Bivariate One Strain Many Compounds Designs Expand the Secondary Metabolite Production Space in Corallococcus coralloides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Assessment of the Biosynthetic Potential
2.2. Bivariate OSMAC Experiments
2.3. Preparation of Biotic Additives
2.4. Extraction and Sample Preparation
2.5. LC-MS Measurement
2.6. Mass Feature Detection and Structure Elucidation
3. Results and Discussion
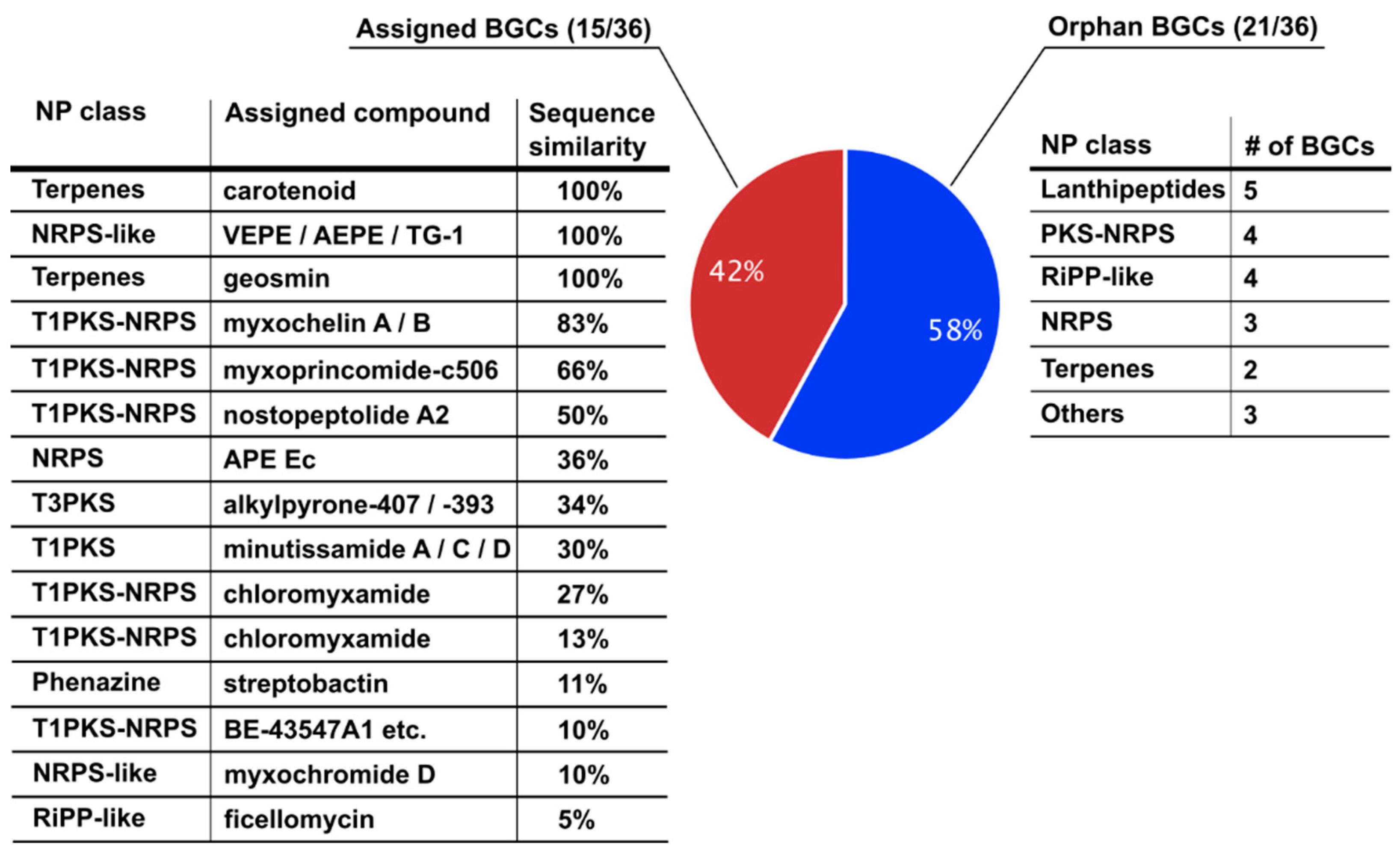
3.1. The Biosynthetic Potential of Corallococcus coralloides DSM2259
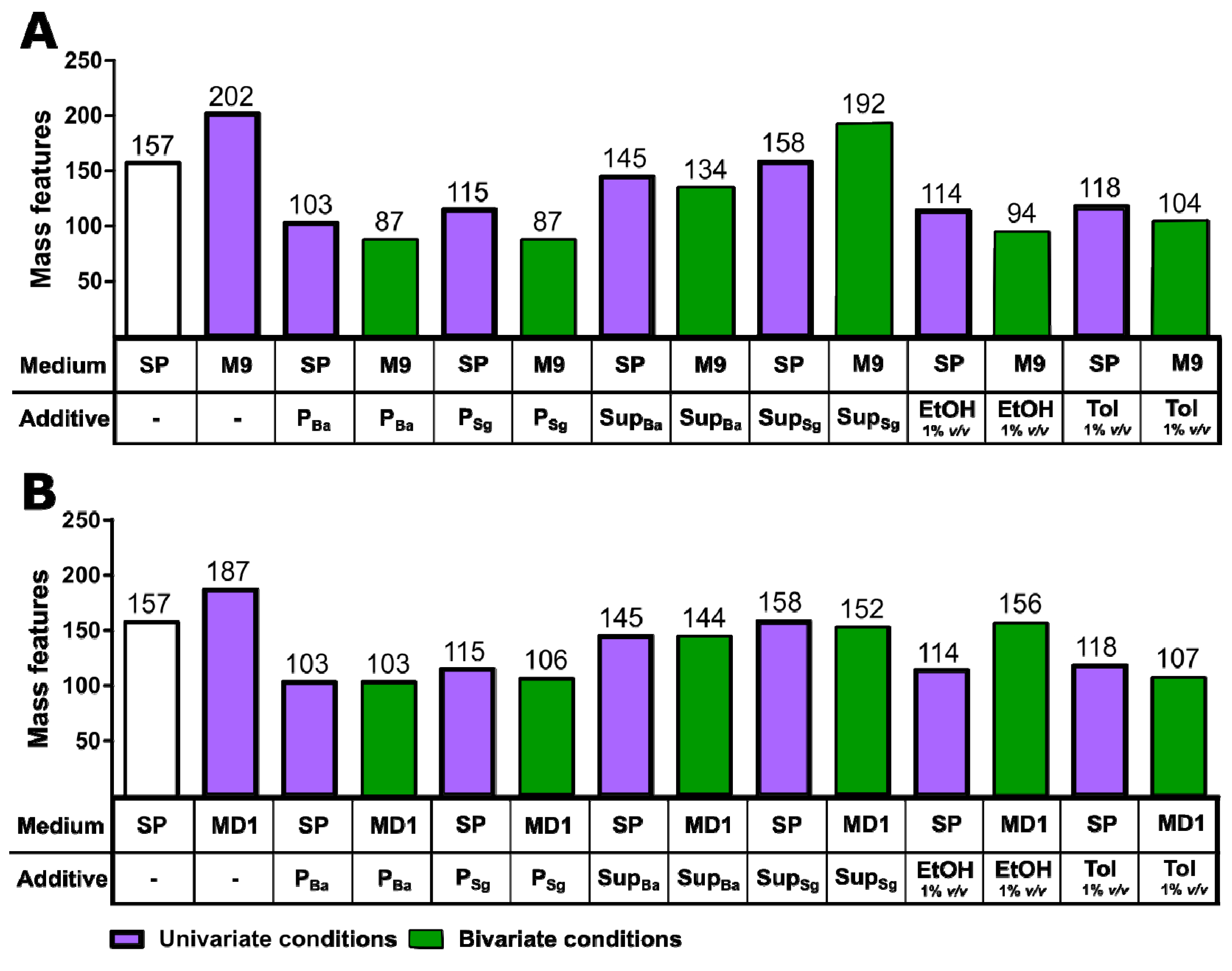
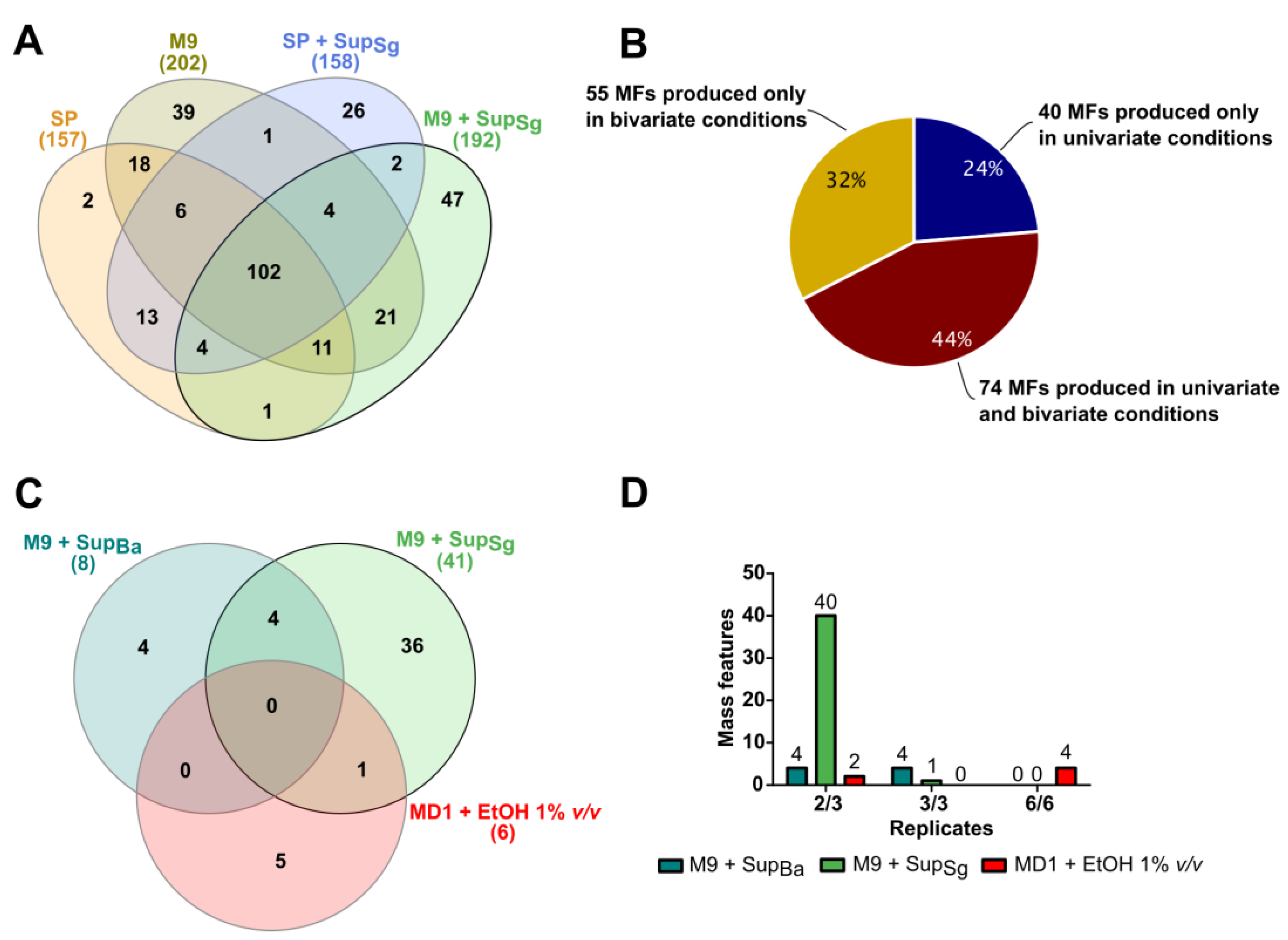
3.2. Discovery of New Secondary Metabolite Mass Features in Uni- and Bivariate OSMAC Screening
3.3. Condition Specificity and Reproducibility of Newly Detected Mass Features in Bivariate OSMAC
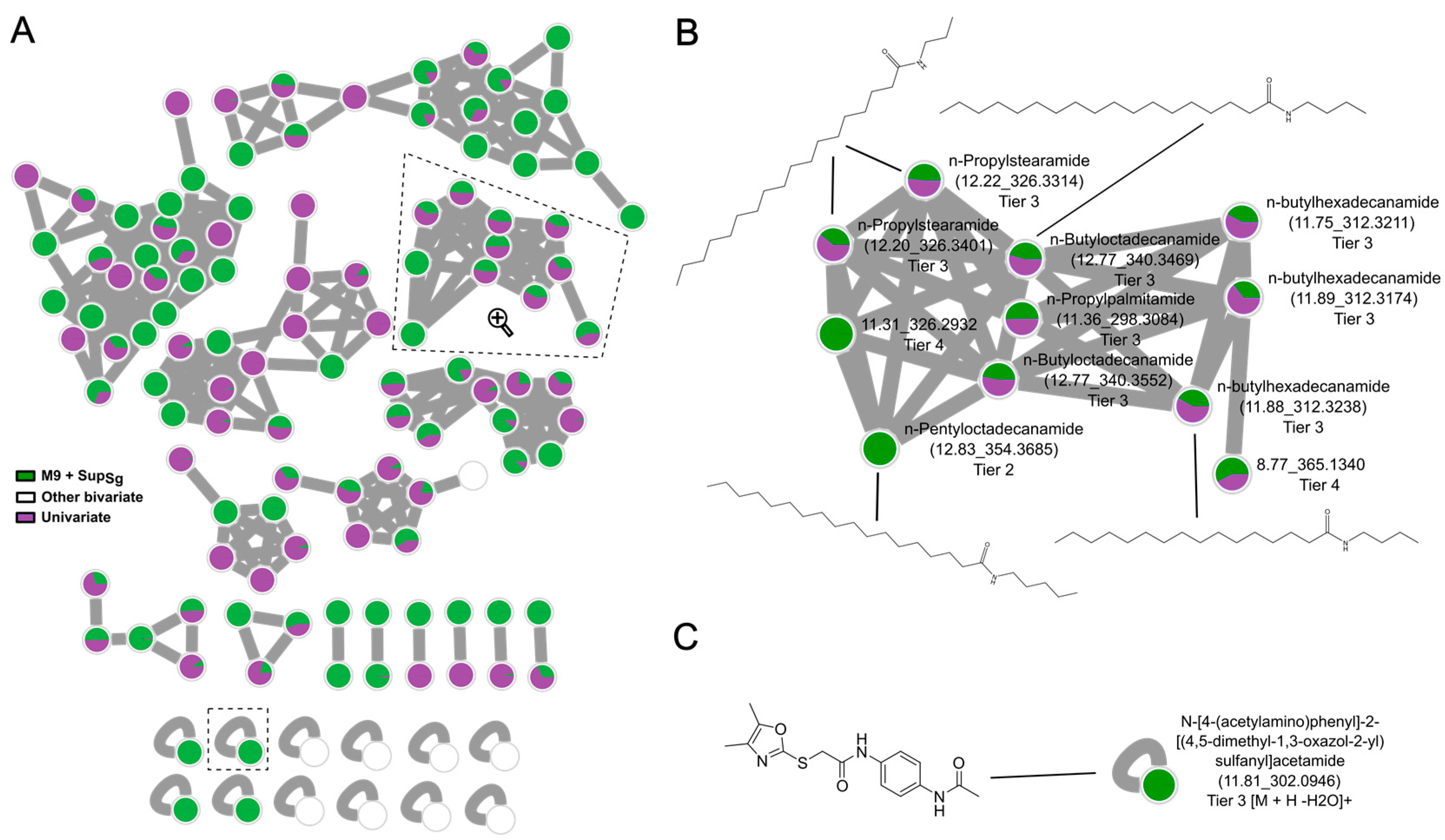
3.4. Identification and Bioinformatics Assignment of Secondary Metabolites in Bivariate OSMAC
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lok, C. Mining the Dark. Nature 2015, 522, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Marcy, Y.; Ouverney, C.; Bik, E.M.; Lösekann, T.; Ivanova, N.; Martin, H.G.; Szeto, E.; Platt, D.; Hugenholtz, P.; Relman, D.A.; et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA 2007, 104, 11889–11894. [Google Scholar] [CrossRef]
- Müller, R.; Wink, J. Future potential for anti-infectives from bacteria—How to exploit biodiversity and genomic potential. Int. J. Med. Microbiol. 2014, 304, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Okada, B.K.; Wu, Y.; Xu, F.; Seyedsayamdost, M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Du, A.-Q.; Zhou, Z.-Y.; Lai, C.; Yu, W.-C.; Yu, J.-B.; Yu, Y.-L.; Chen, J.-W.; Zhang, H.-W.; Xu, X.-W.; et al. An atlas of bacterial secondary metabolite biosynthesis gene clusters. Environ. Microbiol. 2021, 23, 6981–6992. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Müller, R. The impact of bacterial genomics on natural product research. Angew. Chem.-Int. Ed. 2005, 44, 6828–6846. [Google Scholar] [CrossRef] [PubMed]
- Furness, E.; Whitworth, D.E.; Zwarycz, A. Predatory Interactions between Myxobacteria and Their Prey; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 18–19. [Google Scholar]
- Sydney, N.; Swain, M.T.; So, J.M.T.; Hoiczyk, E.; Tucker, N.P.; Whitworth, D.E. The Genetics of Prey Susceptibility to Myxobacterial Predation: A Review, including an Investigation into Pseudomonas aeruginosa Mutations Affecting Predation by Myxococcus xanthus. Microb. Physiol. 2021, 31, 57–66. [Google Scholar] [CrossRef]
- Korp, J.; Gurovic, M.S.V.; Nett, M. Antibiotics from predatory bacteria. Beilstein J. Org. Chem. 2016, 12, 594–607. [Google Scholar] [CrossRef]
- Gregory, K.; Salvador, L.A.; Akbar, S.; Adaikpoh, B.I.; Stevens, D.C. Survey of biosynthetic gene clusters from sequenced myxobacteria reveals unexplored biosynthetic potential. Microorganisms 2019, 7, 181. [Google Scholar] [CrossRef]
- Livingstone, P.G.; Ingleby, O.; Girdwood, S.; Cookson, A.R.; Morphew, R.M.; Whitworth, D.E. Predatory Organisms with Untapped Biosynthetic Potential: Description of Novel Corallococcus species. Appl. Environ. Microbiol. 2020, 86, 2. [Google Scholar] [CrossRef]
- Babadi, Z.K.; Garcia, R.; Ebrahimipour, G.H.; Risdian, C.; Kämpfer, P.; Jarek, M.; Müller, R.; Wink, J. Corallococcus soli sp. Nov., a Soil Myxobacterium Isolated from Subtropical Climate, Chalus County, Iran, and Its Potential to Produce Secondary Metabolites. Microorganisms 2022, 10, 1262. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.; Hubmann, G.; Rosenthal, K.; Lütz, S. Triaging of culture conditions for enhanced secondary metabolite diversity from different bacteria. Biomolecules 2021, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.; Kehraus, S.; Schäberle, T.F.; Neu, E.; Almeida, C.; Roth, M.; König, G.M. Corallorazines from the myxobacterium Corallococcus coralloides. J. Nat. Prod. 2014, 77, 159–163. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, H.J.; Kim, G.W.; Cho, K.; Takahashi, S.; Koshino, H.; Kim, W.G. Isolation of Coralmycins A and B, potent anti-gram negative compounds from the Myxobacteria Corallococcus coralloides M23. J. Nat. Prod. 2016, 79, 2223–2228. [Google Scholar] [CrossRef]
- Erol, Ö.; Schäberle, T.F.; Schmitz, A.; Rachid, S.; Gurgui, C.; El Omari, M.; Lohr, F.; Kehraus, S.; Piel, J.; Müller, R.; et al. Biosynthesis of the myxobacterial antibiotic corallopyronin A. ChemBioChem 2010, 11, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Parales, R.; Singer, M. In silico characterization of a novel putative aerotaxis chemosensory system in the myxobacterium, Corallococcus coralloides. BMC Genom. 2018, 19, 757. [Google Scholar] [CrossRef]
- Livingstone, P.G.; Morphew, R.M.; Whitworth, D.E. Genome Sequencing and Pan-Genome Analysis of 23 Corallococcus spp. Strains reveal unexpected diversity, with particular plasticity of predatory Gene Sets. Front. Microbiol. 2018, 9, 3187. [Google Scholar] [CrossRef] [PubMed]
- Huntley, S.; Zhang, Y.; Treuner-Lange, A.; Kneip, S.; Sensen, C.W.; Søgaard-Andersen, L. Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM 2259. J. Bacteriol. 2012, 194, 3012–3013. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, 339–346. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Dickschat, J.S.; Bode, H.B.; Mahmud, T.; Müller, R.; Schulz, S. A novel type of geosmin biosynthesis in Myxobacteria. J. Org. Chem. 2005, 70, 5174–5182. [Google Scholar] [CrossRef]
- Loureiro, C.; Galani, A.; Gavriilidou, A.; de Mares, M.C.; van der Oost, J.; Medema, M.H.; Sipkema, D.; Sanchez, L.M. Comparative metagenomic analysis of biosynthetic diversity across sponge microbiomes highlights metabolic novelty, conservation, and diversification. mSystems 2022, 7, e0035722. [Google Scholar] [CrossRef]
- Pettit, R.K. Mixed fermentation for natural product drug discovery. Appl. Microbiol. Biotechnol. 2009, 83, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Ozdal, M. A new strategy for the efficient production of pyocyanin, a versatile pigment, in Pseudomonas aeruginosa OG1 via toluene addition. 3 Biotech 2019, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Hanh, T.T.H.; Hang, D.T.T.; Van Minh, C.; Dat, N.T. Anti-inflammatory effects of fatty acids isolated from Chromolaena odorata. Asian Pac. J. Trop. Med. 2011, 4, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Srinivas, V.; Alekhya, G.; Prakash, B. Effect of plant growth-promoting Streptomyces sp. on growth promotion and grain yield in chickpea (Cicer arietinum L). 3 Biotech 2015, 5, 799–806. [Google Scholar] [CrossRef]
- Tanvir, R.; Sajid, I.; Hasnain, S.; Kulik, A.; Grond, S. Rare actinomycetes Nocardia caishijiensis and Pseudonocardia carboxydivorans as endophytes, their bioactivity and metabolites evaluation. Microbiol. Res. 2016, 185, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Hai, Y.; Wei, M.-Y.; Wang, C.-Y.; Gu, Y.-C.; Shao, C.-L. The intriguing chemistry and biology of sulfur-containing natural products from marine microorganisms (1987–2020). Mar. Life Sci. Technol. 2021, 3, 488–518. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Zein, N.; Sinha, A.M.; McGahren, W.J.; Ellestad, G.A. Calicheamicin γ 1 I: An Antitumor antibiotic that cleaves double-stranded DNA site specifically. Science 1988, 240, 1198–1201. [Google Scholar] [CrossRef] [PubMed]
- Seyedsayamdost, M.R. Toward a global picture of bacterial secondary metabolism. J. Ind. Microbiol. Biotechnol. 2019, 46, 301–311. [Google Scholar] [CrossRef]
- Zhang, M.-Z.; Chen, Q.; Mulholland, N.; Beattie, D.; Irwin, D.; Gu, Y.-C.; Yang, G.-F.; Clough, J. Synthesis and fungicidal activity of novel pimprinine analogues. Eur. J. Med. Chem. 2012, 53, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Giddens, A.C.; Boshoff, H.I.; Franzblau, S.G.; Barry, C.E.; Copp, B.R. Antimycobacterial natural products: Synthesis and preliminary biological evaluation of the oxazole-containing alkaloid texaline. Tetrahedron Lett. 2005, 46, 7355–7357. [Google Scholar] [CrossRef]
- Pettit, G.R.; Knight, J.C.; Herald, D.L.; Davenport, R.; Pettit, R.K.; Tucker, B.E.; Schmidt, J.M. Isolation of Labradorins 1 and 2 from Pseudomonas syringae pv. coronafaciens. J. Nat. Prod. 2002, 65, 1793–1797. [Google Scholar] [CrossRef]
- Kumar, D.; Kumar, N.M.; Sundaree, S.; Johnson, E.O.; Shah, K. An expeditious synthesis and anticancer activity of novel 4-(3′-indolyl)oxazoles. Eur. J. Med. Chem. 2010, 45, 1244–1249. [Google Scholar] [CrossRef]
- Bristow, R.L.; Haworth-Duff, A.; Young, I.S.; Myers, P.; Hampson, M.R.; Williams, J.; Maher, S. An automated micro solid phase extraction gas chromatography-mass spectrometry (μSPE-GC-MS) detection method for geosmin and 2-methylisoborneol in drinking water. Sci. Rep. 2023, 13, 1768. [Google Scholar] [CrossRef]
- Muñoz-Dorado, J.; Marcos-Torres, F.J.; García-Bravo, E.; Moraleda-Muñoz, A.; Pérez, J. Myxobacteria: Moving, killing, feeding, and surviving together. Front. Microbiol. 2016, 7, 781. [Google Scholar] [CrossRef] [PubMed]
- Wrótniak-Drzewiecka, W.; Brzezińska, A.J.; Dahm, H.; Ingle, A.P.; Rai, M. Current trends in myxobacteria research. Ann. Microbiol. 2016, 66, 17–33. [Google Scholar] [CrossRef]
- Bader, C.D.; Haack, P.A.; Panter, F.; Krug, D.; Müller, R. Expanding the scope of detectable microbial natural products by complementary analytical methods and cultivation systems. J. Nat. Prod. 2021, 84, 268–277. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lindig, A.; Schwarz, J.; Hubmann, G.; Rosenthal, K.; Lütz, S. Bivariate One Strain Many Compounds Designs Expand the Secondary Metabolite Production Space in Corallococcus coralloides. Microorganisms 2023, 11, 2592. https://doi.org/10.3390/microorganisms11102592
Lindig A, Schwarz J, Hubmann G, Rosenthal K, Lütz S. Bivariate One Strain Many Compounds Designs Expand the Secondary Metabolite Production Space in Corallococcus coralloides. Microorganisms. 2023; 11(10):2592. https://doi.org/10.3390/microorganisms11102592
Chicago/Turabian StyleLindig, Anton, Jenny Schwarz, Georg Hubmann, Katrin Rosenthal, and Stephan Lütz. 2023. "Bivariate One Strain Many Compounds Designs Expand the Secondary Metabolite Production Space in Corallococcus coralloides" Microorganisms 11, no. 10: 2592. https://doi.org/10.3390/microorganisms11102592
APA StyleLindig, A., Schwarz, J., Hubmann, G., Rosenthal, K., & Lütz, S. (2023). Bivariate One Strain Many Compounds Designs Expand the Secondary Metabolite Production Space in Corallococcus coralloides. Microorganisms, 11(10), 2592. https://doi.org/10.3390/microorganisms11102592






