Multi-Omics Profiling of Candida albicans Grown on Solid Versus Liquid Media
Abstract
1. Introduction
1.1. Background Information
1.2. Multi-Omics Approach in Microorganisms
2. Materials and Methods
2.1. Solvents and Reagents
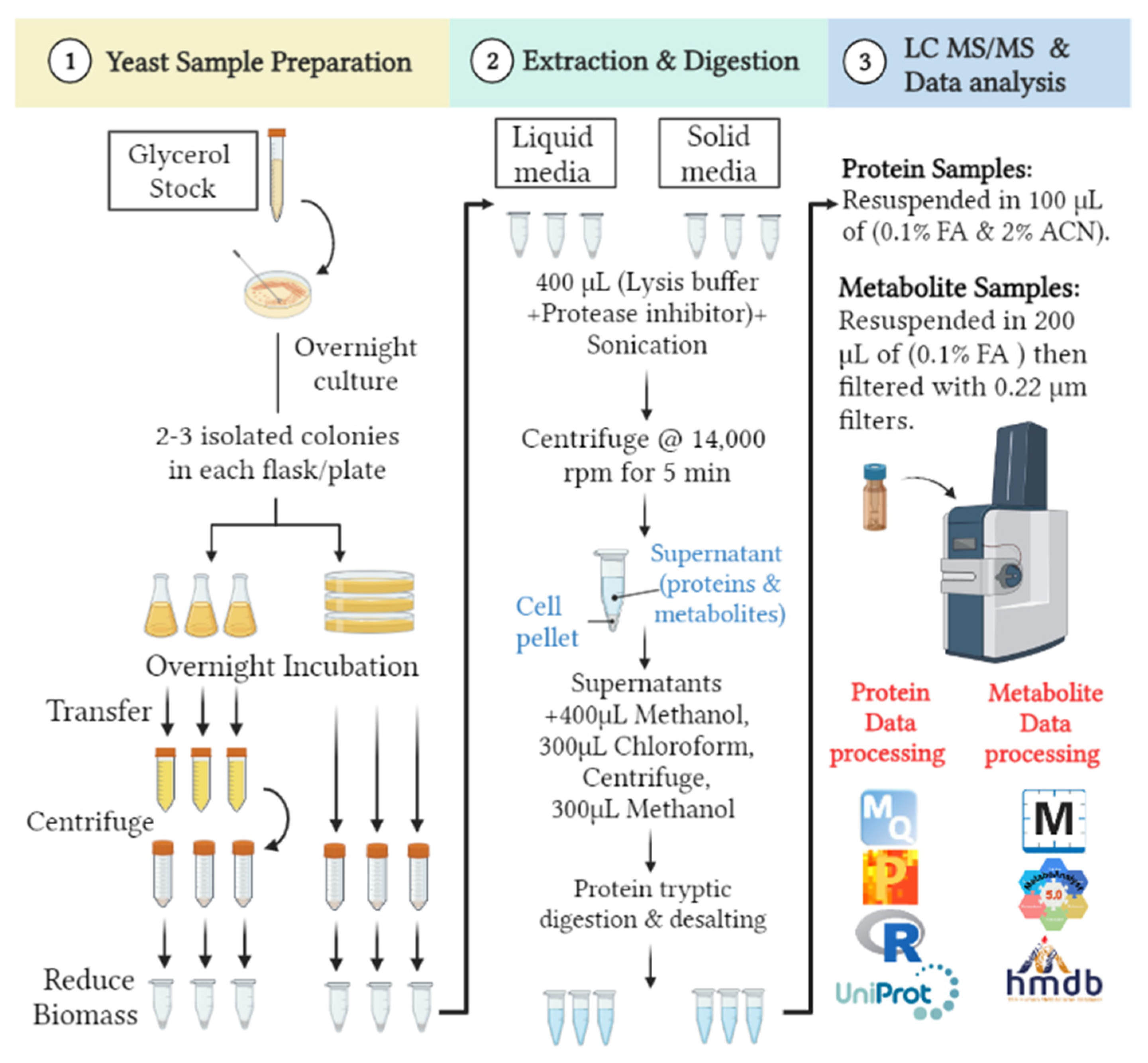
2.2. Yeast Sample Preparation
2.3. Proteomics and Metabolomics Extraction
2.4. In-Solution Protein Tryptic Digestion and Desalting
2.5. Ultra High-Performance Liquid Chromatography-Tandem Mass Spectrometry (UHPLC–MS/MS) Nano-Proteomics
2.6. Ultra-High-Performance Liquid Chromatography Coupled to Electrospray Ionization and Quadrupole Time-of-Flight Mass Spectrometry (UHPLC-ESI-QTOF-MS) Metabolomics
2.7. Bioinformatics Analysis and Statistical Approach
3. Results
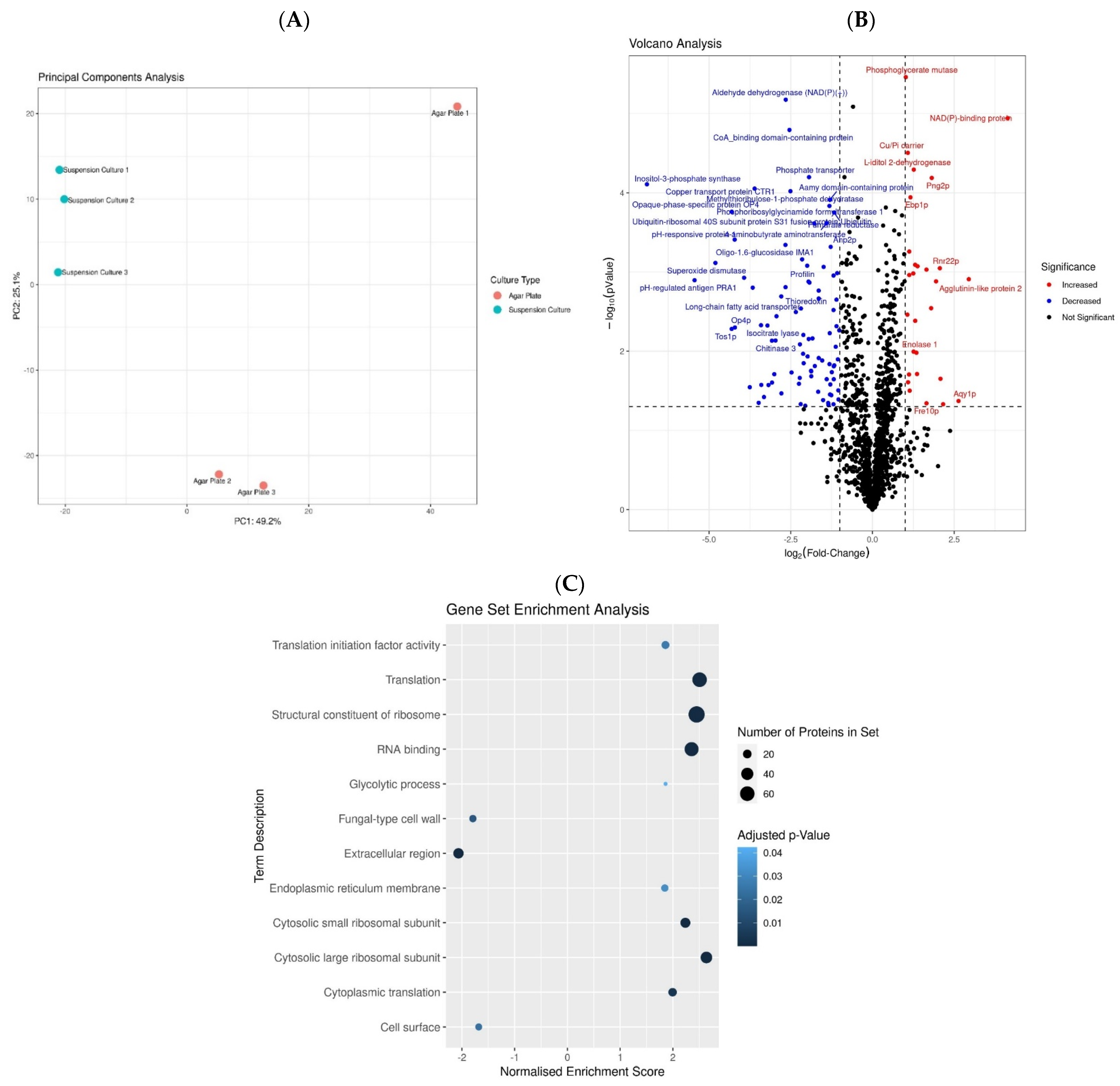
3.1. Proteomics Analysis Reveals That C. albicans Employs Different Protein Machinery to Sustain Growth on Agar Solid and Liquid Media Culture
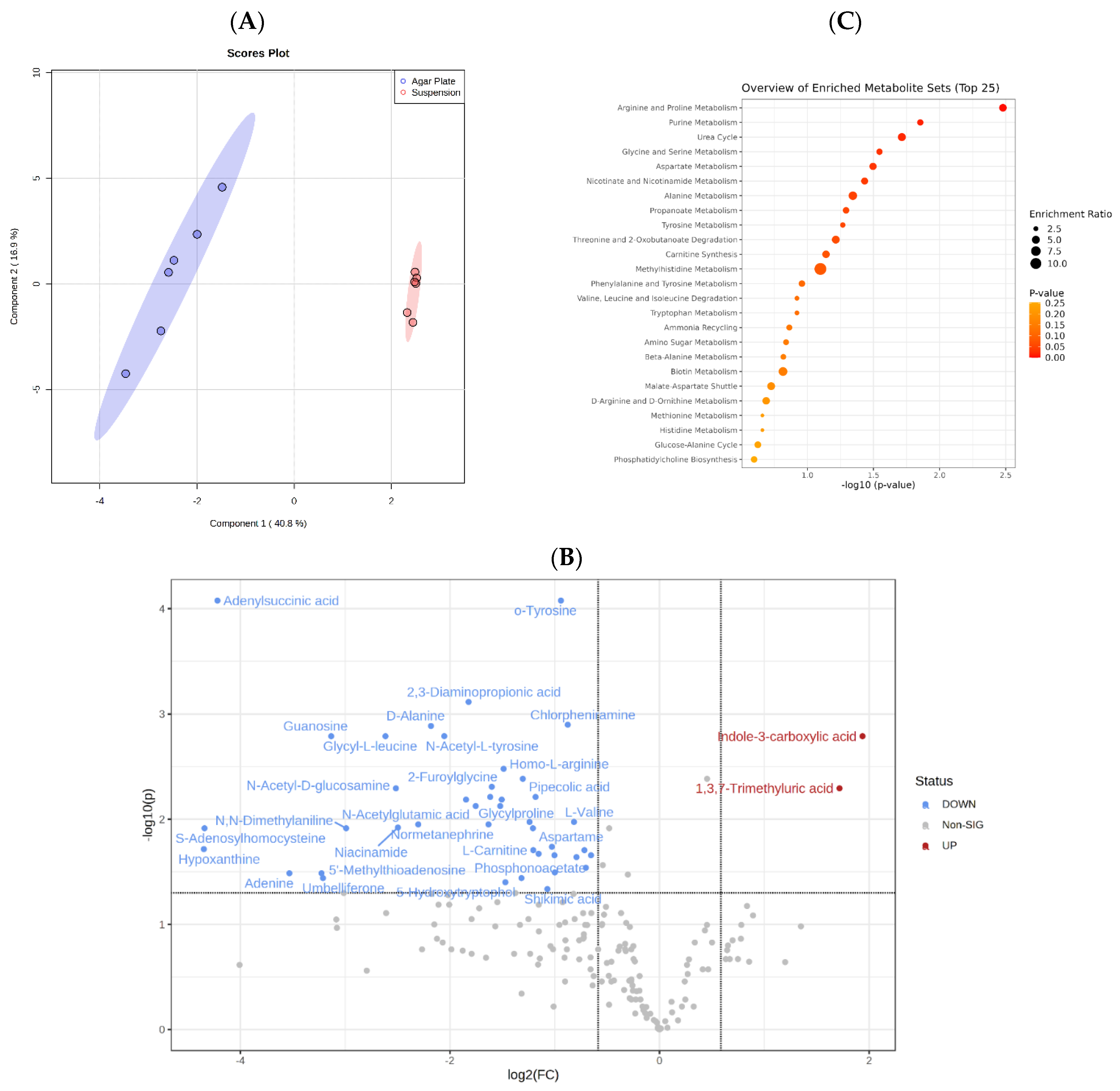
3.2. Metabolomics Analysis Indicates That C. albicans Possesses a Versatile and Robust Metabolism
4. Discussion
| Protein UniProt ID | Protein Name | GO–Biological Process (Source Uniprot) | Physiology and Morphogenesis | References |
|---|---|---|---|---|
| Part I—Proteins with increased abundance in solid agar | ||||
| Q56XX2 | Cell surface mannoprotein M65 | Cell surface, cell adhesion and biofilm formation, response to starvation, filamentous growth | Surface mannoprotein is required for hyphal morphogenesis, surface adherence, and pathogenicity. It plays an important role during biofilm development and maintenance and acts as a major antigen target of host cell-mediated immune response. | [73] |
| P87020 | pH-regulated antigen PRA1 | Cell surface, hyphal cell wall, adhesion of symbiont to host, Zin ion binding. Evasion of host immune response | Cell surface protein is involved in the host–parasite interaction during Candida infection. With MP65, it represents a major component of the biofilm matrix. It sequesters zinc from host tissue and mediates leukocyte adhesion and migration. As a released protein, it controls host complement attack, assisting the fungus in escaping host surveillance. It decreases complement-mediated adhesion, as well as the uptake of C. albicans by human macrophages. | |
| Q5AG89 | Thioredoxin reductase TRR1 | Cell redox homeostasis, cellular response to oxidative stress, fungal biofilm matrix | Belongs to the class-II pyridine nucleotide-disulfide oxidoreductase family and is a good target for potentially board-spectrum antifungal antibodies. | [74] |
| A0A1D8PGF8 | Msb2p | Cell surface, site polarized growth, osmosensor activity, filamentous growth, positive regulation of single-species biofilm formation, signal transduction involved in filamentous growth | Mucin Msb2 regulates the cek1 MAPK pathway. Msb2 shedding occurred differentially in cells grown planktonically or on solid surfaces in the presence of cell wall and osmotic stressors. | [75] |
| Q5AB48 | RBT4 | Cell surface, extracellular space, cholesterol binding | A secreted protein that acts as a virulence factor during infection, such as in posttraumatic corneal infections. It acts as an important antigen in patients with systemic candidiasis and plays a role in protection against phagocyte attack. | [76,77,78] |
| A0A1D8PH78 | Farnesyl pyrophosphate synthase | Farnesyltranstransferase activity | Farnesyl pyrophosphate synthase is part of the second module of the ergosterol biosynthesis pathway that includes the middle steps of the pathway. | [79] |
| Q5A470 | Opaque-phase-specific protein OP4 | Switches between white and opaque phases. | ||
| Q5AIB2 | SCW4 | Cell surface, fungal-type cell wall, carbohydrate metabolic process, cell wall organization | Supports cell wall assembly and integrity. | [80,81] |
| Q5AF37 | Inhibitor I9 domain-containing protein | Secreted protein | Virulence and pathogen interaction— is able to induce cell death in planta. The inhibitor I9 domain was more abundant in secretomes of a wide range of necrotizing fungi relative to biotrophs. | [82] |
| P43076 | pH-responsive protein 1 | Cell septum, cellular bud membrane, cell adhesion, entry into host | Required for apical cell growth and plays a vital role in morphogenesis. It may be integral to the pathogenic ability of the organism. | [83,84] |
| P40953 P40954 | Chitinase 2 Chitinase 3 | Filamentous growth of a population of unicellular organisms in response to starvation | Chitinase is involved in the remodeling of chitin in the fungal cell wall. It plays a role in cell separation. | [85] |
| Q59XU5 | Ras-like protein 1 | Actin fusion focus, cell cortex of cell tip, filamentous growth of a population of unicellular organism in response to heat | Required for the regulation of both the MAP kinase signaling pathway and cAMP signaling pathway. The activation of these pathways contributes to the pathogenicity of cells. Induction of morphological transition from yeast to the polarized filamentous form. | [86] |
| A0A1D8PSE1 | Mlc1p | Cellular bud neck contractile ring | Response to the maintenance of polarized growth. | [87] |
| Q5AHA4 | Predicted GPI-anchored protein 17 | Virulence | Predicted GPI-anchored protein role during fungal host infection | [88,89,90] |
| Q59NP5 | Secreted beta-glucosidase SUN41 | Single-species biofilm formation in or on the host organism, adhesion of symbiont to host | Cell surface beta-glucosidase is involved in cytokinesis, cell wall biogenesis, adhesion to host tissue, and biofilm formation. It plays an important role in the host–pathogen interaction. | [91,92] |
| Q5A786 | Profilin | Actin polymerization and depolymerization | Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents polymerization, whereas it enhances it at low concentrations. | [93,94] |
| Q59NP1 | Copper transport protein CTR1 | Cooper transport | Required for high-affinity copper transport into the cell. It is induced during biofilm formation and contact with macrophages as well as by alkaline pH via RIM101. | [95] |
| A0A1D8PL61 | Midasin | ATP hydrolysis activity, ribosomal large subunit export | A nuclear chaperon required for maturation and nuclear export of pre-60S ribosome subunits. It is essential for ribosome maturation in yeast (Saccharomyces cerevisiae). | [96] |
| A0A1D8PGE0 | Dynein light chain | Dynein intermediate chain binding | Acts as one of several non-catalytic accessory components of the cytoplasmic dynein complex. It may play a role in changing or maintaining the spatial distribution of cytoskeletal structures. | |
| Part II—Proteins with increased abundance in liquid media—virulence, pathogenicity, and host interaction | ||||
| P0CT51 | Blood-induced peptide 1 | Host interaction, virulence factor | Plays an important role in survival in host blood through increased tolerance to stress, such as salt or cycloheximide, which is essential for virulence. | [68] |
| A0A1D8PPK1 | Probable NADPH dehydrogenase | Steroid binding, steroid metabolic process | Oxidoreductase binds mammalian estrogens with high affinity. | [97,98] |
| A0A1D8PP43 | Adh1p | Biological process involved in interaction with the host, biofilm formation | Promotes C. albicans pathogenicity by stimulating oxidative phosphorylation. | [99] |
| P0CU38 | Agglutinin-like protein 2 | Cell adhesion involved in multi-species biofilm formation | A cell surface adhesion protein that mediates both yeast-to-host tissue adherence and yeast aggregation. Plays an important role in the pathogenesis of C. albicans infections. | [100,101,102] |
| O13318 | pH-responsive protein 2 | Hyphal cell wall, fungal-type cell wall | Required for apical cell growth and plays an essential role in morphogenesis. It may be integral to the pathogenic ability of an organism. | |
| Q5AFB4 | GST2p | Cellular response to oxidative stress | Required for nitrogen starvation-induced filamentous growth in C. albicans. | [103] |
| Q59PT0 | V-type proton ATPase subunit B | Autophagy, vacuolar acidification | Plays an important role in resistance to several stresses, as well as in autophagy and virulence. | [104] |
| P10613 | Lanosterol 14-alpha demethylase | Sterol 14-demethylase activity, cell growth mode switching from budding to filamentous | Plays an essential role in the third module ergosterol biosynthesis pathway. | [105,106] |
| Q5AHH4 | Small heat shock protein 21 | Cellular heat acclimation, cellular response to oxidative stress | A heat shock protein required for pathogenicity. Mediates thermotolerance and adaptation to oxidative stress. Plays a role in the capacity of damaging human-derived endothelial and oral epithelial cells during infections. Potentiates resistance to antifungal drugs as well as resistance to killing by human neutrophils. | [107,108] |
| Metabolite Accession Number | Metabolite Name | Role in Physiology and Morphogenesis | References |
|---|---|---|---|
| HMDB00679 | Homocitrulline | Secondary metabolites or non-essential metabolites that may serve a role as defense or signaling molecules. | |
| HMDB06050 | o-Tyrosine | Hydroxy radical biomarker of oxidative damage to protein. | [109] |
| HMDB02064 | N-Acetylputrescine | Cellular processes include cell cycle progression and growth produced by the breakdown of amino acids. Putrescine and cadaverine are primarily responsible for the foul odor of putrefying flesh but also contribute to the odor of bad breath and bacterial vaginosis. | [110] |
| HMDB0000130 | Homogentisic acid | In Pseudomonas, aeruginosa is a component of Pyomelamin, a black-brown negatively charged polymer produced during L-Tyrosine catabolism. Bordetella parapertussis confers a survival advantage during host infection. | [72,111] |
| HMDB0003070 | Shikimic acid | It is an important biochemical intermediate in plants and microorganisms. It is a compound that, when extracted from anise plants, has autoinflammatory effects. Accumulates Candida maltosa in the presence of a growth inhibitor herbicide, glyphosate. | [71,112] |
| HMDB0000070 | Pipecolic acid | Biofilm formation of Enterococcus faecalis and C. albicans. | [113] |
| HMDB0000133 | Guanosine | Fungal-associated infection. | [114] |
| HMDB0004110 | Phosphonoacetate | Physiologically essential metabolites involved in an organism’s growth, development, or reproduction. | |
| HMDB0001539 | Asymmetric dimethylarginine | In C. albicans lacking CaHmt1, asymmetric dimethylarginine and omega-monomethylarginine levels are decreased. | [115] |
| HMDB0001988 | 4-Hydroxycyclohexylcarboxylic acid | Microorganisms possess important biological activities, such as antibacterial, anti-inflammatory, and hypoglycemic effects. | [116] |
| HMDB00819 | Normetanephrine | Normetanephrine is a metabolite of norepinephrine created by the action of catechol-O-methyl transferase on norepinephrine. The latter can influence microbial pathogenesis, the growth and production of virulence factors in enterotoxigenic and enterohemorrhagic strains of Escherichia coli. | [117] |
| HMDB002006 | 2,3-Diaminopropionic acid | N3-(4-Methoxyfumaroyl)-l-2,3-Diaminopropionic acid is a strong inhibitor of the essential fungal enzyme glucosamine-6-phosphate. | [118] |
| HMDB0000206 | N6-Acetyl-l-lysine | A novel 2-oxoglutarate aminotransferase catalyzing the second step of lysine catabolism, the oxidative transamination of the alpha-group of N6-acetyllysine, was identified in Candida maltose. The enzyme was strongly induced in cells grown on L-lysine as the sole carbon source. | [119] |
| HMDB0001173 | 5′-Methylthioadenosine | It has been shown to influence the regulation of gene expression, proliferation, and apoptosis. |
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharifzadeh, A.; Khosravi, A.R.; Shokri, H.; Asadi Jamnani, F.; Hajiabdolbaghi, M.; Ashrafi Tamami, I. Oral microflora and their relation to risk factors in HIV+ patients with oropharyngeal candidiasis. J. Mycol. Med. 2013, 23, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Klingspor, L.; Tortorano, A.M.; Peman, J.; Willinger, B.; Hamal, P.; Sendid, B.; Velegraki, A.; Kibbler, C.; Meis, J.; Sabino, R.; et al. Invasive Candida infections in surgical patients in intensive care units: A prospective, multicentre survey initiated by the European Confederation of Medical Mycology (ECMM) (2006–2008). Clin. Microbiol. Infect. 2015, 21, 87.e1–87.e10. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, A.A.; Harrison, L.H.; Farley, M.M.; Hollick, R.; Stein, B.; Chiller, T.M.; Lockhart, S.R.; Park, B.J. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008-2013: Results from population-based surveillance. PLoS ONE 2015, 10, e0120452. [Google Scholar] [CrossRef]
- Yapar, N. Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 2014, 10, 95–105. [Google Scholar] [CrossRef]
- Kumamoto, C.A. Inflammation and gastrointestinal Candida colonization. Curr. Opin. Microbiol. 2011, 14, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Kashem, S.W.; Igyarto, B.Z.; Gerami-Nejad, M.; Kumamoto, Y.; Mohammed, J.A.; Jarrett, E.; Drummond, R.A.; Zurawski, S.M.; Zurawski, G.; Berman, J.; et al. Candida albicans morphology and dendritic cell subsets determine T helper cell differentiation. Immunity 2015, 42, 356–366. [Google Scholar] [CrossRef]
- Liu, H. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int. J. Med. Microbiol. 2002, 292, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Quindós, G. Epidemiology of candidaemia and invasive candidiasis. A changing face. Rev. Iberoam. Micol. 2014, 31, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, A.; Rupp, S.; Taylor, B.N.; Röllinghoff, M.; Schröppel, K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 2000, 38, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.J.; Köhler, J.R.; DiDomenico, B.; Loebenberg, D.; Cacciapuoti, A.; Fink, G.R.; Nonfilamentous, C. albicans mutants are avirulent. Cell 1997, 90, 939–949. [Google Scholar] [CrossRef]
- van der Meer, J.W.; van de Veerdonk, F.L.; Joosten, L.A.; Kullberg, B.J.; Netea, M.G. Severe candida spp. infections: New insights into natural immunity. Int. J. Antimicrob. Agents 2010, 36 (Suppl. S2), S58–S62. [Google Scholar] [CrossRef] [PubMed]
- Whiteway, M.; Bachewich, C. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 2007, 61, 529–553. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Nguyen, M.H. Finding the “missing 50%” of invasive candidiasis: How nonculture diagnostics will improve understanding of disease spectrum and transform patient care. Clin. Infect. Dis. 2013, 56, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Erwig, L.P.; Gow, N.A. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef]
- Sudbery, P.; Gow, N.; Berman, J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004, 12, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Moyes, D.L.; Wächtler, B.; Hube, B. Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 2011, 13, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Grubb, S.E.; Murdoch, C.; Sudbery, P.E.; Saville, S.P.; Lopez-Ribot, J.L.; Thornhill, M.H. Adhesion of Candida albicans to endothelial cells under physiological conditions of flow. Infect. Immun. 2009, 77, 3872–3878. [Google Scholar] [CrossRef]
- Fradin, C.; De Groot, P.; MacCallum, D.; Schaller, M.; Klis, F.; Odds, F.C.; Hube, B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 2005, 56, 397–415. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, C.; Tang, J. Disruption of the intestinal mucosal barrier in Candida albicans infections. Microbiol. Res. 2013, 168, 389–395. [Google Scholar] [CrossRef]
- MT, A.L.B.; Soares, N.C.; Semreen, M.H.; Cacciatore, S.; Dash, N.R.; Hamad, M.; Mousa, M.K.; Salam, J.S.A.; Al Gharaibeh, M.F.; Zerbini, L.F.; et al. Candida albicans PPG1, a serine/threonine phosphatase, plays a vital role in central carbon metabolisms under filament-inducing conditions: A multi-omics approach. PLoS ONE 2021, 16, e0259588. [Google Scholar] [CrossRef]
- Trevijano-Contador, N.; Rueda, C.; Zaragoza, O. Fungal morphogenetic changes inside the mammalian host. Semin. Cell Dev. Biol. 2016, 57, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Cottier, F.; Mühlschlegel, F.A. Sensing the environment: Response of Candida albicans to the X factor. FEMS Microbiol. Lett. 2009, 295, 1–9. [Google Scholar] [CrossRef][Green Version]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Odds, F.C.; Webster, C.E.; Mayuranathan, P.; Simmons, P.D. Candida concentrations in the vagina and their association with signs and symptoms of vaginal candidosis. J. Med. Vet. Mycol. 1988, 26, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Johnson, A.D. Candida albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Tobudic, S.; Kratzer, C.; Lassnigg, A.; Presterl, E. Antifungal susceptibility of Candida albicans in biofilms. Mycoses 2012, 55, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Biswas, S.; Van Dijck, P.; Datta, A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 2007, 71, 348–376. [Google Scholar] [CrossRef]
- Washburn, M.P.; Yates, J.R. Analysis of the microbial proteome. Curr. Opin. Microbiol. 2000, 3, 292–297. [Google Scholar] [CrossRef]
- Loo, J.A.; Quinn, J.P.; Ryu, S.I.; Henry, K.D.; Senko, M.W.; McLafferty, F.W. High-resolution tandem mass spectrometry of large biomolecules. Proc. Natl. Acad. Sci. USA 1992, 89, 286–289. [Google Scholar] [CrossRef]
- Yates, J.R. Mass spectral analysis in proteomics. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 297–316. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass. Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, G.G.; Goodacre, R. (Eds.) Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis; Springer Science and Business Media LLC: Dordrecht, The Netherlands, 2003. [Google Scholar] [CrossRef]
- Ball, B.; Langille, M.; Geddes-McAlister, J. Fun(gi)omics: Advanced and Diverse Technologies to Explore Emerging Fungal Pathogens and Define Mechanisms of Antifungal Resistance. mBio 2020, 11, e01020-20. [Google Scholar] [CrossRef] [PubMed]
- Fortuin, S.; Soares, N.C. The Integration of Proteomics and Metabolomics Data Paving the Way for a Better Understanding of the Mechanisms Underlying Microbial Acquired Drug Resistance. Front. Med. 2022, 9, 849838. [Google Scholar] [CrossRef] [PubMed]
- Torres-Sangiao, E.; Soares, N.C.; Liu, X. Editorial: Proteomic Approaches to Unravel Mechanisms of Resistance and Immune Evasion of Bacterial Pathogens. Front. Med. 2022, 9, 945264. [Google Scholar] [CrossRef]
- Bataineh, M.T.A.; Cacciatore, S.; Semreen, M.H.; Dash, N.R.; Soares, N.C.; Zhu, X.; Dash, N.R.; Mousa, M.K.; Salam, J.S.A.; Zerbini, L.F.; et al. Exploring the effect of estrogen on Candida albicans hyphal cell wall glycans and ergosterol synthesis. Front. Cell. Infect. Microbiol. 2022, 12, 977157. [Google Scholar] [CrossRef]
- Semreen, M.H.; Soliman, S.S.M.; Saeed, B.Q.; Alqarihi, A.; Uppuluri, P.; Ibrahim, A.S. Metabolic Profiling of Candida auris, a Newly-Emerging Multi-Drug Resistant Candida Species, by GC-MS. Molecules 2019, 24, 399. [Google Scholar] [CrossRef] [PubMed]
- Huertas, B.; Prieto, D.; Pitarch, A.; Gil, C.; Pla, J.; Díez-Orejas, R. Serum Antibody Profile during Colonization of the Mouse Gut by Candida albicans: Relevance for Protection during Systemic Infection. J. Proteome Res. 2017, 16, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Amador-García, A.; Zapico, I.; Borrajo, A.; Malmström, J.; Monteoliva, L.; Gil, C. Extending the Proteomic Characterization of Candida albicans Exposed to Stress and Apoptotic Inducers through Data-Independent Acquisition Mass Spectrometry. mSystems 2021, 6, e0094621. [Google Scholar] [CrossRef] [PubMed]
- Vaz, C.; Pitarch, A.; Gómez-Molero, E.; Amador-García, A.; Weig, M.; Bader, O.; Monteoliva, L.; Gil, C. Mass Spectrometry-Based Proteomic and Immunoproteomic Analyses of the Candida albicans Hyphal Secretome Reveal Diagnostic Biomarker Candidates for Invasive Candidiasis. J. Fungi 2021, 7, 501. [Google Scholar] [CrossRef] [PubMed]
- Cattò, C.; Corte, L.; Roscini, L.; Cardinali, G.; Villa, F.; Cappitelli, F. Metabolomic and Proteomic Changes in Candida albicans Biofilm in Response to Zosteric Acid Treatment. Int. J. Mol. Sci. 2022, 23, 14067. [Google Scholar] [CrossRef] [PubMed]
- Padder, S.A.; Padder, R.A.; Ramzan, A.; Bashir, G.; Tahir, I.; Rehman, R.U.; Shah, A.H. Glucose metabolic reprogramming and modulation in glycerol biosynthesis regulates drug resistance in clinical isolates of Candida. J. Appl. Microbiol. 2023, 134, lxad091. [Google Scholar] [CrossRef]
- Kocharunchitt, C.; King, T.; Gobius, K.; Bowman, J.P.; Ross, T. Global genome response of Escherichia coli O157:H7 Sakai during dynamic changes in growth kinetics induced by an abrupt downshift in water activity. PLoS ONE 2014, 9, e90422. [Google Scholar] [CrossRef]
- Bowman, J.P.; Hages, E.; Nilsson, R.E.; Kocharunchitt, C.; Ross, T. Investigation of the Listeria monocytogenes Scott A acid tolerance response and associated physiological and phenotypic features via whole proteome analysis. J. Proteome Res. 2012, 11, 2409–2426. [Google Scholar] [CrossRef] [PubMed]
- Fortuin, S.; Iradukunda, J.; Nel, A.J.; Blackburn, J.M.; Soares, N.C. Liquid chromatography mass spectrometry-based proteomics of Escherichia coli single colony. MethodsX 2021, 8, 101277. [Google Scholar] [CrossRef]
- Fortuin, S.; Nel, A.J.M.; Blackburn, J.M.; Soares, N.C. Comparison between the proteome of Escherichia coli single colony and during liquid culture. J. Proteom. 2020, 228, 103929. [Google Scholar] [CrossRef] [PubMed]
- Iradukunda, J.; Ganief, T.; Blackburn, J.M.; Soares, N.C. Mass Spectrometry-Based Analysis of Mycobacterial Single-Colony Proteome. Methods Mol. Biol. 2021, 2259, 181–189. [Google Scholar] [CrossRef]
- Kalule, J.B.; Fortuin, S.; Calder, B.; Robberts, L.; Keddy, K.H.; Nel, A.J.M.; Garnett, S.; Nicol, M.; Warner, D.F.; Soares, N.C.; et al. Proteomic comparison of three clinical diarrhoeagenic drug-resistant Escherichia coli isolates grown on CHROMagar™STEC media. J. Proteom. 2018, 180, 25–35. [Google Scholar] [CrossRef]
- Banerjee, M.; Thompson, D.S.; Lazzell, A.; Carlisle, P.L.; Pierce, C.; Monteagudo, C.; López-Ribot, J.L.; Kadosh, D.; Boone, M.E.C.; Senn, H.; et al. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 2008, 19, 1354–1365. [Google Scholar] [CrossRef]
- Zenati, R.A.; Giddey, A.D.; Al-Hroub, H.M.; Hagyousif, Y.A.; El-Huneidi, W.; Bustanji, Y.; Abu-Gharbieh, E.; Alqudah, M.A.Y.; Shara, M.; Abuhelwa, A.Y.; et al. Evaluation of Two Simultaneous Metabolomic and Proteomic Extraction Protocols Assessed by Ultra-High-Performance Liquid Chromatography Tandem Mass Spectrometry. Int. J. Mol. Sci. 2023, 24, 1354. [Google Scholar] [CrossRef] [PubMed]
- Ramagli, L.S. Quantifying protein in 2-D PAGE solubilization buffers. Methods Mol. Biol. 1999, 112, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, G.R. Chromosome-condensed G1 phase yeast cells are tolerant to desiccation stress. Microb. Cell 2022, 9, 42–51. [Google Scholar] [CrossRef]
- Ashrafi, K.; Sinclair, D.; Gordon, J.I.; Guarente, L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1999, 96, 9100–9105. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, P.; Chaturvedi, A.K.; Srinivasan, A.; Banerjee, M.; Ramasubramaniam, A.K.; Köhler, J.R.; Kadosh, D.; Lopez-Ribot, J.L. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010, 6, e1000828. [Google Scholar] [CrossRef]
- Douglas, L.M.; Wang, H.X.; Keppler-Ross, S.; Dean, N.; Konopka, J.B. Sur7 promotes plasma membrane organization and is needed for resistance to stressful conditions and to the invasive growth and virulence of Candida albicans. mBio 2012, 3, e00254-11. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Kobara, M.; Nakaya, T.; Imamura, H.; Fujii, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; et al. Raman Metabolomics of Candida auris Clades: Profiling and Barcode Identification. Int. J. Mol. Sci. 2022, 23, 11736. [Google Scholar] [CrossRef]
- Himmelreich, U.; Somorjai, R.L.; Dolenko, B.; Daniel, H.-M.; Sorrell, T.C. A rapid screening test to distinguish between Candida albicans and Candida dubliniensis using NMR spectroscopy. Fems Microbiol. Lett. 2005, 251, 327–332. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Uppuluri, P.; Chaffin, W.L. Defining Candida albicans stationary phase by cellular and DNA replication, gene expression and regulation. Mol. Microbiol. 2007, 64, 1572–1586. [Google Scholar] [CrossRef]
- Weerasekera, M.M.; Wijesinghe, G.K.; Jayarathna, T.A.; Gunasekara, C.P.; Fernando, N.; Kottegoda, N.; Samaranayake, L.P. Culture media profoundly affect Candida albicans and Candida tropicalis growth, adhesion and biofilm development. Mem. Inst. Oswaldo Cruz 2016, 111, 697–702. [Google Scholar] [CrossRef]
- Benomar, S.; Ranava, D.; Cárdenas, M.L.; Trably, E.; Rafrafi, Y.; Ducret, A.; Hamelin, J.; Lojou, E.; Steyer, J.-P.; Giudici-Orticoni, M.-T. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nat. Commun. 2015, 6, 6283. [Google Scholar] [CrossRef]
- Kapoore, R.V.; Padmaperuma, G.; Maneein, S.; Vaidyanathan, S. Co-culturing microbial consortia: Approaches for applications in biomanufacturing and bioprocessing. Crit. Rev. Biotechnol. 2022, 42, 46–72. [Google Scholar] [CrossRef]
- Sandini, S.; Stringaro, A.; Arancia, S.; Colone, M.; Mondello, F.; Murtas, S.; Girolamo, A.; Mastrangelo, N.; De Bernardis, F. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol. 2011, 11, 106. [Google Scholar] [CrossRef]
- Bergfeld, A.; Dasari, P.; Werner, S.; Hughes, T.R.; Song, W.-C.; Hortschansky, P.; Brakhage, A.A.; Hünig, T.; Zipfel, P.F.; Beyersdorf, N. Direct Binding of the pH-Regulated Protein 1 (Pra1) from Candida albicans Inhibits Cytokine Secretion by Mouse CD4+ T Cells. Front. Microbiol. 2017, 8, 844. [Google Scholar] [CrossRef]
- Aoki, W.; Tatsukami, Y.; Kitahara, N.; Matsui, K.; Morisaka, H.; Kuroda, K.; Ueda, M. Elucidation of potentially virulent factors of Candida albicans during serum adaptation by using quantitative time-course proteomics. J. Proteom. 2013, 91, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; and Cota, E. Candida albicans agglutinin-like sequence (Als) family vignettes: A review of Als protein structure and function. Front. Microbiol. 2016, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Kredich, N.M.; Hershfield, M.S. S-adenosylhomocysteine toxicity in normal and adenosine kinase-deficient lymphoblasts of human origin. Proc. Natl. Acad. Sci. USA 1979, 76, 2450–2454. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Mo, K.; Tian, G.; Zhou, J.; Gong, J.; Li, L.; Huang, X. Shikimic Acid Regulates the NF-κB/MAPK Signaling Pathway and Gut Microbiota to Ameliorate DSS-Induced Ulcerative Colitis. J. Agric. Food Chem. 2023, 71, 8906–8914. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, Y.; Nishida, T.; Nugraha, D.K.; Sugihara, F.; Horiguchi, Y. Melanin Produced by Bordetella parapertussis Confers a Survival Advantage to the Bacterium during Host Infection. mSphere 2021, 6, e0081921. [Google Scholar] [CrossRef]
- Sandini, S.; La Valle, R.; De Bernardis, F.; Macrì, C.; Cassone, A. The 65 kDa mannoprotein gene of Candida albicans encodes a putative beta-glucanase adhesin required for hyphal morphogenesis and experimental pathogenicity. Cell. Microbiol. 2007, 9, 1223–1238. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, F.F.M.; Paredes, V.; de Sousa, H.R.; D’Áurea Moura Á, N.; Riasco-Palacios, J.; Casadevall, A.; Felipe, M.S.S.; Nicola, A.M. Thioredoxin Reductase 1 Is a Highly Immunogenic Cell Surface Antigen in Paracoccidioides spp.; Candida albicans, and Cryptococcus neoformans. Front. Microbiol. 2019, 10, 2930. [Google Scholar] [CrossRef] [PubMed]
- Puri, S.; Kumar, R.; Chadha, S.; Tati, S.; Conti, H.R.; Hube, B.; Cullen, P.J.; Edgerton, M. Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS ONE 2012, 7, e46020. [Google Scholar] [CrossRef]
- Clancy, C.J.; Nguyen, M.L.; Cheng, S.; Huang, H.; Fan, G.; Jaber, R.A.; Wingard, J.R.; Cline, C.; Nguyen, M.H. Immunoglobulin G responses to a panel of Candida albicans antigens as accurate and early markers for the presence of systemic candidiasis. J. Clin. Microbiol. 2008, 46, 1647–1654. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jackson, B.E.; Mitchell, B.M.; Wilhelmus, K.R. Corneal virulence of Candida albicans strains deficient in Tup1-regulated genes. investig. Ophthalmol. Vis. Sci. 2007, 48, 2535–2539. [Google Scholar] [CrossRef]
- Braun, B.R.; Head, W.S.; Wang, M.X.; Johnson, A.D. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 2000, 156, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Song, J.L.; Lyons, C.N.; Holleman, S.; Oliver, B.G.; White, T.C. Antifungal activity of fluconazole in combination with lovastatin and their effects on gene expression in the ergosterol and prenylation pathways in Candida albicans. Med. Mycol. 2003, 41, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.H.; Yun, S.H.; Kim, M.J.; Kim, H.J.; Sung, B.H.; Kim, S.I.; Sohn, J.H. Secretome-based screening of fusion partners and their application in recombinant protein secretion in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2022, 106, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Millet, N.; Latgé, J.P.; Mouyna, I. Members of Glycosyl-Hydrolase Family 17 of A. fumigatus Differentially Affect Morphogenesis. J. Fungi 2018, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Sperschneider, J.; Win, J.; Kidd, B.; Yoshida, K.; Hane, J.; Saunders, D.G.O.; Singh, K.B. Comparative secretome analysis of Rhizoctonia solani isolates with different host ranges reveals unique secretomes and cell death inducing effectors. Sci. Rep. 2017, 7, 10410. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Nett, J.E.; Zarnowski, R.; Ross, K.M.; Sanchez, H.; Cain, M.T.; Hamaker, J.; Mitchell, A.P.; Andes, D.R. A Candida biofilm-induced pathway for matrix glucan delivery: Implications for drug resistance. PLoS Pathog. 2012, 8, e1002848. [Google Scholar] [CrossRef]
- Yuan, X.; Mitchell, B.M.; Hua, X.; Davis, D.A.; Wilhelmus, K.R. The RIM101 signal transduction pathway regulates Candida albicans virulence during experimental keratomycosis. investig. Ophthalmol. Vis. Sci. 2010, 51, 4668–4676. [Google Scholar] [CrossRef][Green Version]
- McCreath, K.J.; Specht, C.A.; Robbins, P.W. Molecular cloning and characterization of chitinase genes from Candida albicans. Proc. Natl. Acad. Sci. USA 1995, 92, 2544–2548. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Summers, E.; Guo, B.; Fink, G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 1999, 181, 6339–6346. [Google Scholar] [CrossRef] [PubMed]
- Glory, A.; van Oostende, C.T.; Geitmann, A.; Bachewich, C. Depletion of the mitotic kinase Cdc5p in Candida albicans results in the formation of elongated buds that switch to the hyphal fate over time in a Ume6p and Hgc1p-dependent manner. Fungal Genet. Biol. 2017, 107, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Valim, C.X.; Basso, L.R., Jr.; dos Reis Almeida, F.B.; Reis, T.F.; Damásio, A.R.; Arruda, L.K.; Martinez, R.; Roque-Barreira, M.C.; Oliver, C.; Jamur, M.C.; et al. Characterization of PbPga1, an antigenic GPI-protein in the pathogenic fungus Paracoccidioides brasiliensis. PLoS ONE 2012, 7, e44792. [Google Scholar] [CrossRef]
- Snelders, E.; Moyrand, F.; Sturny-Leclère, A.; Vernel-Pauillac, F.; Volant, S.; Janbon, G.; Alanio, A. The role of glycosylphosphatidylinositol (gpi) anchored proteins in Cryptococcusneoformans. Microbes Infect. 2022, 24, 105016. [Google Scholar] [CrossRef]
- Aghaei Gharehbolagh, S.; Mafakher, L.; Salehi, Z.; Asgari, Y.; Hashemi, S.J.; Mahmoudi, S.; Nasimi, M.; Rezaie, S. Unveiling the structure of GPI-anchored protein of Malassezia globosa and its pathogenic role in pityriasis versicolor. J. Mol. Model. 2021, 27, 246. [Google Scholar] [CrossRef]
- Norice, C.T.; Smith, F.J., Jr.; Solis, N.; Filler, S.G.; Mitchell, A.P. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 2007, 6, 2046–2055. [Google Scholar] [CrossRef]
- Hiller, E.; Heine, S.; Brunner, H.; Rupp, S. Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryot. Cell 2007, 6, 2056–2065. [Google Scholar] [CrossRef]
- Ostrander, D.B.; Gorman, J.A. Isolation and characterization of the Candida albicans PFY1 gene for profilin. Yeast 1997, 13, 871–880. [Google Scholar] [CrossRef]
- Ueno, K.; Tamura, Y.; Chibana, H. Target validation and ligand development for a pathogenic fungal profilin, using a knock-down strain of pathogenic yeast Candida glabrata and structure-based ligand design. Yeast 2010, 27, 369–378. [Google Scholar] [CrossRef]
- Marvin, M.E.; Williams, P.H.; Cashmore, A.M. The Candida albicans CTR1 gene encodes a functional copper transporter. Microbiology (Reading) 2003, 149 Pt 6, 1461–1474. [Google Scholar] [CrossRef]
- Sosnowski, P.; Urnavicius, L.; Boland, A.; Fagiewicz, R.; Busselez, J.; Papai, G.; Schmidt, H. The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1. Elife 2018, 7, 39163. [Google Scholar] [CrossRef]
- Buckman, J.; Miller, S.M. Binding and reactivity of Candida albicans estrogen binding protein with steroid and other substrates. Biochemistry 1998, 37, 14326–14336. [Google Scholar] [CrossRef] [PubMed]
- Madani, N.D.; Malloy, P.J.; Rodriguez-Pombo, P.; Krishnan, A.V.; Feldman, D. Candida albicans estrogen-binding protein gene encodes an oxidoreductase that is inhibited by estradiol. Proc. Natl. Acad. Sci. USA 1994, 91, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, S.; Zhao, Y.; Zhang, Y.; Lv, Y.; Jiang, Y.; Wang, Y.; Li, D.; Zhang, H. ADH1 promotes Candida albicans pathogenicity by stimulating oxidative phosphorylation. Int. J. Med. Microbiol. 2019, 309, 151330. [Google Scholar] [CrossRef]
- Fanning, S.; Xu, W.; Beaurepaire, C.; Suhan, J.P.; Nantel, A.; Mitchell, A.P. Functional control of the Candida albicans cell wall by catalytic protein kinase A subunit Tpk1. Mol. Microbiol. 2012, 86, 284–302. [Google Scholar] [CrossRef] [PubMed]
- Monroy-Pérez, E.; Sáinz-Espuñes, T.; Paniagua-Contreras, G.; Negrete-Abascal, E.; Rodríguez-Moctezuma, J.R.; Vaca, S. Frequency and expression of ALS and HWP1 genotypes in Candida albicans strains isolated from Mexican patients suffering from vaginal candidosis. Mycoses 2012, 55, e151–e157. [Google Scholar] [CrossRef]
- Aoki, W.; Kitahara, N.; Miura, N.; Morisaka, H.; Kuroda, K.; Ueda, M. Profiling of adhesive properties of the agglutinin-like sequence (ALS) protein family, a virulent attribute of Candida albicans. FEMS Immunol. Med. Microbiol. 2012, 65, 121–124. [Google Scholar] [CrossRef]
- Lee, S.H.; Chung, S.C.; Shin, J.; Oh, K.B. GST2 is required for nitrogen starvation-induced filamentous growth in Candida albicans. J. Microbiol. Biotechnol. 2014, 24, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Rane, H.S.; Bernardo, S.M.; Hayek, S.R.; Binder, J.L.; Parra, K.J.; Lee, S.A. The contribution of Candida albicans vacuolar ATPase subunit V₁B, encoded by VMA2, to stress response, autophagy, and virulence is independent of environmental pH. Eukaryot. Cell 2014, 13, 1207–1221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hargrove, T.Y.; Friggeri, L.; Wawrzak, Z.; Qi, A.; Hoekstra, W.J.; Schotzinger, R.J.; York, J.D.; Guengerich, F.P.; Lepesheva, G.I. Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. [Google Scholar] [CrossRef] [PubMed]
- Shyadehi, A.Z.; Lamb, D.C.; Kelly, S.L.; Kelly, D.E.; Schunck, W.H.; Wright, J.N.; Corina, D.; Akhtar, M. The mechanism of the acyl-carbon bond cleavage reaction catalyzed by recombinant sterol 14 alpha-demethylase of Candida albicans (other names are: Lanosterol 14 alpha-demethylase, P-45014DM, and CYP51). J. Biol. Chem. 1996, 271, 12445–12450. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Hsp21 potentiates antifungal drug tolerance in Candida albicans. PLoS ONE 2013, 8, e60417. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Jacobsen, I.D.; Miramón, P.; Slesiona, S.; Bohovych, I.M.; Brown, A.J.P.; Hube, B. Small but crucial: The novel small heat shock protein Hsp21 mediates stress adaptation and virulence in Candida albicans. PLoS ONE 2012, 7, e38584. [Google Scholar] [CrossRef]
- Orhan, H.; Vermeulen, N.P.; Tump, C.; Zappey, H.; Meerman, J.H. Simultaneous determination of tyrosine, phenylalanine and deoxyguanosine oxidation products by liquid chromatography-tandem mass spectrometry as non-invasive biomarkers for oxidative damage. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2004, 799, 245–254. [Google Scholar] [CrossRef]
- Rocha, R.O.; Wilson, R.A. Essential, deadly, enigmatic: Polyamine metabolism and roles in fungal cells. Fungal Biol. Rev. 2019, 33, 47–57. [Google Scholar] [CrossRef]
- Urbaniak, M.M.; Gazińska, M.; Rudnicka, K.; Płociński, P.; Nowak, M.; Chmiela, M. In Vitro and In Vivo Biocompatibility of Natural and Synthetic Pseudomonas aeruginosa Pyomelanin for Potential Biomedical Applications. Int. J. Mol. Sci. 2023, 24, 7846. [Google Scholar] [CrossRef]
- Bode, R.; Melo, C.; Birnbaum, D. Mode of action of glyphosate in Candida maltosa. Arch. Microbiol. 1984, 140, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Kuemmerle, J.F.; Kellum, J.M. Serotonin neural receptors mediate motilin-induced motility in isolated, vascularly perfused canine jejunum. J. Surg. Res. 1988, 45, 357–362. [Google Scholar] [CrossRef]
- Didehdar, M.; Chegini, Z.; Tabaeian, S.P.; Razavi, S.; Shariati, A. Cinnamomum: The New Therapeutic Agents for Inhibition of Bacterial and Fungal Biofilm-Associated Infection. Front. Cell. Infect. Microbiol. 2022, 12, 930624. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.E.; Zurita-Lopez, C.; Regis, A.; Blum, E.; Conboy, A.; Elf, S.; Clarke, S. Protein arginine methylation in Candida albicans: Role in nuclear transport. Eukaryot. Cell 2007, 6, 1119–1129. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Xin, J.; Chen, X.; Xu, T.; He, J.; Pan, Z.; Zhang, C. Metabolomic profiles of the liquid state fermentation in co-culture of Eurotium amstelodami and Bacillus licheniformis. Front. Microbiol. 2023, 14, 1080743. [Google Scholar] [CrossRef]
- Lyte, M.; Arulanandam, B.; Nguyen, K.; Frank, C.; Erickson, A.; Francis, D. Norepinephrine induced growth and expression of virulence associated factors in enterotoxigenic and enterohemorrhagic strains of Escherichia coli. Adv. Exp. Med. Biol. 1997, 412, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Janiak, A.; Cybulska, B.; Szlinder-Richert, J.; Borowski, E.; Milewski, S. Facilitated diffusion of glucosamine-6-phosphate synthase inhibitors enhances their antifungal activity. Acta Biochim. Pol. 2002, 49, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Bode, R. Characterization of a novel enzyme, N6-acetyl-L-lysine: 2-oxoglutarate aminotransferase, which catalyses the second step of lysine catabolism in Candida maltosa. Antonie Van. Leeuwenhoek 1992, 62, 285–290. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhameed, R.A.; Semreen, M.H.; Hamad, M.; Giddey, A.D.; Sulaiman, A.; Al Bataineh, M.T.; Al-Hroub, H.M.; Bustanji, Y.; Alzoubi, K.H.; Soares, N.C. Multi-Omics Profiling of Candida albicans Grown on Solid Versus Liquid Media. Microorganisms 2023, 11, 2831. https://doi.org/10.3390/microorganisms11122831
Alhameed RA, Semreen MH, Hamad M, Giddey AD, Sulaiman A, Al Bataineh MT, Al-Hroub HM, Bustanji Y, Alzoubi KH, Soares NC. Multi-Omics Profiling of Candida albicans Grown on Solid Versus Liquid Media. Microorganisms. 2023; 11(12):2831. https://doi.org/10.3390/microorganisms11122831
Chicago/Turabian StyleAlhameed, Rouba Abdulsalam, Mohammad H. Semreen, Mohamad Hamad, Alexander D. Giddey, Ashna Sulaiman, Mohammad T. Al Bataineh, Hamza M. Al-Hroub, Yasser Bustanji, Karem H. Alzoubi, and Nelson C. Soares. 2023. "Multi-Omics Profiling of Candida albicans Grown on Solid Versus Liquid Media" Microorganisms 11, no. 12: 2831. https://doi.org/10.3390/microorganisms11122831
APA StyleAlhameed, R. A., Semreen, M. H., Hamad, M., Giddey, A. D., Sulaiman, A., Al Bataineh, M. T., Al-Hroub, H. M., Bustanji, Y., Alzoubi, K. H., & Soares, N. C. (2023). Multi-Omics Profiling of Candida albicans Grown on Solid Versus Liquid Media. Microorganisms, 11(12), 2831. https://doi.org/10.3390/microorganisms11122831






