Abstract
Notch signaling manipulates the function and phenotype of dendritic cells (DCs), as well as the interaction between DCs and CD4+ T cells. However, the role of Notch signaling in Helicobacter pylori (H. pylori) infection remains elusive. Murine bone marrow-derived dendritic cells (BMDCs) were pretreated in the absence or presence of Notch signaling inhibitor DAPT prior to H. pylori stimulation and the levels of Notch components, cytokines and surface markers as well as the differentiation of CD4+ T cells in co-culture were measured using quantitative real-time PCR (qRT-PCR), Western blot, enzyme-linked immunosorbent assay (ELISA) and flow cytometry. Compared with the control, the mRNA expression of all Notch receptors and Notch ligands Dll4 and Jagged1 was up-regulated in H. pylori-stimulated BMDCs. The blockade of Notch signaling by DAPT influenced the production of IL-1β and IL-10 in H. pylori-pulsed BMDCs, and reduced the expression of Notch1, Notch3, Notch4, Dll1, Dll3 and Jagged2. In addition, DAPT pretreatment decreased the expression of maturation markers CD80, CD83, CD86, and major histocompatibility complex class II (MHC-II) of BMDCs, and further skewed Th17/Treg balance toward Treg. Notch signaling regulates the function and phenotype of DCs, thus mediating the differentiation of CD4+ T cells during H. pylori infection.
1. Introduction
Helicobacter pylori (H. pylori) is a micro-aerobic, spiral-shaped Gram-negative bacterium that mainly colonizes human gastric mucosa and easily causes various gastrointestinal diseases such as gastritis, peptic ulcers and even gastric cancer. H. pylori was listed as Class I carcinogen by World Health Organization in 1994 []. At present, over 50% of the world’s population is infected with H. pylori []. Although the development of antibiotics combined with proton pump inhibitors has achieved an excellent therapeutic effect in controlling infection, drug resistance and immune escape [] remain two major problems in the clearance of H. pylori. In general, H. pylori infection induces a robust immune response in the host, but this process is not enough to completely eliminate the pathogen, and can even lead to lifelong infection. Thus, the prevention and treatment of H. pylori infection is still a major public health problem to be settled urgently. Therefore, it is necessary to explore the pathogenesis and immune response mechanisms in H. pylori infection and develop novel therapeutic strategies for controlling the infection.
Innate immunity is the first line of defense against H. pylori. It has been acknowledged that dendritic cells (DCs) are key members of innate immunity and the most powerful antigen-presenting cells (APCs) that act as messengers between innate and adaptive immune responses []. Several studies have demonstrated that DCs play an essential role in the immune responses caused by H. pylori infection [,]. The interaction between DCs and CD4+ T cells induced CD4+ T cells differentiation into Th1, Th2, Th17 and Regulatory T cells (Tregs), which determine the progress and prognosis of diseases [,]. Furthermore, cytokines, such as IL-6, IL-8, IL-12 and IL-23 produced by DCs, were up-regulated after H. pylori stimulation, which formed a micro-environment that influenced the differentiation of CD4+ T cells [,,]. In addition, compared with uninfected individuals, a large amount of immune-tolerogenic DCs were detected in samples from H. pylori-infected mice and human gastric mucosa [,]. Collectively, H. pylori affects the immune responses of CD4+ T cells via altering the function and phenotype of DCs in order to survive. However, the mechanism underlying this process has not been fully elucidated. Therefore, it is of great significance to explore the changes in DC function and phenotype during H. pylori infection, laying the foundation for further studies on the mechanism through which DCs regulates CD4+ T cell differentiation into different T helper (Th) cell subtypes.
Accumulating evidence has proved that Notch signaling is involved in the activation and regulation of immune cells in various diseases [,]. Notch signaling is a key player in modulating the function and phenotype of DCs as well as the interaction between DCs and CD4+ T cells [,]. In mammals, Notch family consists of four Notch receptors (Notch1–Notch4) and five ligands (Delta Like 1 (Dll1), Dll3, Dll4, Jagged1 and Jagged2) []. The binding of ligands to receptors triggers proteolytic cleavage of Notch receptors by γ-secretase, and Notch intracellular domains (NICDs) are subsequently released into cytoplasm, and then migrate into the nucleus to form a transcriptional complex that activates downstream target genes, thus regulating cell proliferation, differentiation, fate determination, diseases development and so on [,]. Previous studies showed that Notch ligands Dll1, Dll4 and Jagged1 were highly expressed in DCs [,,], and Notch signaling was essential in DC maturation and DC-mediated T cell response [,]. Moreover, Notch ligands Dll1 and Jagged1 altered the phenotype of DCs and affected the secretion of cytokines and chemokines by DCs []. Another study revealed that the Delta-like ligands promoted Th1 or Th17 cell polarization, while the Jagged ligands induced Treg or Th2 cell differentiation []. Our previous study confirmed that Notch1 was involved in regulating CD4+ T cell differentiation into Th1 cells, a subtype considered to be protective in H. pylori-infected patients []. In addition, Jagged1 augmented the anti-H. pylori activity of macrophages by increasing the production of pro-inflammatory factors []. However, the detailed mechanisms through which Notch signaling regulates the function and phenotype of DCs and further affects the interaction with CD4+ T cells have not been reported.
In this study, we demonstrated that H. pylori induced the expression of all Notch receptors and Notch ligands Dll4 and Jagged1 in BMDCs. Blockade of Notch signaling with γ-secretase inhibitor (N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester, DAPT) prior to H. pylori infection influenced the expression of Notch components and the production of cytokines in BMDCs. Besides, DAPT pretreatment decreased the expression of maturation markers CD80, CD83, CD86, and MHC-II of BMDCs, and further skewed the Th17/Treg balance toward Treg. Overall, our findings suggested that Notch signaling regulated the function and maturation of DCs and influenced the differentiation of CD4+ T cells during H. pylori infection. These data will provide insight for elucidating immune defense mechanisms in H. pylori infection and developing novel therapies for controlling the infection.
2. Materials and Methods
2.1. H. pylori Culture
H. pylori Sydney strain (SS1) was cultured on H. pylori agar basal medium containing 7% sheep blood, supplemented with 0.33 μg/mL polymyxin B sulfate, 5 μg/mL amphotericin B, 5 µg/mL trimethoprim, 5 μg/mL cefsulodin sodium and 10 μg/mL vancomycin hydrochloride at 37 °C in a micro-aerobic environment for 48 h, and then bacterial colonies were collected and resuspended with sterilized phosphate-buffered saline (PBS) for subsequent experiments.
2.2. Generation of Murine Bone Marrow-Derived DCs
Female C57BL/6 mice (6–8 weeks old) were provided by the Laboratory Animal Center, Southern Medical University. Bone marrow cells isolated from the femora and tibiae of mice were cultured in RPMI-1640 medium supplemented with 20 ng/mL murine GM-CSF (Peprotech, Rocky Hill, NJ, USA), 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin to obtain bone marrow-derived DCs (BMDCs) as previously described [].
2.3. Stimulation of BMDCs with H. pylori
BMDCs were seeded on 6-well plates at a density of 1 × 106/mL and cultured in fresh medium supplemented with 10% fetal bovine serum at 37 °C in 5% CO2. H. pylori was directly added into the medium at a multiplicity of infection (MOI) of 50 CFU/cell as previously described [], and incubated with BMDCs for 24 h. In the control group, an equivalent volume of PBS was added into the uninfected wells. To block Notch signaling, BMDCs were pretreated with DAPT (20 μM) or equal volume of DMSO for 24 h before H. pylori stimulation.
2.4. Co-Culture of BMDCs and CD4+ T Cells
According to the methods of co-culture described previously [,,], splenic CD4+ T cells from syngeneic C57BL/6 mice were selected using anti-CD4-MicroBeads (Miltenyi, Bergisch Gladbach, Germany). BMDCs treated as above were harvested and in the presence of Mitomycin C (20 μg/mL) for 1.5 h followed by washing twice to remove bacteria. Then, BMDCs were collected and co-cultured with CD4+ T cells at a ratio of 1:10 for 72 h.
2.5. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from cells using TRIzol (Takara, Dalian, China). Complementary DNA (cDNA) was generated from 1 μg of total RNA using a cDNA synthesis kit (EZBioscience, Roseville, MN, USA). qRT-PCR was performed to detect target genes. Amplification was performed in a 10 μL reaction volume under the conditions of one cycle of initial denaturation at 95 °C for 5 min, 95 °C for 10 s and 60 °C for 30 s, followed by 40 cycles to amplify target genes using 2× SYBR Green Color qTR-PCR Mix (EZbiosicence, Roseville, MN, USA).Cycle thresholds obtained were normalized to β-Actin, and the detection of target genes was conducted in triplicate. The relative expression of target genes was calculated using the comparative threshold cycle (Ct) method, 2−ΔΔCT []. The sequences of primers are listed in Table 1.

Table 1.
Primer sequences for qRT-PCR in this study.
2.6. Protein Extraction and Western Blot
Western blot was performed to measure the protein level of Notch ligands Dll1, Dll4 and Jagged1 according to a previous study []. Total protein was extracted from cells using RIPA lysis (Genstar, Beijing, China) containing 1 mM Phenylmethylsulfonyl fluoride (PMSF). Subsequently, the concentration of total protein was measured using a Bicinchoninic Acid Kit (Fdbio science, Hangzhou, China). Protein samples were loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at 20 μg, and then transferred onto polyvinylidene difluoride (PVDF) membranes (MerckMillipore, Burlington, MA, USA). The primary and secondary antibodies are listed in Table 2. Finally, Super ECL Detection Reagent (Yeasen Biotechnology Co., Ltd., Shanghai, China) was added to the PVDF membranes, and the protein bands were detected with a visualizer.

Table 2.
Antibodies used for Western blot in this study.
2.7. ELISA
Referring to the method described previously [], culture supernatants of BMDCs were collected and assayed for IL-1β, IL-6, IL-12p70, TNF-α, IL-10, and TGF-β using ELISA kits (CUSABIO, Wuhan, China) according to the manufacturer’s instructions.
2.8. Flow Cytometry
Flow cytometry was used to assess the expression of DC surface markers including CD11c, MHC-II, CD80, CD83, CD86 and Jagged1. BMDCs were resuspended with PBS, treated with FcR blocking reagent, and incubated at 4 °C for 15 min in the dark. Subsequently, BMDCs were incubated with antibodies (BD Biosciences & Biolegend, San Diego, CA, USA) against the surface markers, as above, at 4 °C for 30 min. Finally, samples were detected with a flow cytometer and analyzed using FlowJo vX software.
2.9. Statistical Analysis
The data were analyzed using GraphPad Prism 6.0 software and presented as mean ± SD (mean ± SD, n = 3). All experiments were repeated at least three times for each treatment group. Comparisons between groups were assessed using a two-sample Student’s t-test. Differences were considered statistically significant when p values < 0.05 (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, N.S: no statistical difference).
3. Results
3.1. H. pylori Induced the Expression of All Notch Receptors and Notch Ligands Dll4 and Jagged1 in BMDCs
Our previous studies demonstrated that Notch1 was involved in Th1 cell differentiation [] and Jagged1 enhanced macrophage bactericidal activity in response to H. pylori infection []. To further extend the results, we measured the expression of Notch components in H. pylori-stimulated BMDCs using qRT-PCR. It was observed that all Notch receptors were highly expressed (Figure 1A). As for Notch ligands, H. pylori significantly induced the expression of Dll4 and Jagged1 at the mRNA level, and slightly decreased Dll1, but there were no statistical differences in the genes of Dll3 and Jagged2 (Figure 1B). For DCs, scientists focus on Notch ligands, and recent studies have proved that Dll1, Dll4, and Jagged1 are involved in activating DCs []. Therefore, we detected the protein expression of Dll1, Dll4 and Jagged1 using Western blot. Only Jagged1 was increased remarkably, while Dll1 and Dll4 were not (Figure 1C). Subsequently, we further examined the mRNA expression of Jagged1 in H. pylori-infected BMDCs at different time points and MOI. The expression of Jagged1 was the highest at 16 h and MOI 20. Likewise, the level and percentage of Jagged 1 were elevated, as determined using flow cytometry (Figure 1E,F), while Dll4 was too low to be detected. These results indicate that the expression of Notch components in DCs has been altered by H. pylori, and Jagged1 may play an essential role.
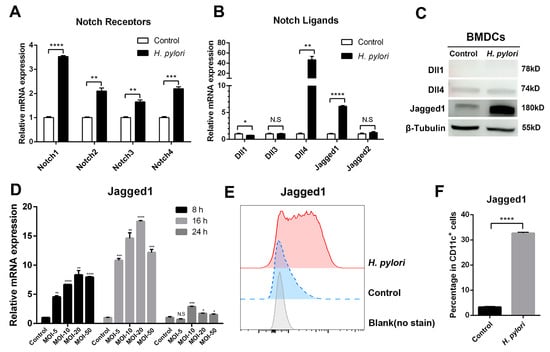
Figure 1.
Expression profile of Notch molecules in BMDCs during H. pylori infection. BMDCs were stimulated with H. pylori (MOI 50) for 24 h. PBS was used as control for H. pylori. qRT-PCR was performed to assess the mRNA expression of (A) Notch receptors and (B) Notch ligands in BMDCs. Relative expression is normalized to β-Actin. (C) Western blot was performed to assess the protein levels of Dll1, Dll4 and Jagged1 in BMDCs. β-Tubulin was used as loading control. (D) BMDCs were stimulated with different MOI (5, 10, 20 and 50) of H. pylori for 8 h, 16 h or 24 h, and qRT-PCR was performed to assess the mRNA level of Jagged1. Flow cytometry was performed to measure the level of Jagged1 on BMDCs stimulated with H. pylori (MOI 20) for 16 h, and (E) the representative histogram plot and (F) the percentage were illustrated. The data are presented as the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, N.S: no statistical difference.
3.2. Notch Signaling Was Involved in the Regulation of Cytokines in H. pylori-Infected BMDCs
As one of the first defenders, DCs influence the local immune micro-environment via secreting cytokines, thus regulating innate and adaptive immune responses. Meanwhile, the expression of cytokines could be affected by Notch components [,]. To explore whether Notch signaling is involved in the expression of cytokines in H. pylori-pulsed BMDCs, cells were pretreated in the absence or presence of Notch signaling inhibitor DAPT, and the expression of pro-inflammatory and anti-inflammatory cytokines was determined using ELISA. It was observed that H. pylori induced the up-regulation of IL-1β and IL-10, and DAPT treatment increased IL-1β but decreased IL-10 (Figure 2A). Since the above experiments have demonstrated that H. pylori induced a change in Notch receptors and ligands in BMDCs, we further detected whether DAPT influences the profile of Notch components using qRT-PCR. It was observed that DAPT pretreatment in BMDCs prior to H. pylori stimulation decreased the levels of Notch1, Notch3, and Notch4, as well as Dll1, Dll3 and Jagged2, whereas Dll4 and Jagged1 were still on the rise (Figure 2B,C). This may explain why DAPT treatment leads to changes in the diversity of cytokine levels. Taken together, the results suggest that Notch signaling is involved in the regulation of cytokines, and different Notch components may exert specific effects in BMDCs during H. pylori infection.
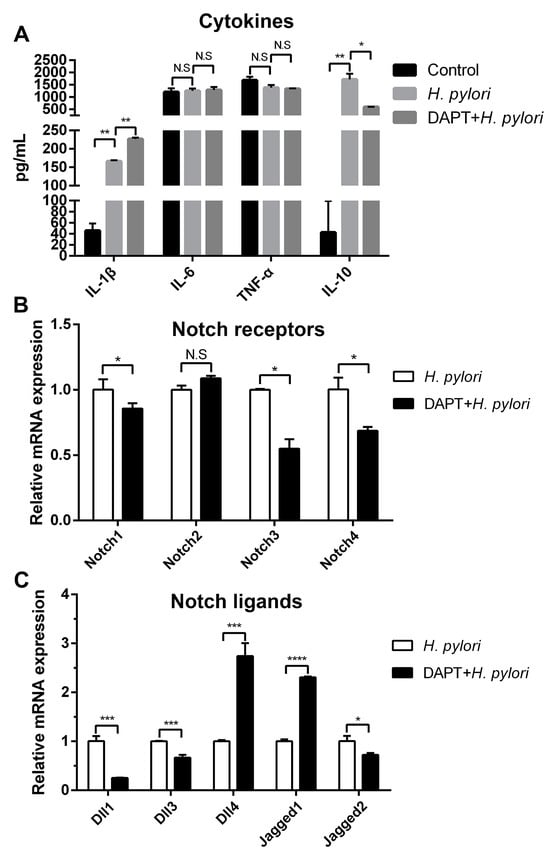
Figure 2.
Inhibition of Notch signaling influenced the production of cytokines in BMDCs during H. pylori infection. BMDCs were pretreated with DAPT (20 μM) for 24 h, and then stimulated with H. pylori (MOI 50) for 24 h. DMSO was used as control for DAPT. (A) ELISA was used to determine the level of IL-1β, IL-6, TNF-α and IL-10. qRT-PCR was performed to examine the mRNA expression of (B) Notch receptors and (C) Notch ligands. Relative expression is normalized to β-Actin. The data are presented as the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01 and *** p < 0.001, **** p < 0.0001, N.S: no statistical difference.
3.3. Notch Signaling Was Involved in Regulating the Phenotype of BMDCs during H. pylori Infection
The expression of maturation markers is up-regulated in tissue-resident immature DCs upon pathogen stimulation []. To verify the effect of Notch signaling on the phenotype of DCs in H. pylori infection, BMDCs were pretreated with or without DAPT prior to H. pylori stimulation, and surface markers were determined using flow cytometry. H. pylori induced the elevation of CD80, CD83, CD86, and MHC-II, whereas DAPT pretreatment resulted in a sharp decrease in those four surface markers (Figure 3), indicating that Notch signaling influences the maturation and phenotype of DCs during H. pylori infection.
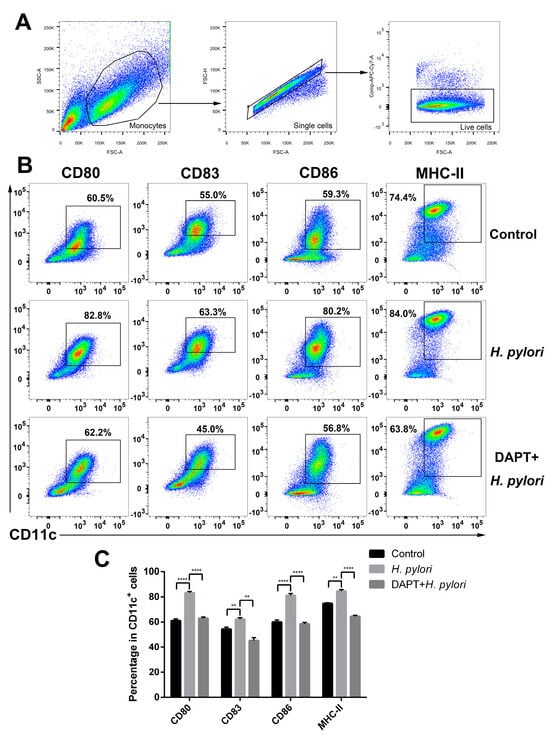
Figure 3.
Inhibition of Notch signaling decreased the expression of maturation markers on BMDCs during H. pylori infection. BMDCs were pretreated with DAPT (20 μM) for 24 h, and then stimulated with H. pylori (MOI 50) for another 24 h. Naïve BMDCs were used as uninfected controls. DMSO was used as a control for DAPT. (A,B) Representative dot plots and the gating strategy of CD11c+ BMDCs are shown. Flow cytometry was performed to evaluate the expression of CD80, CD83, CD86 and MHC-II on BMDCs. (C) Percentages of BMDCs expressing CD80, CD83, CD86 or MHC-II are shown. The data are presented as the mean ± SD of three experiments. ** p < 0.01 and **** p < 0.0001.
3.4. Inhibition of Notch Signaling in BMDCs Shifted the Th17/Treg Balance toward Treg of CD4+ T Cells
Previous studies have shown that Notch signaling is involved in the regulation of DC activation, thereby affecting DC-induced CD4+ T cell differentiation in inflammatory and infectious diseases []. However, whether Notch signaling plays a role in DC-medicated Th differentiation during H. pylori infection has not been reported. To target Notch signaling, BMDCs were pretreated with or without DAPT before H. pylori infection, and then co-cultured with CD4+ T cells generated from syngeneic C57BL/6 mice. The expression of characteristic transcription factors and hallmark cytokines of Th1 (Tbx21, Ifnγ), Th2 (Gata3, Il4), Th17 (Rorγt, Il17A and Il17F) and Treg (Foxp3, Tgfβ and Il10) was determined using qRT-PCR. The results showed that Rorγt, Il17A, and Il17F were simultaneously up-regulated after H. pylori stimulation (Figure 4D), while the transcription factors and cytokines of other Th cells decreased (Figure 4A,B,E). Except for Il17A, DAPT pretreatment partially reversed the changes induced by H. pylori. In addition, the ratio of Tbx21/Gata3 was down-regulated and the ratio of Rorγt/Foxp3 was up-regulated after H. pylori infection, whereas DAPT pretreatment reversed this change (Figure 4C,F). Subsequently, we performed flow cytometry to further detect the differentiation of CD4+ T cells (Figure 5A). In line with the mRNA level, the percentage of Th1, Th2, and Treg was down-regulated in H. pylori-pulsed BMDC group (Figure 5B). DAPT pretreatment elevated the percentage of Treg and had no effect on Th1 and Th2 (Figure 5B), which accounted for the decreased ratio of Th17/Treg (Figure 5C). Taken together, DAPT pretreatment in BMDCs shifted Th17/Treg balance toward Treg, revealing an important role of Notch signaling in CD4+ T cell differentiation during H. pylori infection.
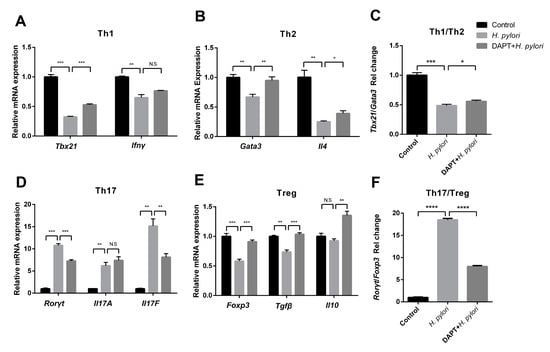
Figure 4.
The mRNA expression profile of Th cells was reversed by DAPT in the co-culture system of H. pylori-infected BMDCs and CD4+ T cells. BMDCs pretreated in the presence or absence of DAPT (20 μM, 24 h) were stimulated with H. pylori (MOI 50, 24 h) and then co-cultured with splenic CD4+ T cells from syngeneic C57BL/6 mice for 72 h. The mRNA expression of characteristic transcription factors and cytokines of (A) Th1 (Tbx21, Ifnγ), (B) Th2 (Gata3, Il4), (D) Th17 (Rorγt, Il17A, Il17F) and (E) Treg (Foxp3, Tgfβ, Il10) was assessed using qRT-PCR. (C) The ratio of Tbx21/Gata3 and (F) Rorγt/Foxp3 is shown. Relative expression is normalized to β-Actin. The data are presented as the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01 and *** p < 0.001, **** p < 0.0001, N.S: no statistical difference.
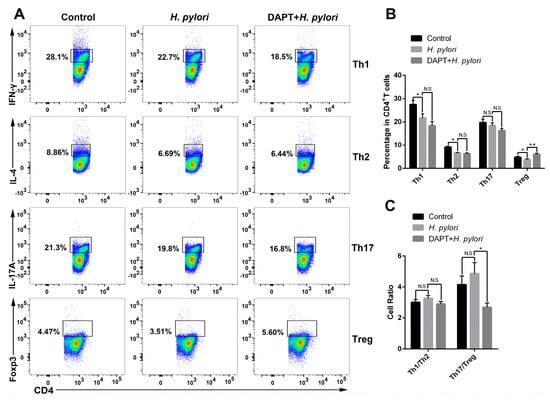
Figure 5.
DAPT-pretreated BMDCs skewed Th17/Treg balance toward Treg. BMDCs pretreated in the presence or absence of DAPT (20 μM, 24 h) were stimulated with H. pylori (MOI 50, 24 h) and then co-cultured with splenic CD4+ T from syngeneic C57BL/6 mice for 72 h. Flow cytometric analyses were performed to measure the differentiation of CD4+ T cells. (A) Representative dot plots of Th1 (IFN-γ), Th2 (IL-4), Th17 (IL-17A) and Treg (Foxp3) are shown. (B) Percentages of Th1, Th2, Th17 and Treg and (C) the ratio of Th1/Th2 and Th17/Treg are shown. The data are presented as the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01, and N.S: no statistical difference.
4. Discussions
In recent decades, Notch signaling has been proven to be involved in the immune system, regulating T cell and B cell development, myeloid lineage commitment [], and Th cell differentiation []. As for DCs, accumulating evidence has shown that Notch signaling affects DC development, maturation, and differentiation in vivo, [,] and is closely related to a variety of pathogen infections []. Meanwhile, Notch components are widely expressed in DCs, and different stimuli induce DCs to produce different expression profiles of receptors and ligands []. In this study, the mRNA level of all Notch receptors was elevated in BMDCs pulsed with H. pylori. As previously demonstrated, different ligands could activate distinct target programs through the same Notch receptor []. Among the five ligands of Notch signaling, each ligand has its own preference for Notch receptors, and the ligation of different ligands leads to different Notch intracellular domains (NICDs)’ translocation into the nucleus, resulting in activation of various downstream target genes []. Thus, different combinations of ligands and receptors have diverse effects. The Notch ligand Dll4 was increased in DCs infected with mycobacteria [] and respiratory syncytial virus (RSV) []. Jagged1 was up-regulated after LPS [] and Curdlan [] stimulation. In accordance, our results showed that Dll4 and Jagged1 were elevated in H. pylori-infected BMDCs at mRNA level, but only Jagged1 showed a remarkable increase at protein level, consistent with our previous study in which the expression of Dll1, Dll4 and Jagged1 was significantly up-regulated in H. pylori-infected macrophages at the RNA level, but only Jagged1 was increased remarkably at the protein level [], indicating that the mRNA and protein expression of Notch ligands are not always consistent. Therefore, we suppose that Jagged1 plays a crucial role in H. pylori infection.
Accumulating evidence has indicated that Notch signaling is involved in the modulation of cytokine secretion, thereby orchestrating the immune response []. Notch signaling was activated in M. bovis BCG pulsed-DCs, and enhanced the expression of pro-inflammatory cytokines TNF-α, IL-1β, and IL-6, whereas Notch signaling inhibitor DAPT hampered the production of TNF-α in plasmacytoid DCs and decreased the level of IL-10 in conventional DCs []. Similarly, DAPT reversed the elevation of TNF-α and IL-6 induced by P. brasiliensis []. In our study, the expression of IL-1β and IL-10 was increased in H. pylori-pulsed BMDCs, whereas DAPT treatment dramatically inhibited the production of IL-10, but further increased the expression of IL-1β, with little impact on TNF-α and IL-6. Unfortunately, there was no detectable IL-12, consistent with a previous study in which H. pylori-secreted factors inhibited IL-12 secretion of DCs []. The failure of TGF-β detection may be due to the low expression of TGF-β and the insufficient detection sensitivity of the kit. Under normal physiological conditions, pro-inflammatory and anti-inflammatory responses are in a state of dynamic balance. However, H. pylori infection breaks the balance, inducing high levels of pro-inflammatory cytokines. Meanwhile, excessive inflammatory responses lead to tissue damage and contribute to the pathogenesis of inflammatory disorders. In order to maintain immune homeostasis, the negative feedback regulation is also activated upon the initiation of pro-inflammatory response, which may explain the up-regulation of both pro-inflammatory cytokine IL-1β and anti-inflammatory cytokine IL-10 in our results. In addition, Gentle et al. found that Jagged1-mediated Notch signaling enhanced the expression of IL-10 and IL-2 in response to LPS. However, Jagged1 inhibited LPS-induced IL-12 secretion via a post-transcriptional mechanism. They further demonstrated that modulation of LPS-induced IL-10 and IL-2 was independent of γ-secretase-mediated canonical Notch signaling, indicating that the mechanisms vary in the regulation of different cytokines []. Thus, we supposed that the regulation of cytokines in H. pylori-infected DCs was dependent on different signals, and the cross-talk of multiple signaling pathways may be involved in the regulation of some cytokines [,]. Furthermore, DAPT pretreatment changed the profile of Notch receptors and ligands in BMDCs, which may partially influence the cytokine profile []. Taken together, these results suggest that Notch signaling is involved in the regulation of cytokines in H. pylori-infected BMDCs.
It has been proved that Notch signaling plays an important role in regulating the phenotype and maturation of DCs in many inflammatory and infectious diseases. For example, induction of Dll4 on DCs via concurrent stimulation of LPS and R848 (Toll like receptor 7/8 agonist) promoted the maturation of DCs characterized with high levels of CD80, CD86, CD40, CD103 and CD11b []. As for Jagged1, there are controversial views. One study showed that Jagged1 promoted the expression of maturation markers on DCs, including MHC-I, MHC-II, CD80, CD83, and CD86 [], while another study suggested that Jagged1 decreased the levels of MHC-II, CD86 and CD40, inducing tolerogenic DCs []. In addition, there was a viewpoint that Jagged1 had no impact on DC maturation, but enhanced the expression of PD-L1 and OX40L (CD252) []. In the current study, H. pylori increased the expression of CD80, CD83, CD86, and MHC-II on BMDCs, in line with previous studies [], while DAPT pretreatment partially reversed the increase in those surface markers. Therefore, we speculate that DAPT pretreatment may invalidate the function of Notch in DCs, resulting in the failure of transforming into fully mature DCs upon antigen stimulation.
DCs are key regulators of directing CD4+ T cell response in H. pylori infection [], and the interaction between DCs and T cells involves multiple complex signals. Accumulating evidence has shown that Notch ligands on DCs, via binding to Notch receptors on T cells, influence T cell differentiation. For example, Dll4 promoted Th17 differentiation during Mycobacteria infection [], and DAPT inhibited Th17 cell response []. In contrast, respiratory syncytial virus infection up-regulated the expression of Dll4 on DCs, resulting in an attenuated Th17 response []. In some studies, Jagged1 has been shown to be immunosuppressive. DCs expressing Jagged1 promote immunologic tolerance by inducing Treg differentiation []. However, another study indicated that DCs mediate Th17 polarization via Jagged1 activation []. Consistent with the latter report, our study showed that Jagged1 was up-regulated in H. pylori-pulsed BMDCs, leading to Th17 cell differentiation and Th1, Th2 and Treg cell impairment. Additionally, the ratio of Tbx21/Gata3 decreased, while the ratio of Rorγt/Foxp3 increased significantly in the co-culture of H. pylori-pulsed BMDCs and CD4+ T cells. However, DAPT pretreatment partially reversed those changes, except for Il17A. Furthermore, we analyzed the differentiation of CD4+ T cells via flow cytometry, and found that DAPT pretreated-BMDCs induced the differentiation of Treg, shifting the Th17/Treg balance to Treg.
In summary, our study demonstrated for the first time that Notch signaling plays an important role in the regulation of DC function and phenotype during H. pylori infection. Jagged1 may be a crucial regulator in this process, but its role in DCs and whether it is protective or pathogenic remains to be confirmed. As previously described, inhibition of Notch signaling may provide novel strategies for the prevention and treatment of infectious and inflammatory diseases, such as tuberculosis [], chronic hepatitis [] and Behcet’s disease []. However, extensive blockade of Notch signaling has obvious side effects, since Notch components are expressed in a variety of cells and tissues. We should pay more attention to this problem when exploring Notch signaling as a target for controlling H. pylori infection in the future.
Author Contributions
Q.L., Y.L. and Y.N. designed the experiments. Q.L., C.C. and Y.H. conducted the experiments. W.M. and S.R. conducted the data analysis. Q.L. and Y.L. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (No. 81971903), the Basic and Applied basic Research Project of Guangdong Province (No.2023A1515010084), the National Key Research and Development Project of China (No. 2019YFA0903802), and the Innovative Experiment Program of College Students of Guangdong Province, China (S202212121162 and S202312121157).
Data Availability Statement
Data are available on request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zamani, M.; Ebrahimtabar, F.; Zamani, V.; Miller, W.H.; Alizadeh-Navaei, R.; Shokri-Shirvani, J.; Derakhshan, M.H. Systematic review with meta-analysis: The worldwide prevalence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2018, 47, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef]
- Kao, J.Y.; Zhang, M.; Miller, M.J.; Mills, J.C.; Wang, B.; Liu, M.; Eaton, K.A.; Zou, W.; Berndt, B.E.; Cole, T.S.; et al. Helicobacter pylori Immune Escape Is Mediated by Dendritic Cell–Induced Treg Skewing and Th17 Suppression in Mice. Gastroenterology 2010, 138, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Constantino, J.; Gomes, C.; Falcão, A.; Neves, B.M.; Cruz, M.T. Dendritic cell-based immunotherapy: A basic review and recent advances. Immunol. Res. 2017, 65, 798–810. [Google Scholar] [CrossRef] [PubMed]
- Shiu, J.; Blanchard, T.G. Dendritic cell function in the host response to Helicobacter pylori infection of the gastric mucosa. Pathog. Dis. 2013, 67, 46–53. [Google Scholar] [CrossRef]
- Owyang, S.Y.; Zhang, M.; El-Zaatari, M.; Eaton, K.A.; Bishu, S.; Hou, G.; Grasberger, H.; Kao, J.Y. Dendritic cell-derived TGF-β mediates the induction of mucosal regulatory T-cell response to Helicobacter infection essential for maintenance of immune tolerance in mice. Helicobacter 2020, 25, e12763. [Google Scholar] [CrossRef]
- Blosse, A.; Lehours, P.; Wilson, K.T.; Gobert, A.P. Helicobacter: Inflammation, immunology, and vaccines. Helicobacter 2018, 23 (Suppl. S1), e12517. [Google Scholar] [CrossRef]
- Arnold, I.C.; Zhang, X.; Artola-Boran, M.; Fallegger, A.; Sander, P.; Johansen, P.; Müller, A. BATF3-dependent dendritic cells drive both effector and regulatory T-cell responses in bacterially infected tissues. PLoS Pathog. 2019, 15, e1007866. [Google Scholar] [CrossRef]
- Khamri, W.; Walker, M.M.; Clark, P.; Atherton, J.C.; Thursz, M.R.; Bamford, K.B.; Lechler, R.I.; Lombardi, G. Helicobacter pylori Stimulates Dendritic Cells to Induce Interleukin-17 Expression from CD4+T Lymphocytes. Infect. Immun. 2010, 78, 845–853. [Google Scholar] [CrossRef]
- Andres, S.; Schmidt, H.-M.A.; Mitchell, H.; Rhen, M.; Maeurer, M.; Engstrand, L. Helicobacter pylori defines local immune response through interaction with dendritic cells. FEMS Immunol. Med. Microbiol. 2011, 61, 168–178. [Google Scholar] [CrossRef]
- Fehlings, M.; Drobbe, L.; Moos, V.; Viveros, P.R.; Hagen, J.; Beigier-Bompadre, M.; Pang, E.; Belogolova, E.; Churin, Y.; Schneider, T.; et al. Comparative Analysis of the Interaction of Helicobacter pylori with Human Dendritic Cells, Macrophages, and Monocytes. Infect. Immun. 2012, 80, 2724–2734. [Google Scholar] [CrossRef] [PubMed]
- Helmin-Basa, A.; Wiese-Szadkowska, M.; Szaflarska-Popławska, A.; Kłosowski, M.; Januszewska, M.; Bodnar, M.; Marszałek, A.; Gackowska, L.; Michalkiewicz, J. Relationship between Helicobacter pylori Infection and Plasmacytoid and Myeloid Dendritic Cells in Peripheral Blood and Gastric Mucosa of Children. Mediat. Inflamm. 2019, 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Skokos, D.; Nussenzweig, M.C. CD8− DCs induce IL-12–independent Th1 differentiation through Delta 4 Notch-like ligand in response to bacterial LPS. J. Exp. Med. 2007, 204, 1525–1531. [Google Scholar] [CrossRef] [PubMed]
- Dua, B.; Upadhyay, R.; Natrajan, M.; Arora, M.; Narayanaswamy, B.K.; Joshi, B. Notch signaling induces lymphoproliferation, T helper cell activation and Th1/Th2 differentiation in leprosy. Immunol. Lett. 2019, 207, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Goruganthu, M.U.L.; Shanker, A.; Dikov, M.M.; Carbone, D.P. Specific Targeting of Notch Ligand-Receptor Interactions to Modulate Immune Responses: A Review of Clinical and Preclinical Findings. Front. Immunol. 2020, 11, 1958. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.C.; Gonçales, R.A.; Zambuzi, F.A.; Frantz, F.G. Notch signaling pathway in infectious diseases: Role in the regulation of immune response. Inflamm. Res. 2021, 70, 261–274. [Google Scholar] [CrossRef]
- Lai, E.C. Notch signaling: Control of cell communication and cell fate. Development 2004, 131, 965–973. [Google Scholar] [CrossRef]
- Amsen, D.; Helbig, C.; Backer, R.A. Notch in T Cell Differentiation: All Things Considered. Trends Immunol. 2015, 36, 802–814. [Google Scholar] [CrossRef]
- Bugeon, L.; Gardner, L.M.; Rose, A.; Gentle, M.; Dallman, M.J. Cutting Edge: Notch Signaling Induces a Distinct Cytokine Profile in Dendritic Cells That Supports T Cell-Mediated Regulation and IL-2-Dependent IL-17 Production. J. Immunol. 2008, 181, 8189–8193. [Google Scholar] [CrossRef]
- Ting, H.-A.; Schaller, M.A.; Nagata, D.E.d.A.; Rasky, A.J.; Maillard, I.P.; Lukacs, N.W. Notch Ligand Delta-like 4 Promotes Regulatory T Cell Identity in Pulmonary Viral Infection. J. Immunol. 2017, 198, 1492–1502. [Google Scholar] [CrossRef]
- Lee, C.-C.; Lin, C.-L.; Leu, S.-J.; Lee, Y.-L. Overexpression of Notch ligand Delta-like-1 by dendritic cells enhances their immunoregulatory capacity and exerts antiallergic effects on Th2-mediated allergic asthma in mice. Clin. Immunol. 2018, 187, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Gabrilovich, D. Notch signaling in differentiation and function of dendritic cells. Immunol. Res. 2007, 41, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ohishi, K.; Varnum-Finney, B.; Serda, R.E.; Anasetti, C.; Bernstein, I.D. The Notch ligand, Delta-1, inhibits the differentiation of monocytes into macrophages but permits their differentiation into dendritic cells. Blood 2001, 98, 1402–1407. [Google Scholar] [CrossRef]
- Pérez-Cabezas, B.; Naranjo-Gómez, M.; Bastos-Amador, P.; Requena-Fernández, G.; Pujol-Borrell, R.; Borràs, F.E. Ligation of Notch Receptors in Human Conventional and Plasmacytoid Dendritic Cells Differentially Regulates Cytokine and Chemokine Secretion and Modulates Th Cell Polarization. J. Immunol. 2011, 186, 7006–7015. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wen, J.; Chen, C.; Luo, M.; Hu, B.; Wu, D.; Ye, J.; Lin, Y.; Ning, L.; Ning, Y.; et al. Notch 1 Is Involved in CD4+ T Cell Differentiation into Th1 Subtype During Helicobacter pylori Infection. Front. Cell Infect. Microbiol. 2020, 10, 575271. [Google Scholar] [CrossRef]
- Wen, J.; Chen, C.; Luo, M.; Liu, X.; Guo, J.; Wei, T.; Gu, X.; Gu, S.; Ning, Y.; Li, Y. Notch Signaling Ligand Jagged1 Enhances Macrophage-Mediated Response to Helicobacter pylori. Front. Microbiol. 2021, 12, 692832. [Google Scholar] [CrossRef]
- Rizzuti, D.; Ang, M.; Sokollik, C.; Wu, T.; Abdullah, M.; Greenfield, L.; Fattouh, R.; Reardon, C.; Tang, M.; Diao, J.; et al. Helicobacter pylori Inhibits Dendritic Cell Maturation via Interleukin-10-Mediated Activation of the Signal Transducer and Activator of Transcription 3 Pathway. J. Innate Immun. 2014, 7, 199–211. [Google Scholar] [CrossRef]
- Oertli, M.; Sundquist, M.; Hitzler, I.; Engler, D.B.; Arnold, I.C.; Reuter, S.; Maxeiner, J.; Hansson, M.; Taube, C.; Quiding-Järbrink, M.; et al. DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori–specific immune tolerance, and asthma protection. J. Clin. Investig. 2012, 122, 1082–1096. [Google Scholar] [CrossRef]
- Zhang, M.; Berndt, B.E.; Eaton, K.A.; Rathinavelu, S.; Pierzchala, A.; Kao, J.Y. Helicobacter pylori-Pulsed Dendritic Cells Induce H. pylori-Specific Immunity in Mice. Helicobacter 2008, 13, 200–208. [Google Scholar] [CrossRef]
- Hickey, A.; Stamou, P.; Udayan, S.; Ramón-Vázquez, A.; Esteban-Torres, M.; Bottacini, F.; Woznicki, J.A.; Hughes, O.; Melgar, S.; Ventura, M.; et al. Bifidobacterium breve Exopolysaccharide Blocks Dendritic Cell Maturation and Activation of CD4+ T Cells. Front. Microbiol. 2021, 12, 653587. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Meng, S.; Chang, W.; Fan, S.; Xie, J.; Guo, F.; Yang, Y.; Qiu, H.; Liu, L. Mesenchymal stem cells activate Notch signaling to induce regulatory dendritic cells in LPS-induced acute lung injury. J. Transl. Med. 2020, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, H.; Xu, H.; Zeng, W.; Qiu, Y.; Tan, C.; Tang, S.; Zhang, J. Dendritic Cells Promote Treg Expansion but Not Th17 Generation in Response to Talaromyces marneffei Yeast Cells. Infect. Drug Resist. 2020, 13, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Vanderbeck, A.; Maillard, I. Notch signaling at the crossroads of innate and adaptive immunity. J. Leukoc. Biol. 2020, 109, 535–548. [Google Scholar] [CrossRef]
- Gentle, M.E.; Rose, A.; Bugeon, L.; Dallman, M.J. Noncanonical Notch Signaling Modulates Cytokine Responses of Dendritic Cells to Inflammatory Stimuli. J. Immunol. 2012, 189, 1274–1284. [Google Scholar] [CrossRef]
- Milinkovic, I.; Krasavcevic, A.D.; Nikolic, N.; Aleksic, Z.; Carkic, J.; Jezdic, M.; Jankovic, S.; Milasin, J. Notch down-regulation and inflammatory cytokines and RANKL overexpression involvement in peri-implant mucositis and peri-implantitis: A cross-sectional study. Clin. Oral Implant. Res. 2021, 32, 1496–1505. [Google Scholar] [CrossRef]
- Lutz, M.B.; Schuler, G. Immature, semi-mature and fully mature dendritic cells: Which signals induce tolerance or immunity? Trends Immunol. 2002, 23, 445–449. [Google Scholar] [CrossRef]
- Ohishi, K.; Katayama, N.; Shiku, H.; Varnumfinney, B.; Bernstein, I. Notch signalling in hematopoiesis. Semin. Cell Dev. Biol. 2003, 14, 143–150. [Google Scholar] [CrossRef]
- Osborne, B.A.; Minter, L.M. Notch signalling during peripheral T-cell activation and differentiation. Nat. Rev. Immunol. 2006, 7, 64–75. [Google Scholar] [CrossRef]
- Fasnacht, N.; Huang, H.-Y.; Koch, U.; Favre, S.; Auderset, F.; Chai, Q.; Onder, L.; Kallert, S.; Pinschewer, D.D.; MacDonald, H.R.; et al. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J. Exp. Med. 2014, 211, 2265–2279. [Google Scholar] [CrossRef]
- Nandagopal, N.; Santat, L.A.; LeBon, L.; Sprinzak, D.; Bronner, M.E.; Elowitz, M.B. Dynamic Ligand Discrimination in the Notch Signaling Pathway. Cell 2018, 172, 869–880.e19. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.-T.; Cheng, H.-T.; Chang, L.-W.; Ohtsuka, T.; Kageyama, R.; Stormo, G.D.; Kopan, R. Target Selectivity of Vertebrate Notch Proteins.Collaboration between discrete domains and csl-binding site architecture determines activation probability. J. Biol. Chem. 2006, 281, 5106–5119. [Google Scholar] [CrossRef]
- Schaller, M.A.; Allen, R.M.; Kimura, S.; Day, C.L.; Kunkel, S.L. Systemic Expression of Notch Ligand Delta-Like 4 during Mycobacterial Infection Alters the T Cell Immune Response. Front. Immunol. 2016, 7, 527. [Google Scholar] [CrossRef] [PubMed]
- Schaller, M.A.; Neupane, R.; Rudd, B.D.; Kunkel, S.L.; Kallal, L.E.; Lincoln, P.; Lowe, J.B.; Man, Y.; Lukacs, N.W. Notch ligand Delta-like 4 regulates disease pathogenesis during respiratory viral infections by modulating Th2 cytokines. J. Exp. Med. 2007, 204, 2925–2934. [Google Scholar] [CrossRef] [PubMed]
- Liotta, F.; Frosali, F.; Querci, V.; Mantei, A.; Filì, L.; Maggi, L.; Mazzinghi, B.; Angeli, R.; Ronconi, E.; Santarlasci, V.; et al. Human immature myeloid dendritic cells trigger a TH2-polarizing program via Jagged-1/Notch interaction. J. Allergy Clin. Immunol. 2008, 121, 1000–1005.e8. [Google Scholar] [CrossRef]
- Higashi, T.; Hashimoto, K.; Takagi, R.; Mizuno, Y.; Okazaki, Y.; Tanaka, Y.; Matsushita, S. Curdlan Induces DC-Mediated Th17 Polarization via Jagged1 Activation in Human Dendritic Cells. Allergol. Int. 2010, 59, 161–166. [Google Scholar] [CrossRef]
- Cao, Q.; Lu, J.; Kaur, C.; Sivakumar, V.; Li, F.; Cheah, P.S.; Dheen, S.T.; Ling, E. Expression of Notch-1 receptor and its ligands Jagged-1 and Delta-1 in amoeboid microglia in postnatal rat brain and murine BV-2 cells. Glia 2008, 56, 1224–1237. [Google Scholar] [CrossRef]
- Holla, S.; Stephen-Victor, E.; Prakhar, P.; Sharma, M.; Saha, C.; Udupa, V.; Kaveri, S.V.; Bayry, J.; Balaji, K.N. Mycobacteria-responsive sonic hedgehog signaling mediates programmed death-ligand 1- and prostaglandin E2-induced regulatory T cell expansion. Sci. Rep. 2016, 6, 24193. [Google Scholar] [CrossRef]
- Jannuzzi, G.P.; de Almeida, J.R.F.; dos Santos, S.S.; de Almeida, S.R.; Ferreira, K.S. Notch Signaling is Required for Dendritic Cell Maturation and T Cell Expansion in Paracoccidioidomycosis. Mycopathologia 2018, 183, 739–749. [Google Scholar] [CrossRef]
- Kao, J.Y.; Rathinavelu, S.; Eaton, K.A.; Bai, L.; Zavros, Y.; Takami, M.; Pierzchala, A.; Merchant, J.L. Helicobacter pylori-secreted factors inhibit dendritic cell IL-12 secretion: A mechanism of ineffective host defense. Am. J. Physiol. Liver Physiol. 2006, 291, G73–G81. [Google Scholar] [CrossRef]
- Foldi, J.; Chung, A.Y.; Xu, H.; Zhu, J.; Outtz, H.H.; Kitajewski, J.; Li, Y.; Hu, X.; Ivashkiv, L.B. Autoamplification of Notch Signaling in Macrophages by TLR-Induced and RBP-J–Dependent Induction of Jagged1. J. Immunol. 2010, 185, 5023–5031. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, K.; Meng, L.; Mochizuki, I.; Tong, Q.; He, S.; Liu, Y.; Purushe, J.; Fung, H.; Zaidi, M.R.; Zhang, Y.; et al. Programming of donor T cells using allogeneic δ-like ligand 4–positive dendritic cells to reduce GVHD in mice. Blood 2016, 127, 3270–3280. [Google Scholar] [CrossRef] [PubMed]
- Weijzen, S.; Velders, M.P.; Elmishad, A.G.; Bacon, P.E.; Panella, J.R.; Nickoloff, B.J.; Miele, L.; Kast, W.M. The Notch Ligand Jagged-1 Is Able to Induce Maturation of Monocyte-Derived Human Dendritic Cells. J. Immunol. 2002, 169, 4273–4278. [Google Scholar] [CrossRef]
- Lin, C.; Huang, H.; Hsieh, C.; Fan, C.; Lee, Y. Jagged1-expressing adenovirus-infected dendritic cells induce expansion of Foxp3+ regulatory T cells and alleviate T helper type 2-mediated allergic asthma in mice. Immunology 2018, 156, 199–212. [Google Scholar] [CrossRef]
- Kranzer, K.; Eckhardt, A.; Aigner, M.; Knoll, G.; Deml, L.; Speth, C.; Lehn, N.; Rehli, M.; Schneider-Brachert, W. Induction of Maturation and Cytokine Release of Human Dendritic Cells by Helicobacter pylori. Infect. Immun. 2004, 72, 4552–4560. [Google Scholar] [CrossRef]
- Ito, T.; Schaller, M.; Hogaboam, C.M.; Standiford, T.J.; Sandor, M.; Lukacs, N.W.; Chensue, S.W.; Kunkel, S.L. TLR9 regulates the mycobacteria-elicited pulmonary granulomatous immune response in mice through DC-derived Notch ligand delta-like 4. J. Clin. Investig. 2008, 119, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xue, H.; Qi, R.; Wang, Y.; Yuan, L. Effect of γ-secretase inhibitor on Th17 cell differentiation and function of mouse psoriasis-like skin inflammation. J. Transl. Med. 2018, 16, 59. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, W.; Li, J.; Yan, G.; Li, C.; Jin, N.; Chen, J.; Gao, C.; Ma, P.; Xu, S.; et al. Overexpression of Jagged-1 combined with blockade of CD40 pathway prolongs allograft survival. Immunol. Cell Biol. 2014, 93, 213–217. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, H.; Yu, L.; Wu, C.; Luo, X.; Sun, H.; Ding, J. Down-regulation of Notch signaling pathway reverses the Th1/Th2 imbalance in tuberculosis patients. Int. Immunopharmacol. 2018, 54, 24–32. [Google Scholar] [CrossRef]
- Qin, L.; Zhou, Y.-C.; Wu, H.-J.; Zhuo, Y.; Wang, Y.-P.; Si, C.-Y.; Qin, Y.-M. Notch Signaling Modulates the Balance of Regulatory T Cells and T Helper 17 Cells in Patients with Chronic Hepatitis C. DNA Cell Biol. 2017, 36, 311–320. [Google Scholar] [CrossRef]
- Qi, J.; Yang, Y.; Hou, S.; Qiao, Y.; Wang, Q.; Yu, H.; Zhang, Q.; Cai, T.; Kijlstra, A.; Yang, P. Increased Notch pathway activation in Behçet’s disease. Rheumatology 2014, 53, 810–820. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).