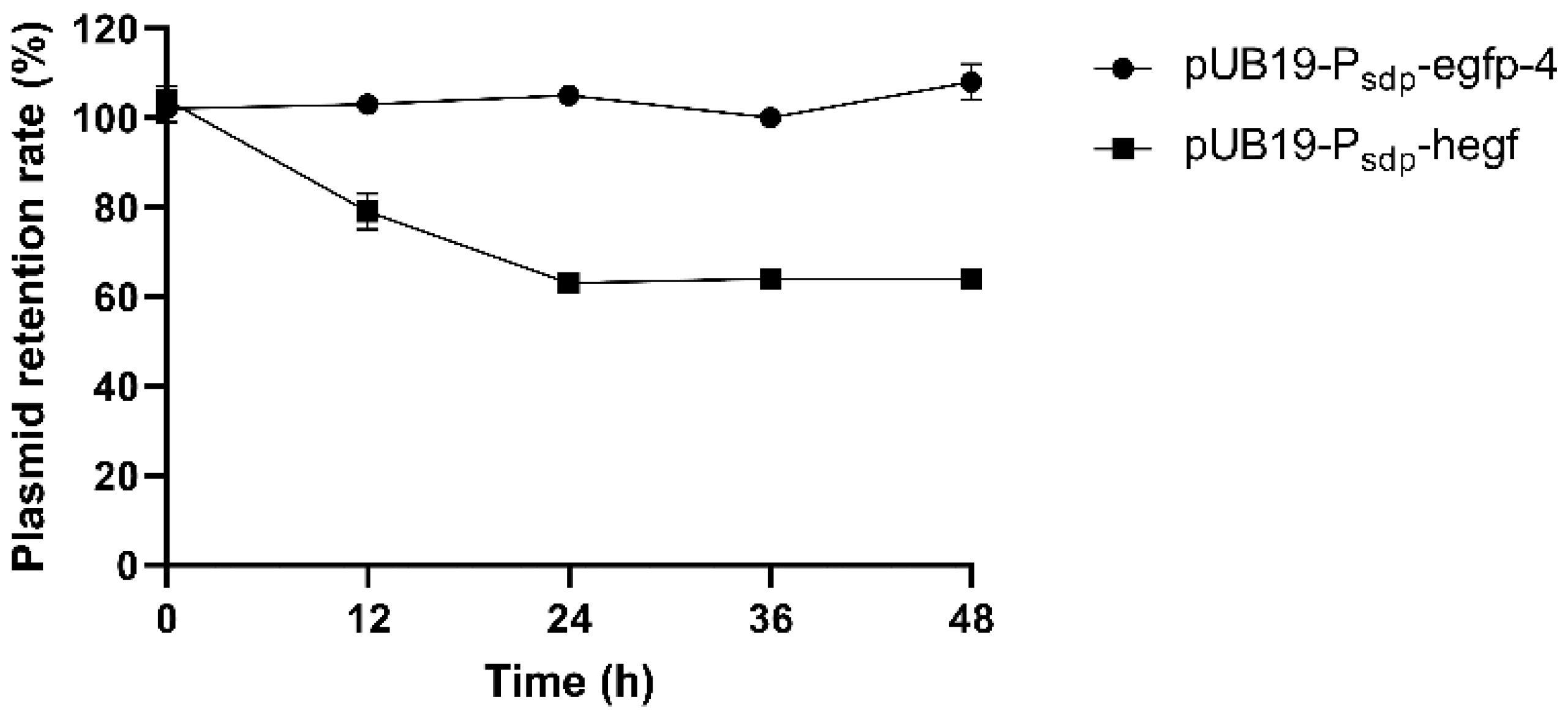
2.9. Plasmid Stability
2.10. Statistical Analysis
3. Results
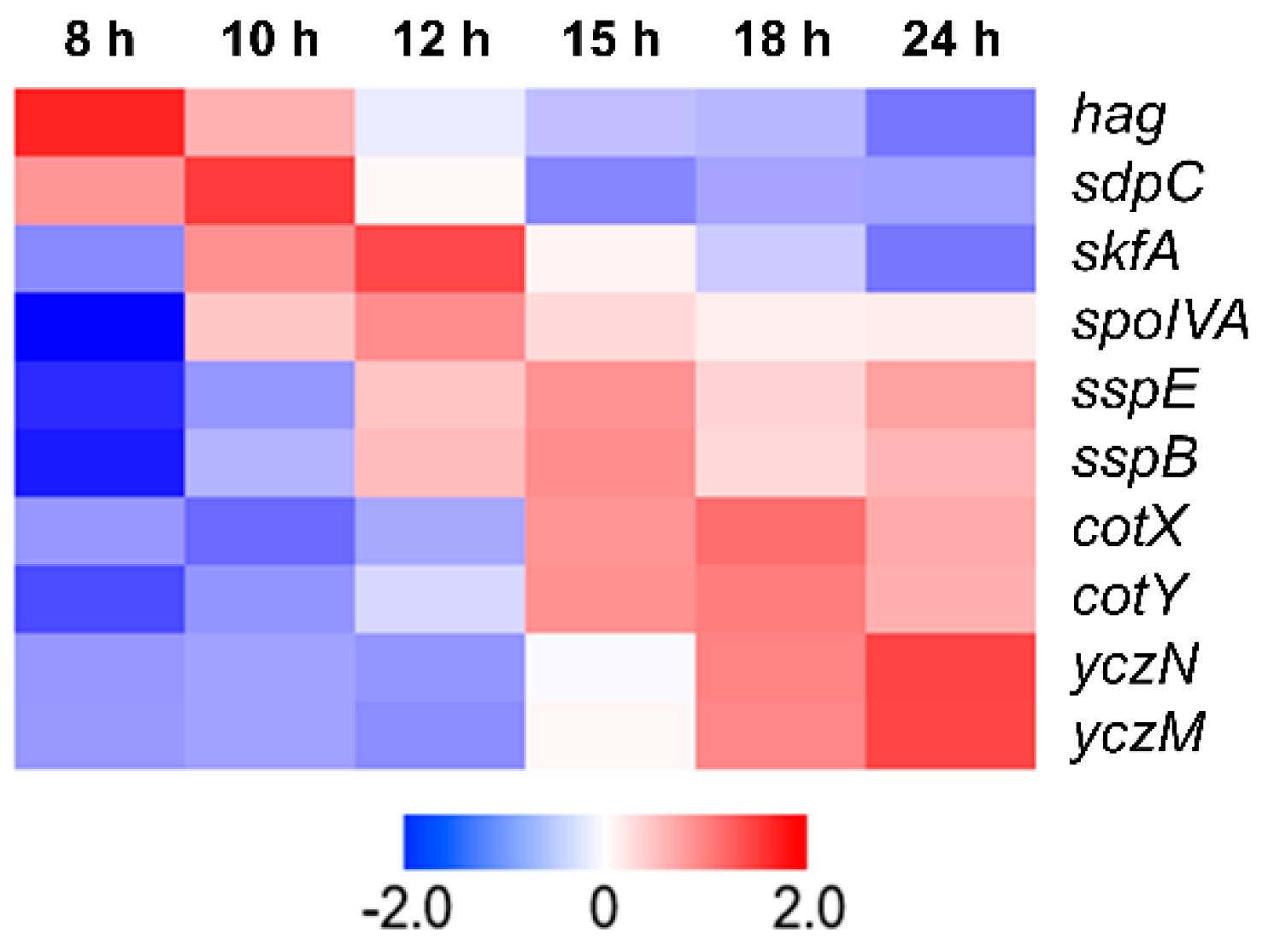
3.1. Selection of Strong Promoter Based on Transcriptome Analysis Data
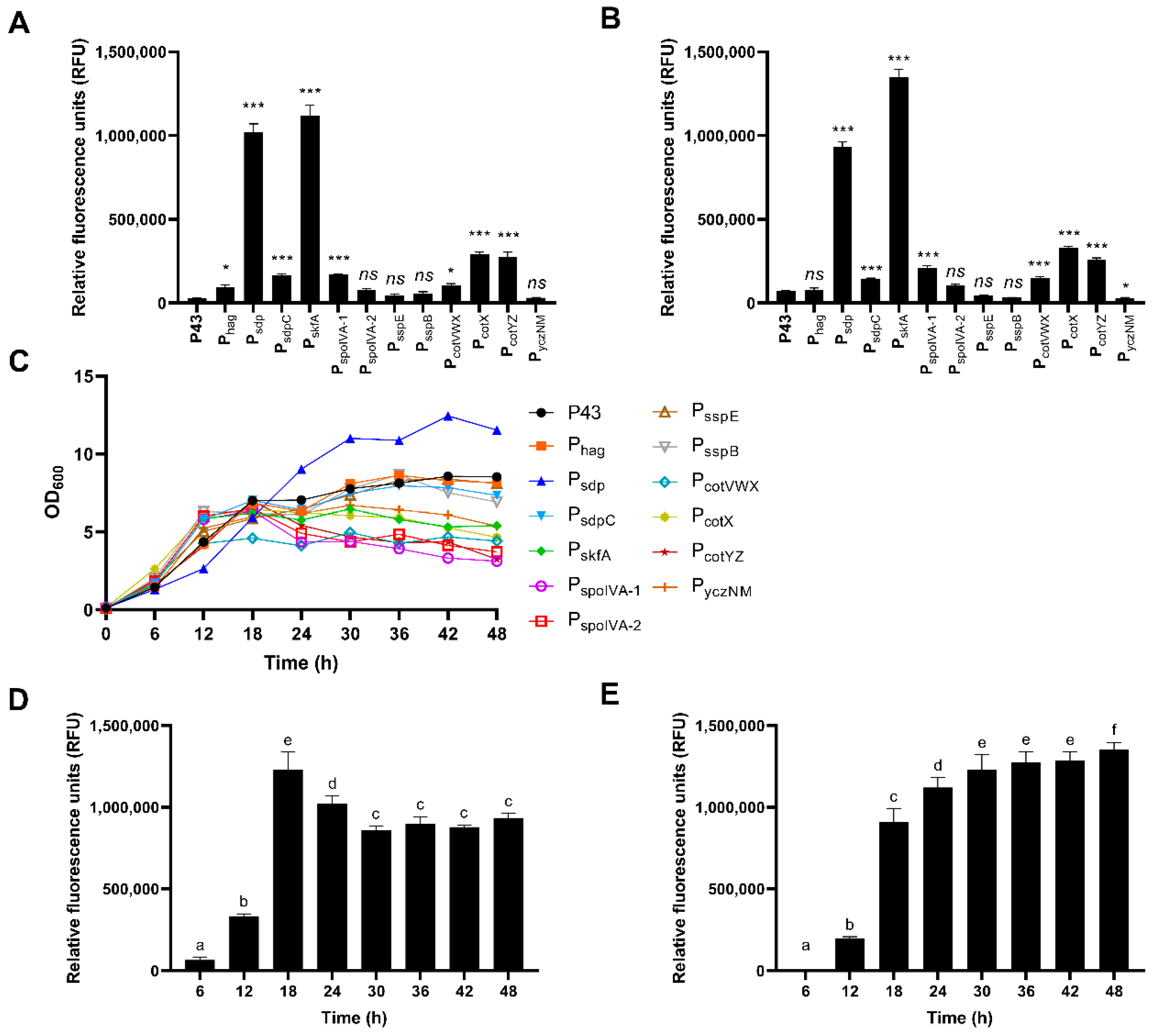
3.2. The Effect of Selected Promoters on the Expression of Enhanced Green Fluorescent Protein (eGFP)
3.3. Expression Cassette Engineering
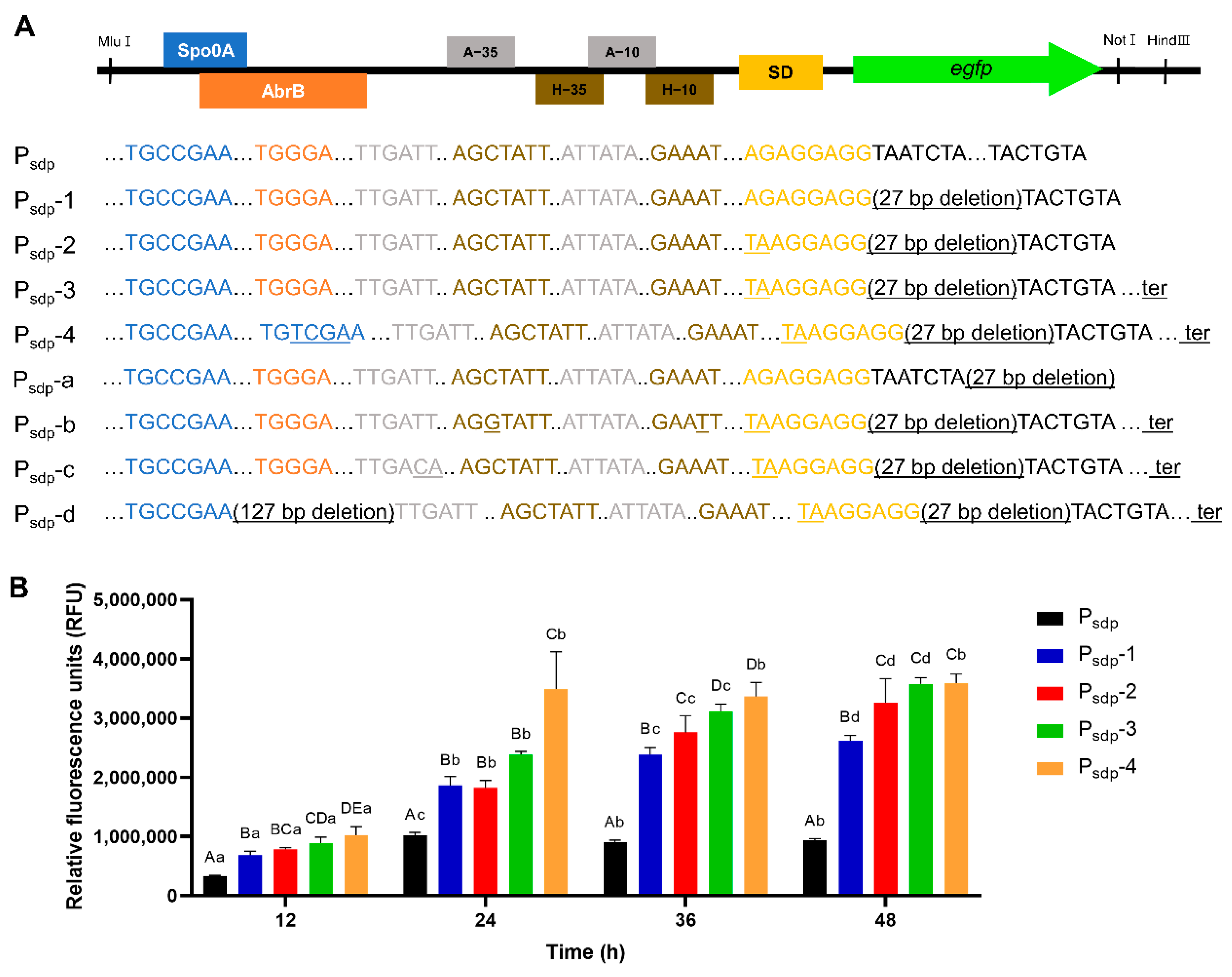
3.3.1. Engineering of Psdp Expression Cassette
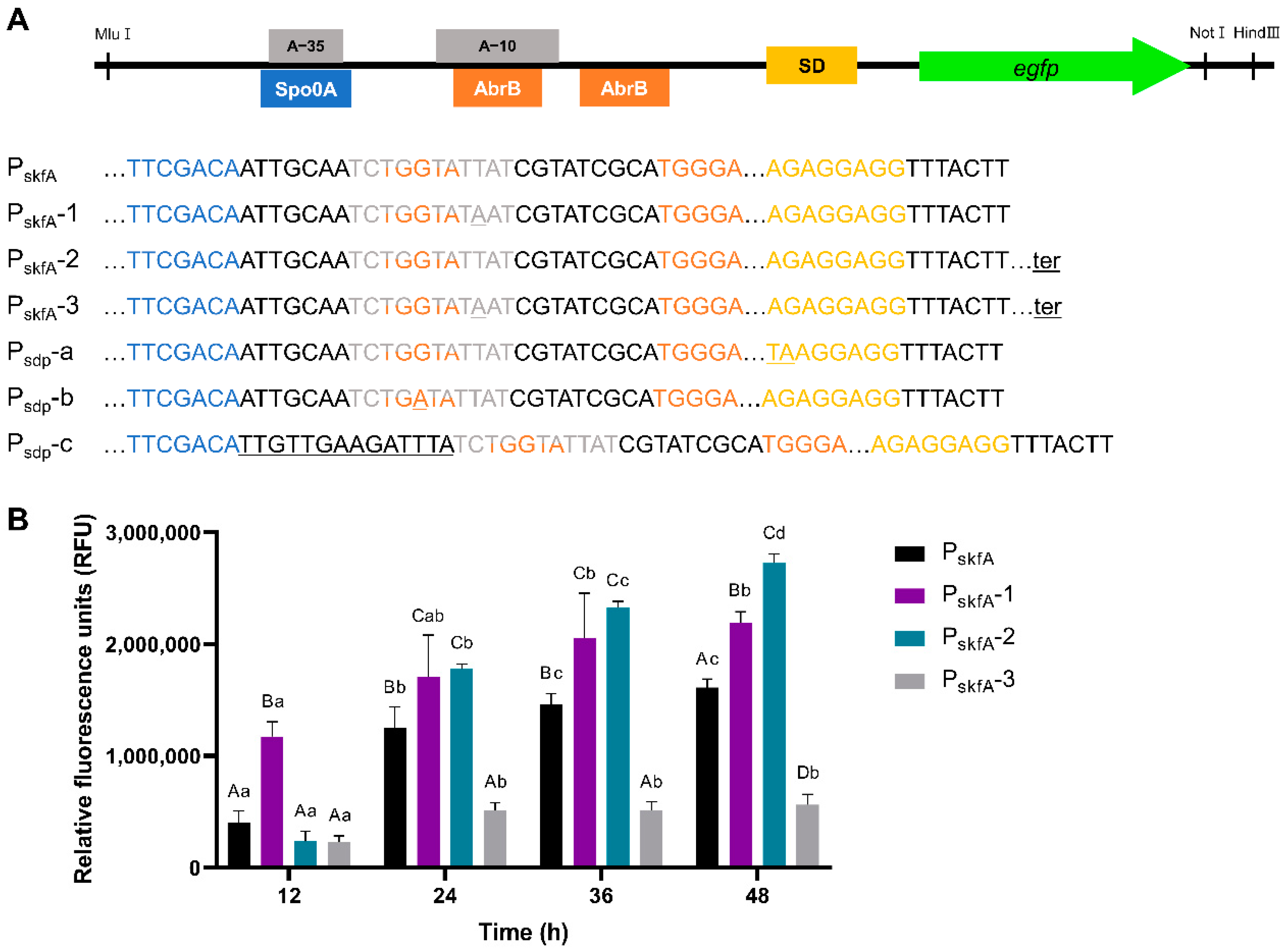
3.3.2. Engineering of PskfA Expression Cassette
3.4. Human Epidermal Growth Factor (hEGF) Expression Using Developed Expression Cassette
3.5. Stability of Recombinant Plasmids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xie, W.; Wu, Q.; Kuang, Z.; Cong, J.; Zhang, Q.; Huang, Y.; Su, Z.; Xiang, Q. Temperature-Controlled Expression of a Recombinant Human-like Collagen I Peptide in Escherichia coli. Bioengineering 2023, 10, 926. [Google Scholar] [CrossRef]
- Al-Janabi, S.S.; Shawky, H.; El-Waseif, A.A.; Farrag, A.A.; Abdelghany, T.M.; El-Ghwas, D.E. Stable, efficient, and cost-effective system for the biosynthesis of recombinant bacterial cellulose in Escherichia coli DH5α platform. J. Genet. Eng. Biotechnol. 2022, 20, 90. [Google Scholar] [CrossRef]
- Shahzadi, I.; Al-Ghamdi, M.A.; Nadeem, M.S.; Sajjad, M.; Ali, A.; Khan, J.A.; Kazmi, I. Scale-up fermentation of Escherichia coli for the production of recombinant endoglucanase from Clostridium thermocellum. Sci. Rep. 2021, 11, 7145. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jiang, C.; Tan, H.; Zhao, X.; Li, K.; Yin, H. Characterization of recombinant E. coli expressing a novel fucosidase from Bacillus cereus 2–8 belonging to GH95 family. Protein Expr. Purif. 2021, 186, 105897. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 2015, 14, 41. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K.; Mukhopadhyay, R.; Mukhija, R.; Krishnan, A.; Garg, L.; Panda, A.K. Optimization of inclusion body solubilization and renaturation of recombinant human growth hormone from Escherichia coli. Protein Expr. Purif. 2000, 18, 182–192. [Google Scholar] [CrossRef]
- Aida, Y.; Pabst, M.J. Removal of endotoxin from protein solutions by phase separation using Triton X-114. J. Immunol. Methods 1990, 132, 191–195. [Google Scholar] [CrossRef]
- Mamat, U.; Wilke, K.; Bramhill, D.; Schromm, A.B.; Lindner, B.; Kohl, T.A.; Corchero, J.L.; Villaverde, A.; Schaffer, L.; Head, S.R. Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb. Cell Fact. 2015, 14, 81. [Google Scholar] [CrossRef]
- Feizollahzadeh, S.; Kouhpayeh, S.; Rahimmansh, I.; Khanahmad, H.; Sabzehei, F.; Ganjalikhani-Hakemi, M.; Andalib, A.; Hejazi, Z.; Rezaei, A. The increase in protein and plasmid yields of E. coli with optimized concentration of ampicillin as selection marker. Iran. J. Biotechnol. 2017, 15, 128. [Google Scholar] [CrossRef]
- Food and Drug Administration. Carbohydrase and protease enzyme preparations derived from Bacillus subtilis or Bacillus amyloliquefaciens; Affirmation of GRAS status as direct food ingredients. Fed. Regist. 1999, 64, 19887–19895. [Google Scholar]
- Dong, H.; Zhang, D. Current development in genetic engineering strategies of Bacillus species. Microb. Cell Fact. 2014, 13, 63. [Google Scholar] [CrossRef]
- Mu, L.; Wen, J. Engineered Bacillus subtilis 168 produces l-malate by heterologous biosynthesis pathway construction and lactate dehydrogenase deletion. World J. Microbiol. Biotechnol. 2013, 29, 33–41. [Google Scholar] [CrossRef]
- Yan, X.; Yu, H.-J.; Hong, Q.; Li, S.-P. Cre/lox system and PCR-based genome engineering in Bacillus subtilis. Appl. Environ. Microbiol. 2008, 74, 5556–5562. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Hair, S.M.; Fujita, M.; Igoshin, O.A.; Tabor, J.J. An engineered B. subtilis inducible promoter system with over 10 000-fold dynamic range. ACS Synth. Biol. 2019, 8, 1673–1678. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.T.; Shaw, J.-F.; Chao, Y.-P.; David Ho, T.-H.; Yu, S.-M. Construction of chromosomally located T7 expression system for production of heterologous secreted proteins in Bacillus subtilis. J. Agric. Food Chem. 2010, 58, 5392–5399. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Fu, Y.; Ren, Z.; Gou, H.; Xu, C. Screening and application of inducible promoters in Ruminiclostridium papyrosolvens. Lett. Appl. Microbiol. 2020, 71, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, T.; Ji, W.; Wang, Q.; Zhang, H.; Chen, G.Q.; Lou, C.; Ouyang, Q. Engineering of core promoter regions enables the construction of constitutive and inducible promoters in Halomonas sp. Biotechnol. J. 2016, 11, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhang, X.; Xiong, Z.; Xia, Y.; Wu, Y.; Ai, L.; Xu, H.; Tian, Y.; Yang, Y.; Wang, G. Characterization of endogenous constitutive promoters from Lactobacillus salivarius for finely-tuning gene expression. Food Biosci. 2022, 50, 101980. [Google Scholar] [CrossRef]
- Xiao, X.; Ouyang, L.; Qi, J.; Chu, J. Exploration and characterization of hypoxia-inducible endogenous promoters in Aspergillus niger. Appl. Microbiol. Biotechnol. 2021, 105, 5529–5539. [Google Scholar] [CrossRef]
- Sun, Z.; Yue, Z.; Yang, X.; Hao, X.; Song, M.; Li, L.; Chen, C.; Chu, C.; Li, C. Efficient phytase secretion and phytate degradation by recombinant Bifidobacterium longum JCM 1217. Front. Microbiol. 2019, 10, 796. [Google Scholar] [CrossRef]
- Lu, Y.; Zheng, X.; Wang, Y.; Zhang, L.; Wang, L.; Lei, Y.; Zhang, T.; Zheng, P.; Sun, J. Evaluation of Aspergillus niger six constitutive strong promoters by fluorescent-auxotrophic selection coupled with flow cytometry: A case for citric acid production. J. Fungi 2022, 8, 568. [Google Scholar] [CrossRef] [PubMed]
- Rud, I.; Jensen, P.R.; Naterstad, K.; Axelsson, L. A synthetic promoter library for constitutive gene expression in Lactobacillus plantarum. Microbiology 2006, 152, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Cui, W.; Suo, F.; Miao, S.; Hao, W.; Chen, Q.; Guo, J.; Liu, Z.; Zhou, L.; Zhou, Z. Development of a novel strategy for robust synthetic bacterial promoters based on a stepwise evolution targeting the spacer region of the core promoter in Bacillus subtilis. Microb. Cell Fact. 2019, 18, 96. [Google Scholar] [CrossRef] [PubMed]
- Sohoni, S.V.; Fazio, A.; Workman, C.T.; Mijakovic, I.; Lantz, A.E. Synthetic promoter library for modulation of actinorhodin production in Streptomyces coelicolor A3(2). PLoS ONE 2014, 9, e99701. [Google Scholar] [CrossRef] [PubMed]
- Mordaka, P.M.; Heap, J.T. Stringency of synthetic promoter sequences in Clostridium revealed and circumvented by tuning promoter library mutation rates. ACS Synth. Biol. 2018, 7, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Siegl, T.; Tokovenko, B.; Myronovskyi, M.; Luzhetskyy, A. Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng. 2013, 19, 98–106. [Google Scholar] [CrossRef]
- Liu, X.; Yang, H.; Zheng, J.; Ye, Y.; Pan, L. Identification of strong promoters based on the transcriptome of Bacillus licheniformis. Biotechnol. Lett. 2017, 39, 873–881. [Google Scholar] [CrossRef]
- Li, S.; Wang, J.; Li, X.; Yin, S.; Wang, W.; Yang, K. Genome-wide identification and evaluation of constitutive promoters in streptomycetes. Microb. Cell Fact. 2015, 14, 172. [Google Scholar] [CrossRef]
- Dou, W.; Zhu, Q.; Zhang, M.; Jia, Z.; Guan, W. Screening and evaluation of the strong endogenous promoters in Pichia pastoris. Microb. Cell Fact. 2021, 20, 156. [Google Scholar] [CrossRef]
- Laoteng, K.; Anantayanon, J.; Chutrakul, C.; Panchanawaporn, S.; Jeennor, S. Transcriptome-based Mining of the Constitutive Promoters for Tuning Gene Expression in Aspergillus oryzae. J. Microbiol. 2023, 61, 199–210. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Wang, B.; Pan, L. High-level extracellular protein expression in Bacillus subtilis by optimizing strong promoters based on the transcriptome of Bacillus subtilis and Bacillus megaterium. Protein Expr. Purif. 2018, 151, 72–77. [Google Scholar] [CrossRef]
- Wu, X.-C.; Lee, W.; Tran, L.; Wong, S. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J. Bacteriol. 1991, 173, 4952–4958. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.-Z.; Doi, R.H. Overlapping promoters transcribed by Bacillus subtilis σ55 and σ37 RNA polymerase holoenzymes during growth and stationary phases. J. Biol. Chem. 1984, 259, 8619–8625. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.-S.; Kim, M.-J.; Hong, K. Novel sinIR promoter for Bacillus subtilis DB104 recombinant protein expression system. J. Appl. Biol. Chem. 2023, 66, 128–137. [Google Scholar] [CrossRef]
- Portela, R.M.; Vogl, T.; Ebner, K.; Oliveira, R.; Glieder, A. Pichia pastoris alcohol oxidase 1 (AOX1) core promoter engineering by high resolution systematic mutagenesis. Biotechnol. J. 2018, 13, 1700340. [Google Scholar] [CrossRef]
- Shabbir Hussain, M.; Gambill, L.; Smith, S.; Blenner, M.A. Engineering promoter architecture in oleaginous yeast Yarrowia lipolytica. ACS Synth. Biol. 2016, 5, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Pan, J.-G.; Park, S.-H.; Choi, S.-K. Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis. J. Biotechnol. 2010, 149, 16–20. [Google Scholar] [CrossRef]
- Cheng, J.; Guan, C.; Cui, W.; Zhou, L.; Liu, Z.; Li, W.; Zhou, Z. Enhancement of a high efficient autoinducible expression system in Bacillus subtilis by promoter engineering. Protein Expr. Purif. 2016, 127, 81–87. [Google Scholar] [CrossRef]
- González-Pastor, J.E. Cannibalism: A social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 2011, 35, 415–424. [Google Scholar] [CrossRef]
- Engelberg-Kulka, H.; Hazan, R. Cannibals defy starvation and avoid sporulation. Science 2003, 301, 467–468. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Carpenter, G.; Cohen, S. Human epidermal growth factor and the proliferation of human fibroblasts. J. Cell. Physiol. 1976, 88, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Shakhakarmi, K.; Seo, J.-E.; Lamichhane, S.; Thapa, C.; Lee, S. EGF, a veteran of wound healing: Highlights on its mode of action, clinical applications with focus on wound treatment, and recent drug delivery strategies. Arch. Pharm. Res. 2023, 46, 299–322. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Lai, C.Y.N.; Sivakumar, T.; Wang, H.; Ng, K.L.; Lam, C.C.; Wong, W. Novel strategy for expression of authentic and bioactive human basic fibroblast growth factor in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2018, 102, 7061–7069. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, P.; Shi, S.; Li, X.; Shi, D.; Zhou, Z.; Li, Z.; Xiao, Y. Expression of gallus epidermal growth factor (gEGF) with food-grade Lactococcus lactis expression system and its biological effects on broiler chickens. Biomolecules 2021, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.-S.; Jeong, H.-E.; Moon, S.-Y.; Shin, S.-H.; Hong, K.-W. Time-Course Transcriptome Analysis of Bacillus subtilis DB104 during Growth. Microorganisms 2023, 11, 1928. [Google Scholar] [CrossRef]
- Vesuna, F.; Winnard, P., Jr.; Raman, V. Enhanced green fluorescent protein as an alternative control reporter to Renilla luciferase. Anal. Biochem. 2005, 342, 345. [Google Scholar] [CrossRef]
- Chang, A.Y.; Chau, V.; Landas, J.A.; Pang, Y. Preparation of calcium competent Escherichia coli and heat-shock transformation. JEMI Methods 2017, 1, 22–25. [Google Scholar]
- Vojcic, L.; Despotovic, D.; Martinez, R.; Maurer, K.-H.; Schwaneberg, U. An efficient transformation method for Bacillus subtilis DB104. Appl. Microbiol. Biotechnol. 2012, 94, 487–493. [Google Scholar] [CrossRef]
- Kawamura, F.; Doi, R.H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J. Bacteriol. 1984, 160, 442–444. [Google Scholar] [CrossRef]
- Li, L.; Qu, W.; Jin, M.; Di, W.; Zeng, R. Extracellular expression of agarase rAgaM1 in Bacillus subtilis and its ability for neoagaro-oligosaccharide production. J. Basic Microbiol. 2019, 59, 359–367. [Google Scholar] [CrossRef]
- Sierro, N.; Makita, Y.; de Hoon, M.; Nakai, K. DBTBS: A database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008, 36, D93–D96. [Google Scholar] [CrossRef]
- Höfler, C.; Heckmann, J.; Fritsch, A.; Popp, P.; Gebhard, S.; Fritz, G.; Mascher, T. Cannibalism stress response in Bacillus subtilis. Microbiology 2016, 162, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, P.; Mäder, U.; Dervyn, E.; Rochat, T.; Leduc, A.; Pigeonneau, N.; Bidnenko, E.; Marchadier, E.; Hoebeke, M.; Aymerich, S. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 2012, 335, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.-C.; Han, L.-L.; Lu, Y.-B.; Feng, H. Construction of a high-expression system in Bacillus through transcriptomic profiling and promoter engineering. Microorganisms 2020, 8, 1030. [Google Scholar] [CrossRef]
- Vellanoweth, R.L.; Rabinowitz, J.C. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 1992, 6, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Volkenborn, K.; Kuschmierz, L.; Benz, N.; Lenz, P.; Knapp, A.; Jaeger, K.-E. The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis. Microb. Cell Fact. 2020, 19, 154. [Google Scholar] [CrossRef] [PubMed]
- Shine, J.; Dalgarno, L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 1974, 71, 1342–1346. [Google Scholar] [CrossRef]
- Wei, Y.; Silke, J.R.; Xia, X. Elucidating the 16S rRNA 3′ boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data. Sci. Rep. 2017, 7, 17639. [Google Scholar] [CrossRef]
- Cambray, G.; Guimaraes, J.C.; Mutalik, V.K.; Lam, C.; Mai, Q.-A.; Thimmaiah, T.; Carothers, J.M.; Arkin, A.P.; Endy, D. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 2013, 41, 5139–5148. [Google Scholar] [CrossRef]
- Cetnar, D.P.; Salis, H.M. Systematic quantification of sequence and structural determinants controlling mRNA stability in bacterial operons. ACS Synth. Biol. 2021, 10, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Curran, K.A.; Karim, A.S.; Gupta, A.; Alper, H.S. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab. Eng. 2013, 19, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liu, L.; Li, X.; Liu, D.; Yuan, Y. Engineering yeast artificial core promoter with designated base motifs. Microb. Cell Fact. 2020, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Ruth, C.; Pitzer, J.; Kickenweiz, T.; Glieder, A. Synthetic core promoters for Pichia pastoris. ACS Synth. Biol. 2014, 3, 188–191. [Google Scholar] [CrossRef]
- Ji, C.-H.; Kim, J.-P.; Kang, H.-S. Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters. ACS Synth. Biol. 2018, 7, 1946–1955. [Google Scholar] [CrossRef]
- Strauch, M.A.; Bobay, B.G.; Cavanagh, J.; Yao, F.; Wilson, A.; Le Breton, Y. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J. Bacteriol. 2007, 189, 7720–7732. [Google Scholar] [CrossRef]
- Blom, E.-J.; Ridder, A.N.; Lulko, A.T.; Roerdink, J.B.; Kuipers, O.P. Time-resolved transcriptomics and bioinformatic analyses reveal intrinsic stress responses during batch culture of Bacillus subtilis. PLoS ONE 2011, 6, e27160. [Google Scholar] [CrossRef]
- Fujita, M.; González-Pastor, J.E.; Losick, R. High-and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 2005, 187, 1357–1368. [Google Scholar] [CrossRef]
- Albano, M.; Smits, W.K.; Ho, L.T.; Kraigher, B.; Mandic-Mulec, I.; Kuipers, O.P.; Dubnau, D. The Rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J. Bacteriol. 2005, 187, 2010–2019. [Google Scholar] [CrossRef]
- Molle, V.; Fujita, M.; Jensen, S.T.; Eichenberger, P.; González-Pastor, J.E.; Liu, J.S.; Losick, R. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003, 50, 1683–1701. [Google Scholar] [CrossRef]
- Cha, H.J.; Chae, H.J.; Choi, S.S.; Yoo, Y.J. Production and secretion patterns of cloned glucoamylase in plasmid-harboring and chromosome-integrated recombinant yeasts employing an SUC2 promoter. Appl. Biochem. Biotechnol. 2000, 87, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Hanittinan, O.; Oo, Y.; Chaotham, C.; Rattanapisit, K.; Shanmugaraj, B.; Phoolcharoen, W. Expression optimization, purification and in vitro characterization of human epidermal growth factor produced in Nicotiana benthamiana. Biotechnol. Rep. 2020, 28, e00524. [Google Scholar] [CrossRef] [PubMed]
| Strains and Plasmids | Description | Resource |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F− Φ80lacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17 (rK−, mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Lab stock |
| Bacillus subtilis DB104 | his nprR2 nprE18 ΔaprA3 | Kawamura and Doi [50] |
| Plasmids | ||
| pUB19-Psin2-egfp | Ap r, Km r, E. coli-Bacillus shuttle vector pUB19 derivative, egfp | Lab stock |
| pET-15b | Ap r, E. coli expression vector | Lab stock |
| pUB19-P43-egfp | pUB19 with P43 in front of egfp | This study |
| pUB19-Phag-egfp | pUB19 with Phag in front of egfp | This study |
| pUB19-Psdp-egfp | pUB19 with Psdp in front of egfp | This study |
| pUB19-PsdpC-egfp | pUB19 with PsdpC in front of egfp | This study |
| pUB19-PskfA-egfp | pUB19 with PskfA in front of egfp | This study |
| pUB19-PspoIVA-1-egfp | pUB19 with PspoIVA-1 in front of egfp | This study |
| pUB19-PspoIVA-2-egfp | pUB19 with PspoIVA-2 in front of egfp | This study |
| pUB19-PsspE-egfp | pUB19 with PsspE in front of egfp | This study |
| pUB19-PsspB-egfp | pUB19 with PsspB in front of egfp | This study |
| pUB19-PcotX-egfp | pUB19 with PcotX in front of egfp | This study |
| pUB19-PcotVWX-egfp | pUB19 with PcotVWX in front of egfp | This study |
| pUB19-PcotYZ-egfp | pUB19 with PcotYZ in front of egfp | This study |
| pUB19-PyczNM-egfp | pUB19 with PyczNM in front of egfp | This study |
| pUB19-Psdp-egfp-1 | pUB19-Psdp-egfp derivative, Psdp-1 expression cassette | This study |
| pUB19-Psdp-egfp-2 | pUB19-Psdp-egfp-1 derivative, Psdp-2 expression cassette | This study |
| pUB19-Psdp-egfp-3 | pUB19-Psdp-egfp-2 derivative, Psdp-3 expression cassette | This study |
| pUB19-Psdp-egfp-4 | pUB19-Psdp-egfp-3 derivative, Psdp-4 expression cassette | This study |
| pUB19-Psdp-egfp-a | pUB19-Psdp-egfp derivative, Psdp-a expression cassette | This study |
| pUB19-Psdp-egfp-b | pUB19-Psdp-egfp-3 derivative, Psdp-b expression cassette | This study |
| pUB19-Psdp-egfp-c | pUB19-Psdp-egfp-3 derivative, Psdp-c expression cassette | This study |
| pUB19-Psdp-egfp-d | pUB19-Psdp-egfp-3 derivative, Psdp-d expression cassette | This study |
| pUB19-PskfA-egfp-1 | pUB19-PskfA-egfp derivative, PskfA-1 expression cassette | This study |
| pUB19-PskfA-egfp-2 | pUB19-PskfA-egfp derivative, PskfA-2 expression cassette | This study |
| pUB19-PskfA-egfp-3 | pUB19-PskfA-egfp-2 derivative, PskfA-3 expression cassette | This study |
| pUB19-PskfA-egfp-a | pUB19-PskfA-egfp derivative, PskfA-a expression cassette | This study |
| pUB19-PskfA-egfp-b | pUB19-PskfA-egfp derivative, PskfA-b expression cassette | This study |
| pUB19-PskfA-egfp-c | pUB19-PskfA-egfp derivative, PskfA-c expression cassette | This study |
| pUB19-Psdp-hegf | pUB19-Psdp-egfp-4 derivative, hegf instead of egfp | This study |
| Expression Cassette | Description | Engineered From |
|---|---|---|
| Psdp-1 | Optimizing of spacer between start codon and SD (35 bp to 8 bp) | Psdp |
| Psdp-2 | Application of consensus SD sequence | Psdp-1 |
| Psdp-3 | Terminator insertion after structure gene | Psdp-2 |
| Psdp-4 | Substitution of AbrB binding site to Spo0A | Psdp-3 |
| Psdp-a | Optimizing of spacer between start codon and SD (35 bp to 8 bp) | Psdp |
| Psdp-b | Application of consensus sequence of σH-dependent promoter | Psdp-3 |
| Psdp-c | Application of consensus sequence of σA-dependent promoter | Psdp-3 |
| Psdp-d | AbrB binding site deletion | Psdp-3 |
| Expression Cassette | Description | Engineered From |
|---|---|---|
| PskfA-1 | Application of consensus sequence of σA-dependent promoter | PskfA |
| PskfA-2 | Terminator insertion after structure gene | PskfA |
| PskfA-3 | Terminator insertion after structure gene | PskfA-2 |
| PskfA-a | Application of consensus SD sequence | PskfA |
| PskfA-b | Modification of AbrB binding site to its reverse sequence | PskfA |
| PskfA-c | Optimizing of spacer between −10 and −35 (7 bp to 14 bp) | PskfA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jun, J.-S.; Jeong, H.-E.; Hong, K.-W. Exploring and Engineering Novel Strong Promoters for High-Level Protein Expression in Bacillus subtilis DB104 through Transcriptome Analysis. Microorganisms 2023, 11, 2929. https://doi.org/10.3390/microorganisms11122929
Jun J-S, Jeong H-E, Hong K-W. Exploring and Engineering Novel Strong Promoters for High-Level Protein Expression in Bacillus subtilis DB104 through Transcriptome Analysis. Microorganisms. 2023; 11(12):2929. https://doi.org/10.3390/microorganisms11122929
Chicago/Turabian StyleJun, Ji-Su, Hyang-Eun Jeong, and Kwang-Won Hong. 2023. "Exploring and Engineering Novel Strong Promoters for High-Level Protein Expression in Bacillus subtilis DB104 through Transcriptome Analysis" Microorganisms 11, no. 12: 2929. https://doi.org/10.3390/microorganisms11122929
APA StyleJun, J.-S., Jeong, H.-E., & Hong, K.-W. (2023). Exploring and Engineering Novel Strong Promoters for High-Level Protein Expression in Bacillus subtilis DB104 through Transcriptome Analysis. Microorganisms, 11(12), 2929. https://doi.org/10.3390/microorganisms11122929






