Changes in Microbial Communities Using Pigs as a Model for Postmortem Interval Estimation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Data Analysis and Processing
3. Results
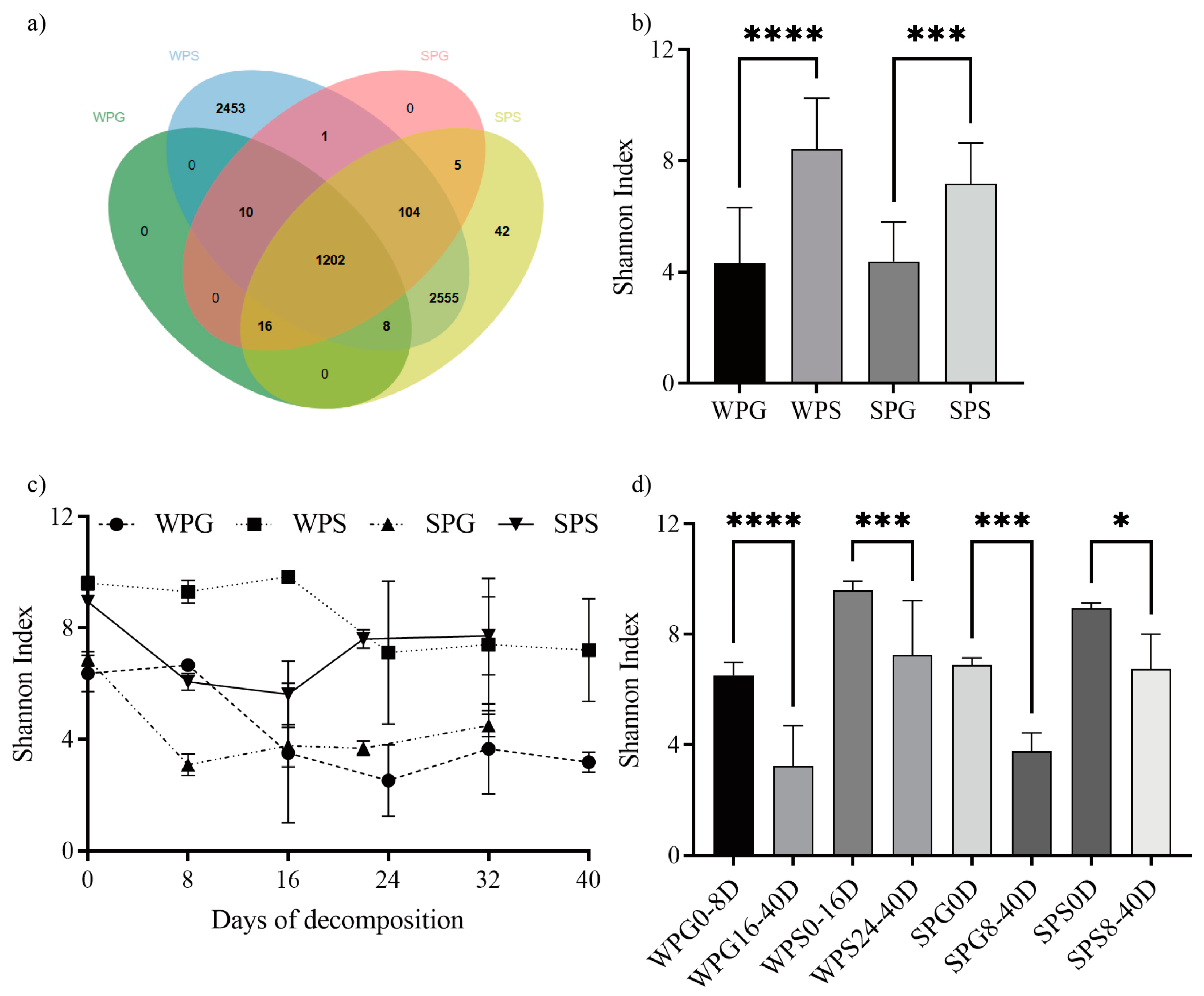
3.1. Bacterial Community Structure Overview
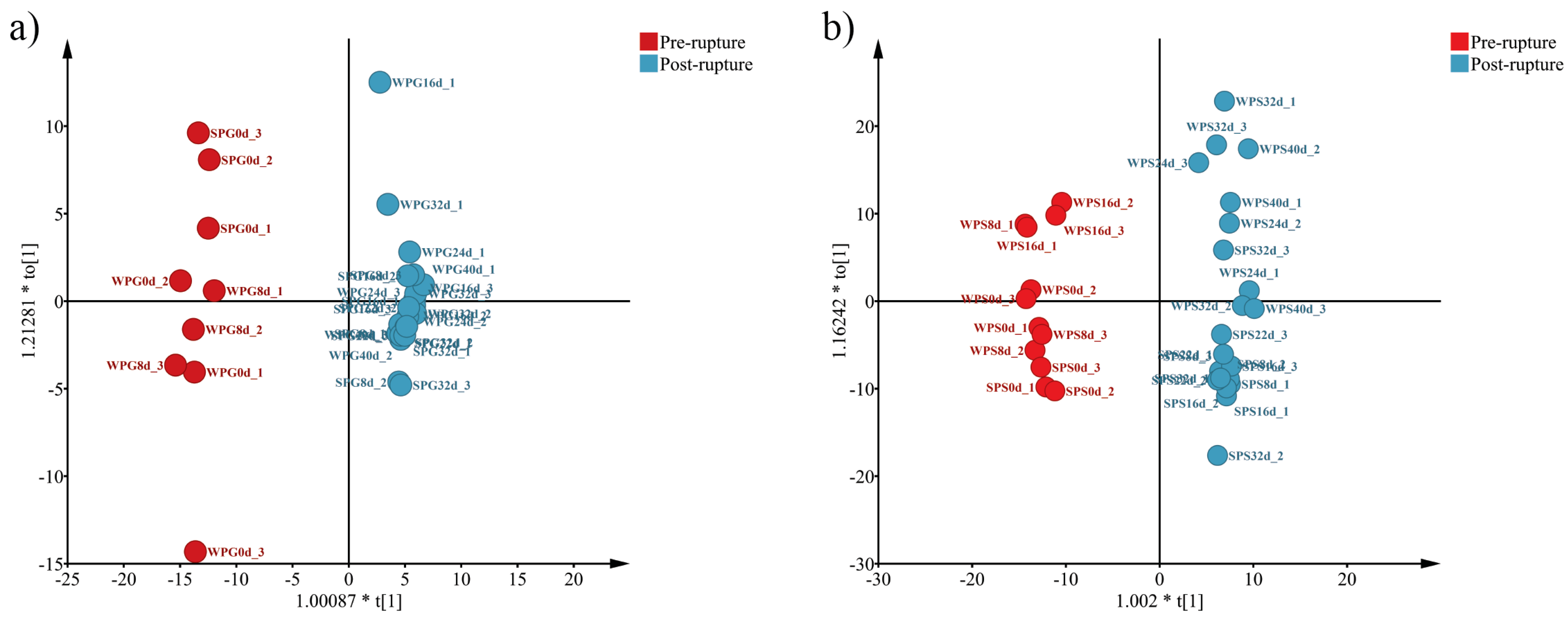
3.2. Alpha Diversity and Beta Diversity
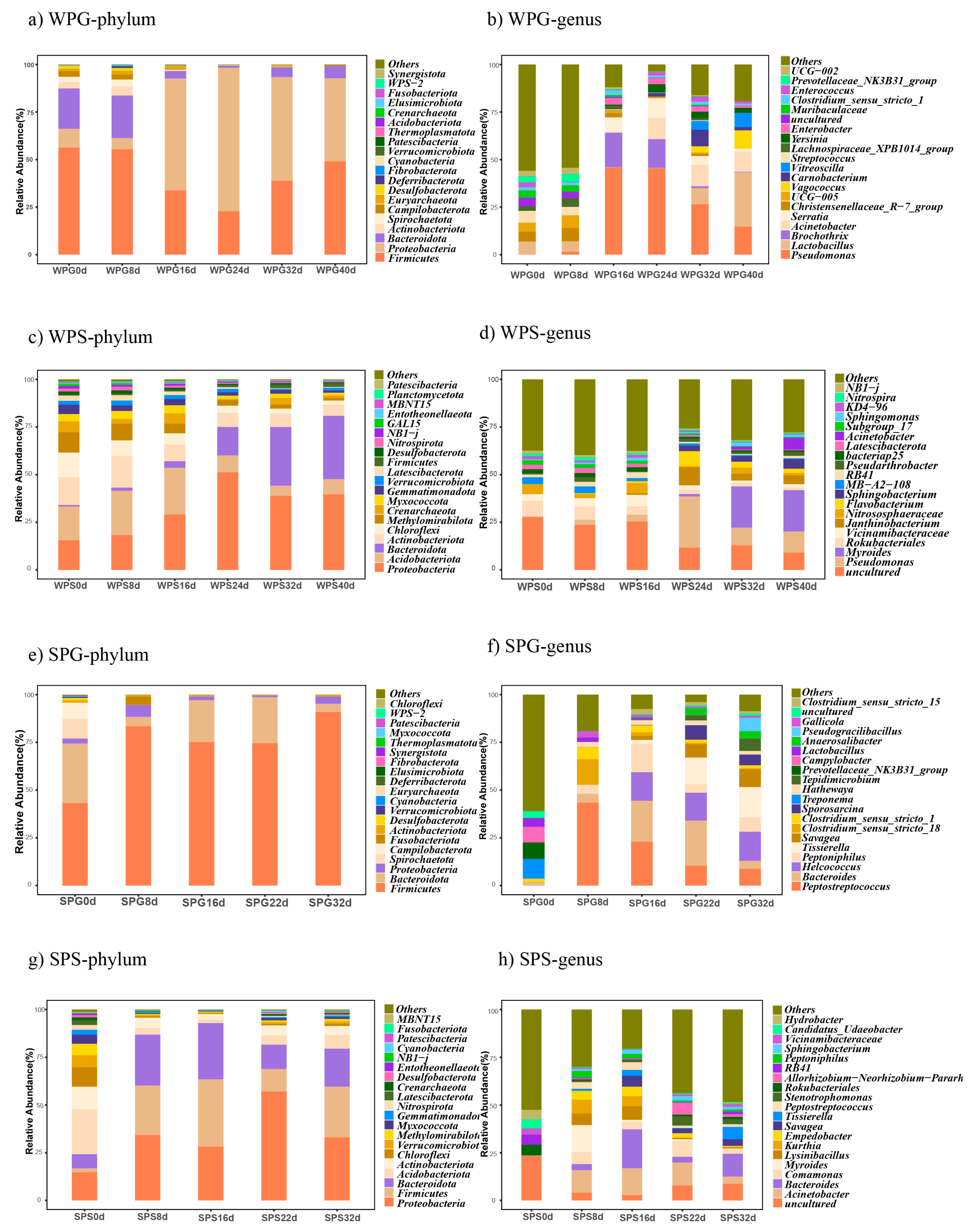
3.3. Taxonomic Analysis
3.4. Utilizing Bacterial Communities to Predict the PMI Based on the Model of RF
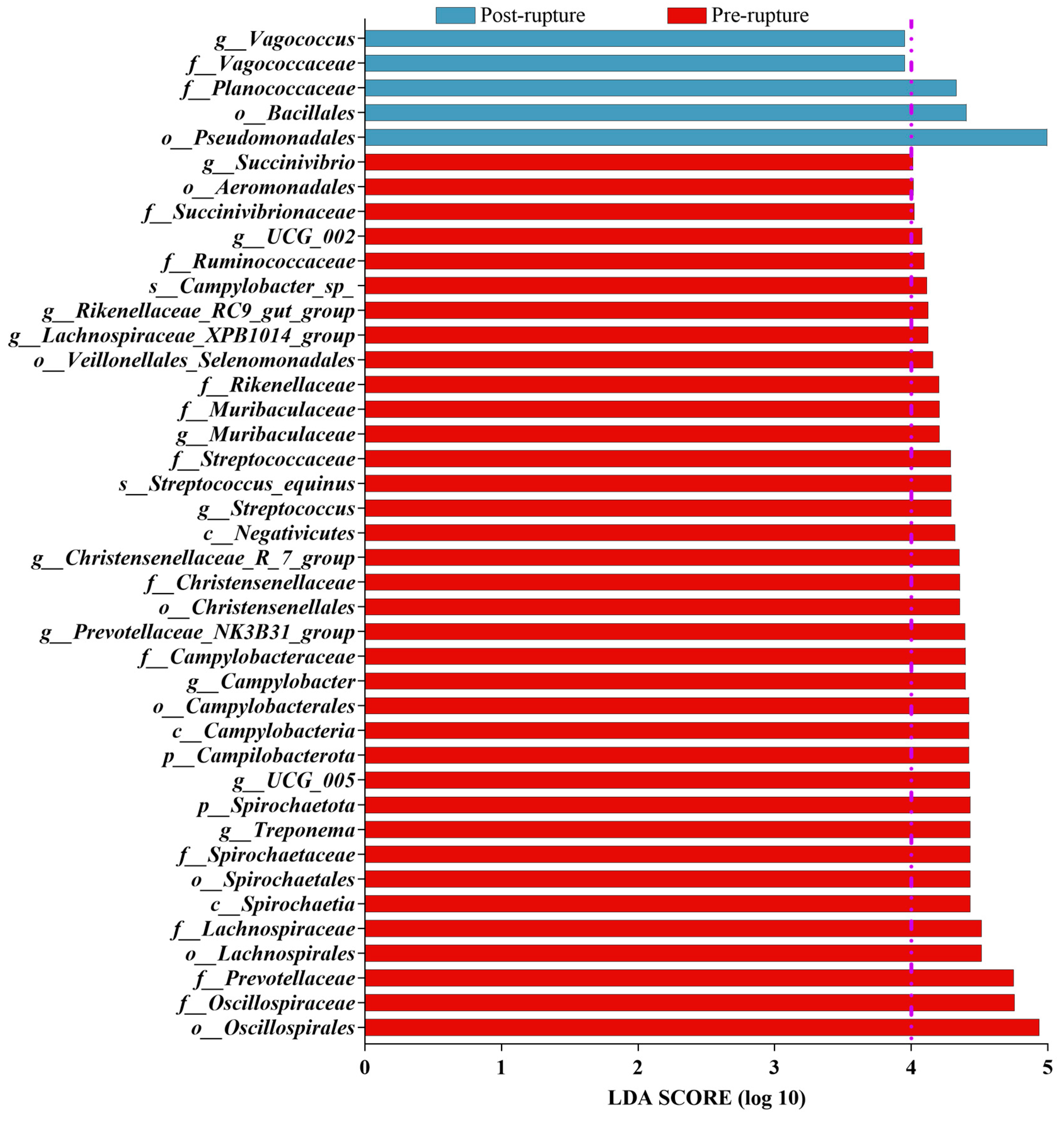
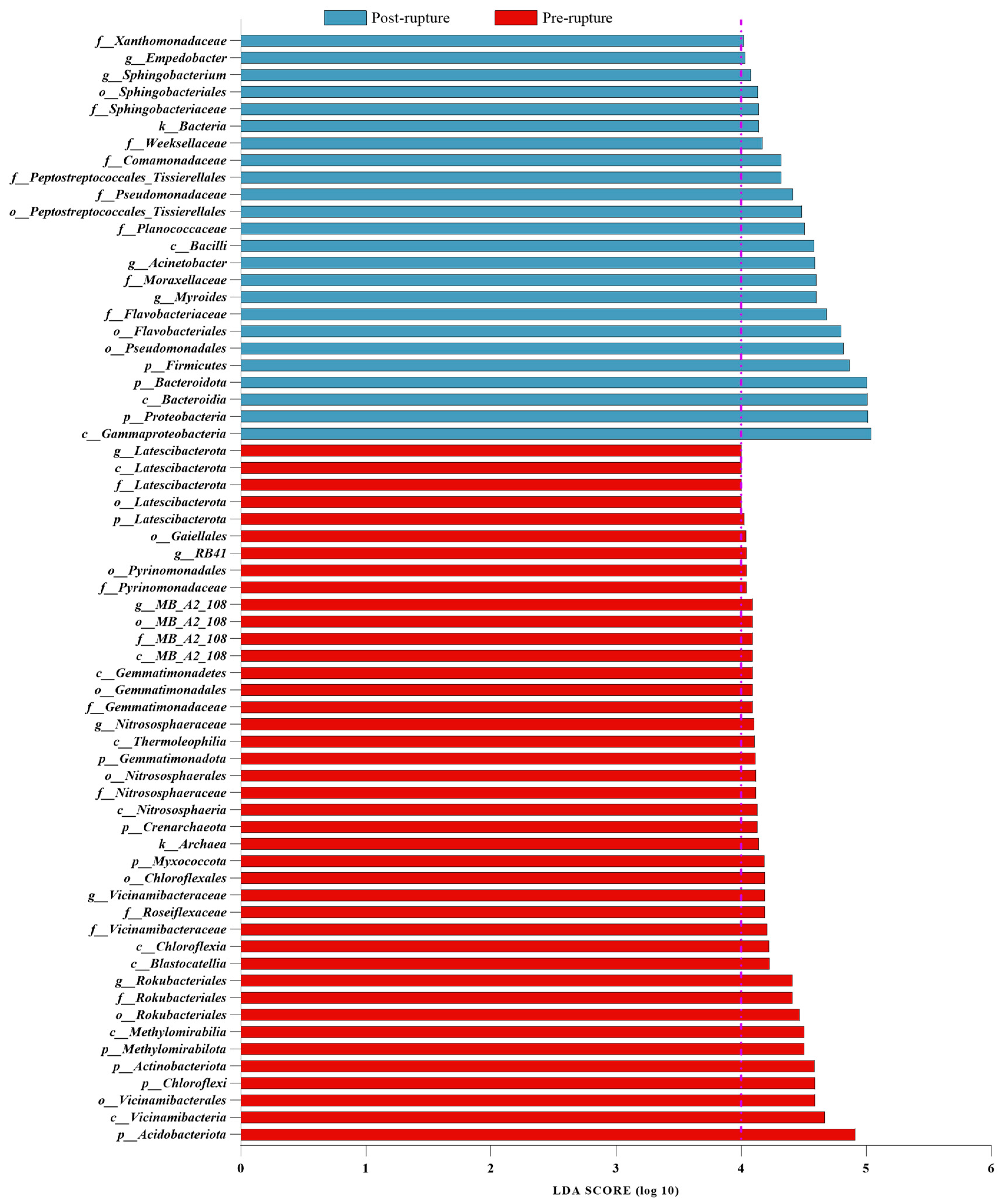
3.5. Bacterial Difference between Pre-Rupture and Post-Rupture Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Henssge, C.; Madea, B. Estimation of the time since death. Forensic Sci. Int. 2007, 165, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Wegener Parfrey, L.; Gonzalez, A.; Lauber, C.L.; Knights, D.; Ackermann, G.; Humphrey, G.C.; Gebert, M.J.; Van Treuren, W.; Berg-Lyons, D.; et al. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. Elife 2013, 2, e01104. [Google Scholar] [CrossRef] [PubMed]
- Melvin, J.R., Jr.; Cronholm, L.S.; Simson, L.R., Jr.; Isaacs, A.M. Bacterial transmigration as an indicator of time of death. J. Forensic Sci. 1984, 29, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Carter, D.O.; Yellowlees, D.; Tibbett, M. Cadaver decomposition in terrestrial ecosystems. Naturwissenschaften 2007, 94, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Laplace, K.; Baccino, E.; Peyron, P.-A. Estimation of the time since death based on body cooling: A comparative study of four temperature-based methods. Int. J. Leg. Med. 2021, 135, 2479–2487. [Google Scholar] [CrossRef]
- Yuan, H.; Wang, Z.; Wang, Z.; Zhang, F.; Guan, D.; Zhao, R. Trends in forensic microbiology: From classical methods to deep learning. Front. Microbiol. 2023, 14, 1163741. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, P.; Zeng, K.; Yuan, H.; Wang, Z.; Li, X.; Yuan, H.; Du, S.; Guan, D.; Wang, L.; et al. Postmortem submersion interval estimation of cadavers recovered from freshwater based on gut microbial community succession. Front. Microbiol. 2022, 13, 988297. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, F.; Wang, L.; Yuan, H.; Guan, D.; Zhao, R. Advances in artificial intelligence-based microbiome for PMI estimation. Front. Microbiol. 2022, 13, 1034051. [Google Scholar] [CrossRef]
- Zhao, X.; Zhong, Z.; Hua, Z. Estimation of the post-mortem interval by modelling the changes in oral bacterial diversity during decomposition. J. Appl. Microbiol. 2022, 133, 3451–3464. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, F.; Zeng, K.; Dong, W.; Yuan, H.; Wang, Z.; Liu, J.; Pan, J.; Zhao, R.; Guan, D. Microbial communities in the liver and brain are informative for postmortem submersion interval estimation in the late phase of decomposition: A study in mouse cadavers recovered from freshwater. Front. Microbiol. 2022, 13, 1052808. [Google Scholar] [CrossRef]
- Tozzo, P.; Amico, I.; Delicati, A.; Toselli, F.; Caenazzo, L. Post-Mortem Interval and Microbiome Analysis through 16S rRNA Analysis: A Systematic Review. Diagnostics 2022, 12, 2641. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, Q.; Zhang, K.; Wu, H.; Wang, G.; Cai, W.; Yu, K.; Sun, Q.; Fan, S.; Wang, Z. Analysis of Postmortem Intestinal Microbiota Successional Patterns with Application in Postmortem Interval Estimation. Microb. Ecol. 2022, 84, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Niu, X.; Qi, R.Q.; Liu, M. Advances in microbial metagenomics and artificial intelligence analysis in forensic identification. Front. Microbiol. 2022, 13, 1046733. [Google Scholar] [CrossRef]
- Emmons, A.L.; Mundorff, A.Z.; Hoeland, K.M.; Davoren, J.; Keenan, S.W.; Carter, D.O.; Campagna, S.R.; DeBruyn, J.M. Postmortem Skeletal Microbial Community Composition and Function in Buried Human Remains. mSystems 2022, 7, e0004122. [Google Scholar] [CrossRef]
- Hyde, E.R.; Haarmann, D.P.; Lynne, A.M.; Bucheli, S.R.; Petrosino, J.F. The living dead: Bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PLoS ONE 2013, 8, e77733. [Google Scholar] [CrossRef] [PubMed]
- Pechal, J.L.; Crippen, T.L.; Benbow, M.E.; Tarone, A.M.; Dowd, S.; Tomberlin, J.K. The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int. J. Leg. Med. 2014, 128, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Burcham, Z.M.; Pechal, J.L.; Schmidt, C.J.; Bose, J.L.; Rosch, J.W.; Benbow, M.E.; Jordan, H.R. Bacterial Community Succession, Transmigration, and Differential Gene Transcription in a Controlled Vertebrate Decomposition Model. Front. Microbiol. 2019, 10, 745. [Google Scholar] [CrossRef] [PubMed]
- Dash, H.R.; Das, S. Microbial community signatures for estimation of postmortem time intervals. Adv. Appl. Microbiol. 2022, 118, 91–113. [Google Scholar] [CrossRef]
- Deel, H.; Emmons, A.L.; Kiely, J.; Damann, F.E.; Carter, D.O.; Lynne, A.; Knight, R.; Xu, Z.Z.; Bucheli, S.; Metcalf, J.L. A Pilot Study of Microbial Succession in Human Rib Skeletal Remains during Terrestrial Decomposition. mSphere 2021, 6, e0045521. [Google Scholar] [CrossRef]
- Cartozzo, C.; Singh, B.; Swall, J.; Simmons, T. Postmortem submersion interval (PMSI) estimation from the microbiome of sus scrofa bone in a freshwater lake. J. Forensic Sci. 2021, 66, 1334–1347. [Google Scholar] [CrossRef]
- Cartozzo, C.; Simmons, T.; Swall, J.; Singh, B. Postmortem submersion interval (PMSI) estimation from the microbiome of Sus scrofa bone in a freshwater river. Forensic Sci. Int. 2021, 318, 110480. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Li, W.J.; Wang, Y.F.; An, G.S.; Lu, X.J.; Du, Q.X.; Li, J.; Sun, J.H. Estimating Postmortem Interval Using Intestinal Microbiota Diversity Based on 16S rRNA High-throughput Sequencing Technology. Fa Yi Xue Za Zhi 2021, 37, 621–626. [Google Scholar] [CrossRef]
- Pittner, S.; Bugelli, V.; Benbow, M.E.; Ehrenfellner, B.; Zissler, A.; Campobasso, C.P.; Oostra, R.J.; Aalders, M.C.G.; Zehner, R.; Lutz, L.; et al. The applicability of forensic time since death estimation methods for buried bodies in advanced decomposition stages. PLoS ONE 2020, 15, e0243395. [Google Scholar] [CrossRef]
- Lutz, H.; Vangelatos, A.; Gottel, N.; Osculati, A.; Visona, S.; Finley, S.J.; Gilbert, J.A.; Javan, G.T. Effects of Extended Postmortem Interval on Microbial Communities in Organs of the Human Cadaver. Front. Microbiol. 2020, 11, 569630. [Google Scholar] [CrossRef] [PubMed]
- Kaszubinski, S.F.; Receveur, J.P.; Wydra, B.; Smiles, K.; Wallace, J.R.; Babcock, N.J.; Weatherbee, C.R.; Benbow, M.E. Cold Case Experiment Demonstrates the Potential Utility of Aquatic Microbial Community Assembly in Estimating a Postmortem Submersion Interval. J. Forensic Sci. 2020, 65, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Fu, X.; Liao, H.; Hu, Z.; Long, L.; Yan, W.; Ding, Y.; Zha, L.; Guo, Y.; Yan, J.; et al. Potential use of bacterial community succession for estimating post-mortem interval as revealed by high-throughput sequencing. Sci. Rep. 2016, 6, 24197. [Google Scholar] [CrossRef]
- Robinson, J.M.; Pasternak, Z.; Mason, C.E.; Elhaik, E. Forensic Applications of Microbiomics: A Review. Front. Microbiol. 2020, 11, 608101. [Google Scholar] [CrossRef]
- Johnson, H.R.; Trinidad, D.D.; Guzman, S.; Khan, Z.; Parziale, J.V.; DeBruyn, J.M.; Lents, N.H. A Machine Learning Approach for Using the Postmortem Skin Microbiome to Estimate the Postmortem Interval. PLoS ONE 2016, 11, e0167370. [Google Scholar] [CrossRef]
- Zhang, Q.; Widmer, G.; Tzipori, S. A pig model of the human gastrointestinal tract. Gut Microbes 2013, 4, 193–200. [Google Scholar] [CrossRef]
- Xiao, L.; Estellé, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.Ø.; Kjeldsen, N.J.; Liu, C.; et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016, 1, 16161. [Google Scholar] [CrossRef]
- Weiss, S.; Carter, D.O.; Metcalf, J.L.; Knight, R. Carcass mass has little influence on the structure of gravesoil microbial communities. Int. J. Leg. Med. 2016, 130, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Song, Y.; Mao, D.; Cao, Y.; Qiu, B.; Gui, P.; Wang, H.; Zhao, X.; Huang, Z.; Sun, L.; et al. Predicting the Postmortem Interval Based on Gravesoil Microbiome Data and a Random Forest Model. Microorganisms 2022, 11, 56. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liang, X.-R.; Zhou, S.-D.; Dang, L.-H.; Li, J.; An, G.-S.; Ren, K.; Jin, Q.-Q.; Liang, X.-H.; Cao, J.; et al. Exploring postmortem succession of rat intestinal microbiome for PMI based on machine learning algorithms and potential use for humans. Forensic Sci. Int. Genet. 2023, 66, 102904. [Google Scholar] [CrossRef]
- de Wit, R.; Bouvier, T. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 2006, 8, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Carter, D.O.; Yellowlees, D.; Tibbett, M. Moisture can be the dominant environmental parameter governing cadaver decomposition in soil. Forensic Sci. Int. 2010, 200, 60–66. [Google Scholar] [CrossRef]
- Procopio, N.; Ghignone, S.; Williams, A.; Chamberlain, A.; Mello, A.; Buckley, M. Metabarcoding to investigate changes in soil microbial communities within forensic burial contexts. Forensic Sci. Int. Genet. 2019, 39, 73–85. [Google Scholar] [CrossRef]
- Li, H.; Zhang, S.; Liu, R.; Yuan, L.; Wu, D.; Yang, E.; Yang, H.; Ullah, S.; Ishaq, H.M.; Liu, H.; et al. Potential use of molecular and structural characterization of the gut bacterial community for postmortem interval estimation in Sprague Dawley rats. Sci. Rep. 2021, 11, 225. [Google Scholar] [CrossRef]
- Valseth, K.; Nesbø, C.L.; Easterday, W.R.; Turner, W.C.; Olsen, J.S.; Stenseth, N.C.; Haverkamp, T.H.A. Temporal dynamics in microbial soil communities at anthrax carcass sites. BMC Microbiol. 2017, 17, 206. [Google Scholar] [CrossRef]
- Sundararaman, A.; Srinivasan, S.; Lee, S.-S. Vagococcus humatus sp. nov., isolated from soil beneath a decomposing pig carcass. Int. J. Syst. Evol. Microbiol. 2017, 67, 330–335. [Google Scholar] [CrossRef]
- Cho, H.-W.; Eom, Y.-B. Forensic Analysis of Human Microbiome in Skin and Body Fluids Based on Geographic Location. Front. Cell. Infect. Microbiol. 2021, 11, 695191. [Google Scholar] [CrossRef]
- Giampaoli, S.; Berti, A.; Valeriani, F.; Gianfranceschi, G.; Piccolella, A.; Buggiotti, L.; Rapone, C.; Valentini, A.; Ripani, L.; Romano Spica, V. Molecular identification of vaginal fluid by microbial signature. Forensic Sci. Int. Genet. 2012, 6, 559–564. [Google Scholar] [CrossRef] [PubMed]


| Time | Sample | Raw Sequence Number | Clean Sequence Number | OTUs |
|---|---|---|---|---|
| Summer | Rectum | 670007 | 652672 | 1338 |
| Grave soil | 663546 | 643916 | 3932 | |
| Winter | Rectum | 800908 | 772761 | 1236 |
| Grave soil | 858300 | 827585 | 6333 |
| Stage | Sample | Phylum | Class | Order | Family | Genus |
|---|---|---|---|---|---|---|
| Pre-rupture | rectum | Spirochaetota | Spirochaetia | Oscillospirales | Oscillospiraceae | Treponema |
| Campilobacterota | Campylobacteria | Lachnospirales | Prevotellaceae | UCG_005 | ||
| Desulfobacterota | Negativicutes | Spirochaetales | Lachnospiraceae | Campylobacter | ||
| Deferribacterota | Desulfovibrionia | Campylobacterales | Spirochaetaceae | Prevotellaceae_NK3B31_group | ||
| Deferribacteres | Christensenellales | Campylobacteraceae | Christensenellaceae_R_7_group | |||
| soil | Acidobacteriota | KD4_96 | Frankiales | Nitrospiraceae | Subgroup_25 | |
| Chloroflexi | MB_A2_108 | Latescibacterota | Rokubacteriales | MND1 | ||
| Actinobacteriota | TK10 | Haliangiales | MB_A2_108 | Subgroup_5 | ||
| Methylomirabilota | NB1_j | Subgroup_25 | Vicinamibacteraceae | KD4_96 | ||
| Myxococcota | Vicinamibacteria | Nitrospirales | Haliangiaceae | 11_24 | ||
| Post-rupture | rectum | - | - | Pseudomonadales | Vagococcaceae | Vagococcus |
| Burkholderiales | Planococcaceae | |||||
| Bacillales | ||||||
| soil | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Flavobacteriaceae | Myroides | |
| Bacteroidota | Bacteroidia | Flavobacteriales | Moraxellaceae | Acinetobacter | ||
| Firmicutes | Bacilli | Peptostreptococcales_Tissierellales | Planococcaceae | Sphingobacterium | ||
| Sphingobacteriales | Pseudomonadaceae | Empedobacter | ||||
| Enterobacterales | Peptostreptococcales_Tissierellales | Stenotrophomonas |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Zhang, X.; Hu, S.; Nie, H.; Gui, P.; Zhong, Z.; Guo, Y.; Zhao, X. Changes in Microbial Communities Using Pigs as a Model for Postmortem Interval Estimation. Microorganisms 2023, 11, 2811. https://doi.org/10.3390/microorganisms11112811
Yang F, Zhang X, Hu S, Nie H, Gui P, Zhong Z, Guo Y, Zhao X. Changes in Microbial Communities Using Pigs as a Model for Postmortem Interval Estimation. Microorganisms. 2023; 11(11):2811. https://doi.org/10.3390/microorganisms11112811
Chicago/Turabian StyleYang, Fan, Xiangyan Zhang, Sheng Hu, Hao Nie, Peng Gui, Zengtao Zhong, Yadong Guo, and Xingchun Zhao. 2023. "Changes in Microbial Communities Using Pigs as a Model for Postmortem Interval Estimation" Microorganisms 11, no. 11: 2811. https://doi.org/10.3390/microorganisms11112811
APA StyleYang, F., Zhang, X., Hu, S., Nie, H., Gui, P., Zhong, Z., Guo, Y., & Zhao, X. (2023). Changes in Microbial Communities Using Pigs as a Model for Postmortem Interval Estimation. Microorganisms, 11(11), 2811. https://doi.org/10.3390/microorganisms11112811






