Rapid Detection and Quantification of Viable Cells of Pectobacterium brasiliense Using Propidium Monoazide Combined with Real-Time PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Cultivation, and DNA Extraction
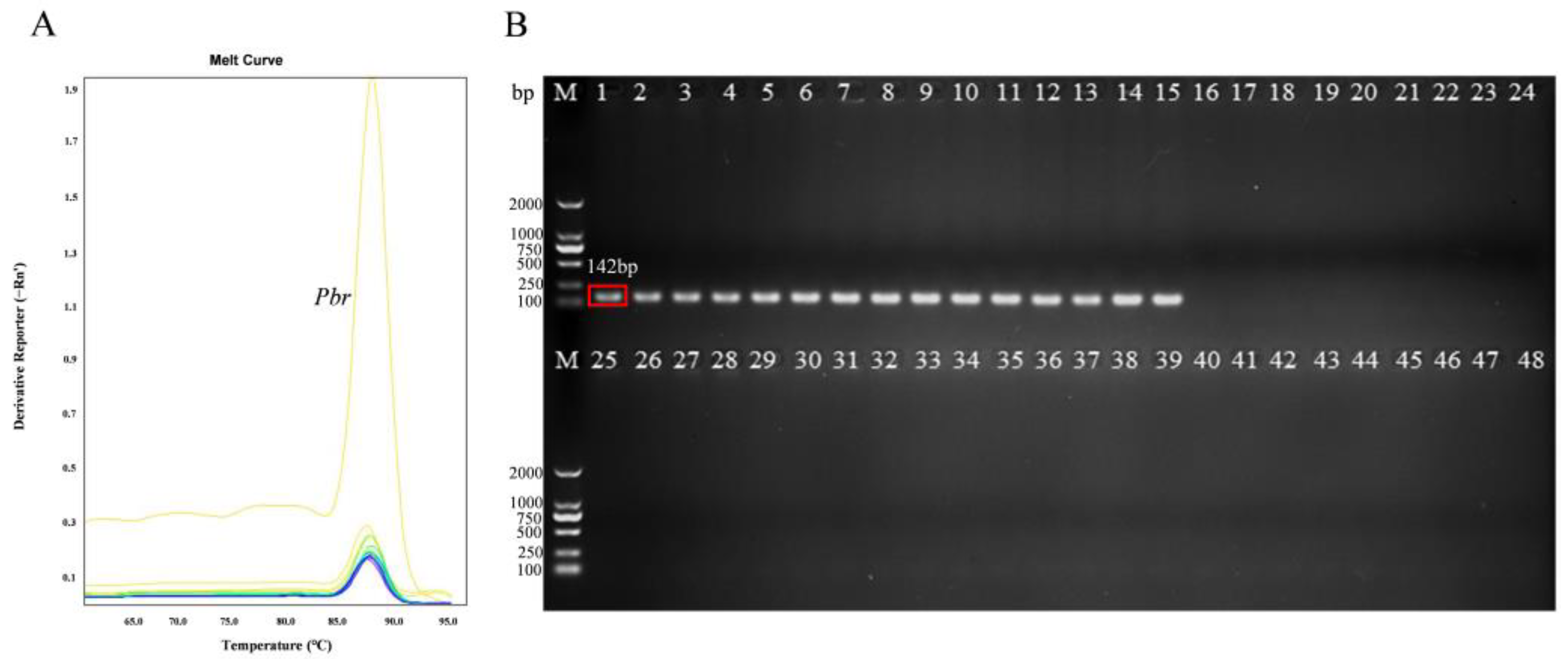
2.2. Primer Design and Specificity Testing
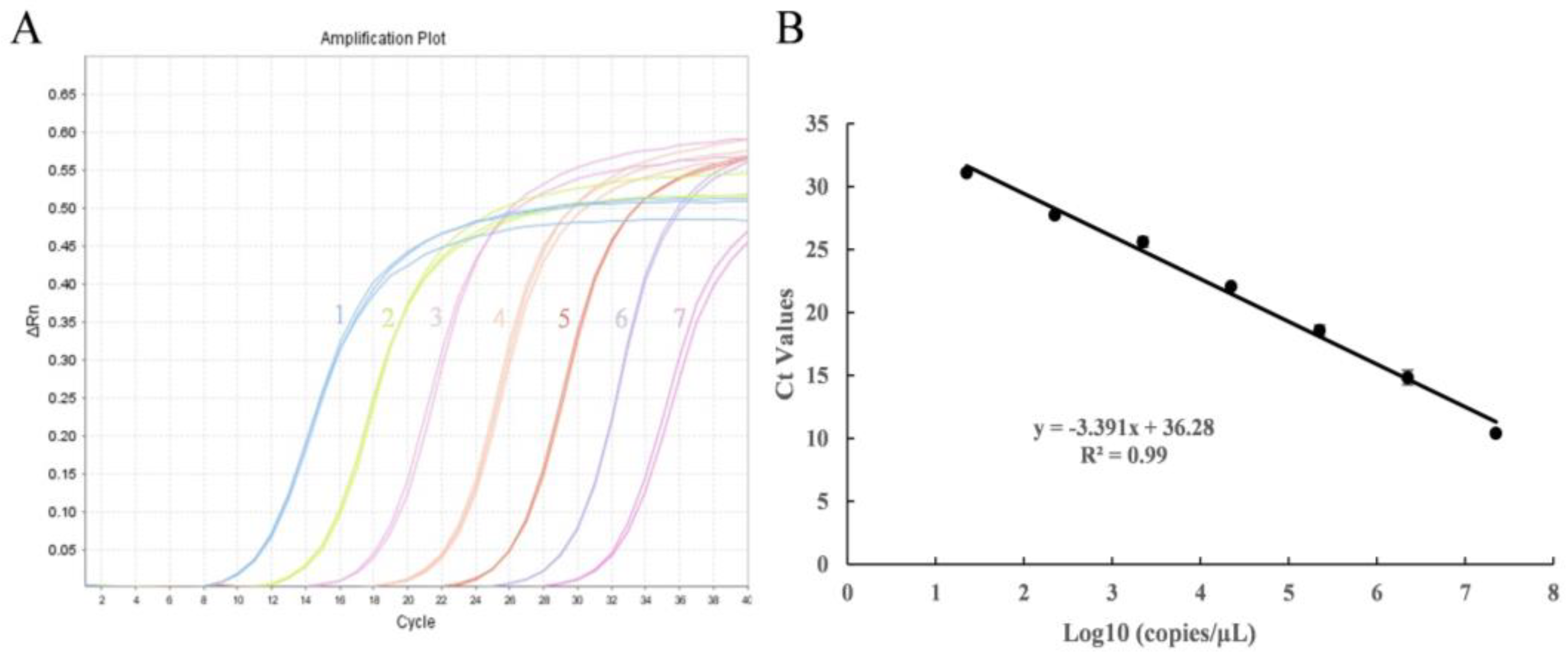
2.3. Primer Sensitivity Testing and Quantification Standards
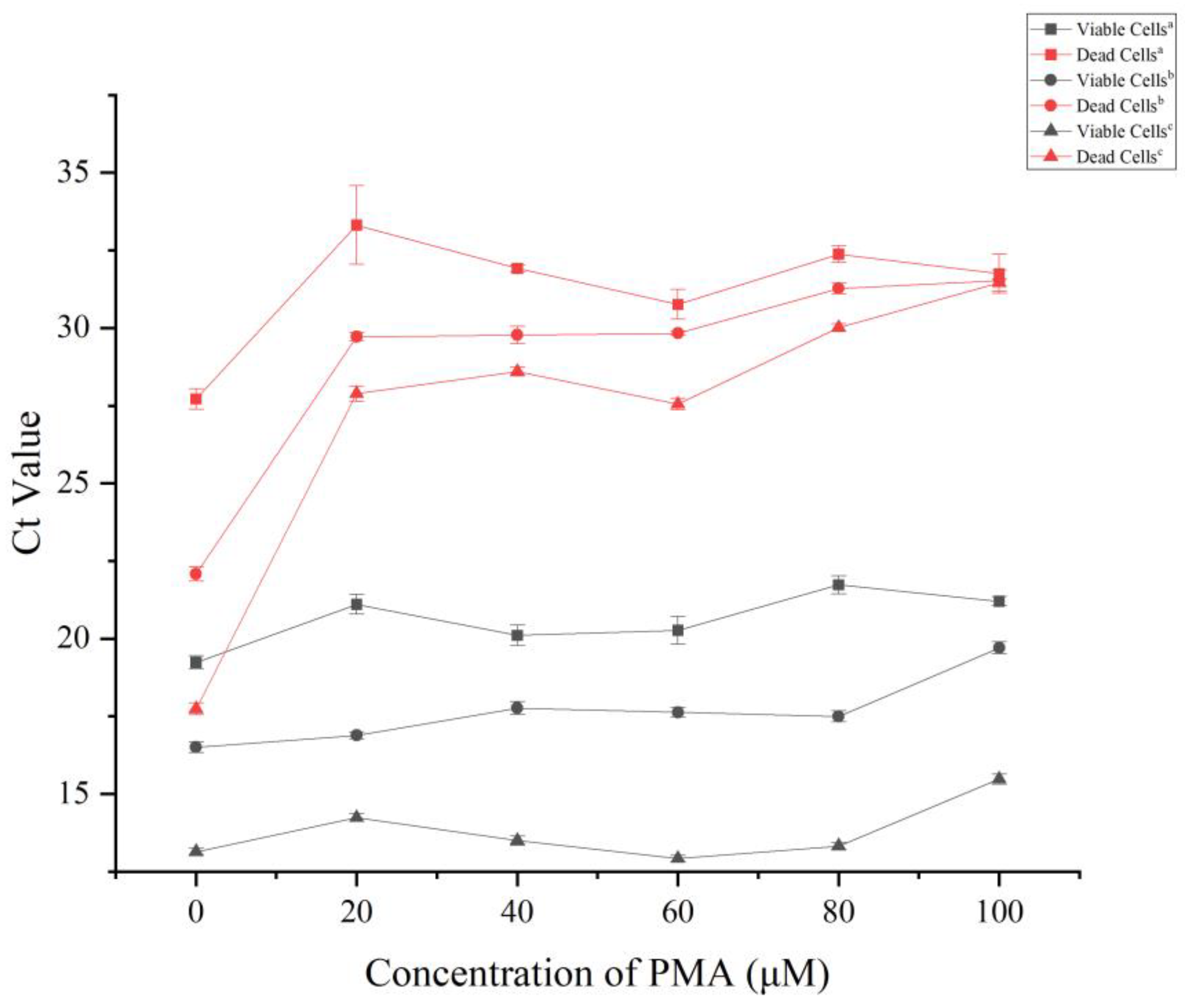
2.4. Optimization of the PMA Pretreatment Concentration
2.5. The PMA-qPCR Assay Using Different Ratios of Viable and Dead Cells
2.6. Analysis of Artificially Contaminated Cucumber Seeds via the PMA-qPCR Assay
2.7. Analysis of Pbr Mortality in Cucumber Seeds after Artificial Warm Broth Soaking Based on the PMA-qPCR Assay
2.8. Detection of Viable Pbr in Potato Tuber and Cucumber Seed Samples Collected from Different Regions of China Based on the PMA-qPCR Method
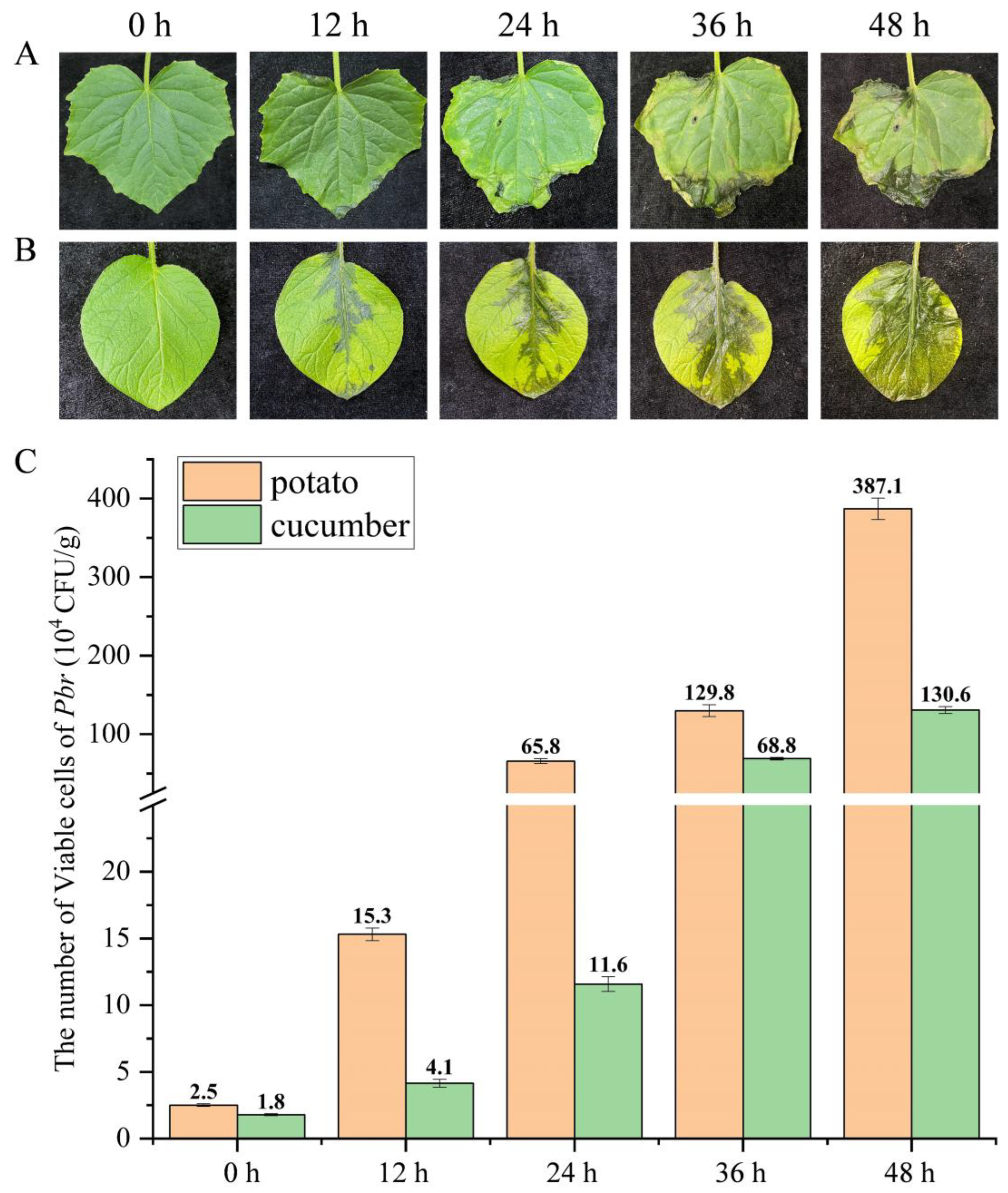
2.9. Dynamic Changes in Viable Pbr Amounts on Cucumber and Potato Leaves Detected via the PMA-qPCR Assay
2.10. Statistical Analysis
3. Results
3.1. Specificity of the Primers
3.2. Sensitivity and Standard Curve of Real-Time PCR
3.3. Optimization of PMA Pretreatment Conditions
3.4. The PMA-qPCR Viable Cell Assay
3.5. Detection of Viable Pbr Carried on Potato Tubers and Cucumber Seeds Based on the PMA-qPCR Assay
3.6. Effect of Warm Broth Soaking on the Activity of Pathogenic Bacteria
3.7. Dynamic Changes in Pbr on Potato and Cucumber Leaves Detected via PMA-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carstens, A.B.; Djurhuus, A.M.; Kot, W.; Jacobs-Sera, D.; Hatfull, G.F.; Hansen, L.H. Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani. Viruses 2018, 10, 621. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.O. The Changing Face of Bacterial Soft-Rot Diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Davis, M.J.; Brlansky, R.H. Quantification of live candidatus liberibacter asiaticus populations using real-time pcr and propidium monoazide. Plant Dis. 2017, 97, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Degefu, Y. Co-occurrence of latent Dickeya and Pectobacterium species in potato seed tuber samples from northern Finland: Co-colonization of latent Dickeya and Pectobacterium species in potato seed lots. Agric. Food Sci. 2021, 30, 1–7. [Google Scholar] [CrossRef]
- Merwe, J.J.V.D.; Coutinho, T.A.; Korsten, L.; Waals, J.E.V.D. Pectobacterium carotovorum subsp. brasiliensis causing blackleg on potatoes in South Africa. Eur. J. Plant Pathol. 2010, 126, 175–185. [Google Scholar] [CrossRef]
- Naas, H.; Sebaihia, M.; Orfei, B.; Rezzonico, F.; Buonaurio, R.; Moretti, C. Pectobacterium carotovorum subsp. brasiliense and Pectobacterium carotovorum subsp. carotovorum as causal agents of potato soft rot in Algeria. Eur. J. Plant Pathol. 2018, 151, 1027–1034. [Google Scholar] [CrossRef]
- Khojasteh, M.; Taghavi, S.M.; Khodaygan, P.; Hamzehzarghani, H.; Chen, G.; Bragard, C.; Koebnik, R.; Osdaghi, E. Molecular Typing Reveals High Genetic Diversity of Xanthomonas translucens Strains Infecting Small-Grain Cereals in Iran. Appl. Environ. Microbiol. 2019, 85, e01518-19. [Google Scholar] [CrossRef]
- Lukianova, A.A.; Evseev, P.V.; Stakheev, A.A.; Kotova, I.B.; Zavriev, S.K.; Ignatov, A.N.; Miroshnikov, K.A. Quantitative Real-Time PCR Assay for the Detection of Pectobacterium parmentieri, a Causal Agent of Potato Soft Rot. Plants 2021, 10, 1880. [Google Scholar] [CrossRef]
- Meng, X.; Chai, A.; Shi, Y.; Xie, X.; Ma, Z.; Li, B. Emergence of Bacterial Soft Rot in Cucumber Caused by Pectobacterium carotovorum subsp. brasiliense in China. Plant Dis. 2017, 101, 279–287. [Google Scholar] [CrossRef]
- Yuan, L.; Zhao, Y.; Xie, H.; Shi, Y.; Xie, X.; Chai, A.; Li, L.; Li, B. Selection and evaluation of suitable reference genes for quantitative gene expression analysis during infection of Cucumis sativus with Pectobacterium brasiliense. J. Appl. Microbiol. 2022, 132, 3717–3734. [Google Scholar] [CrossRef]
- Horvath, D.M.; Stall, R.E.; Jones, J.B.; Pauly, M.H.; Vallad, G.E.; Doug, D.; Staskawicz, B.J.; Scott, J.W.; Sunghun, P. Transgenic Resistance Confers Effective Field Level Control of Bacterial Spot Disease in Tomato. PLoS ONE 2012, 7, e42036. [Google Scholar] [CrossRef] [PubMed]
- Bartola, M.; Byrne, S.; Mullins, E. Characterization of Potato Virus Y Isolates and Assessment of Nanopore Sequencing to Detect and Genotype Potato Viruses. Viruses 2020, 12, 478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Feng, T.; Du, R.; Tian, X.; Wang, Y.; Zhang, X.H. Heterologous Expression of the Marine-Derived Quorum Quenching Enzyme MomL Can Expand the Antibacterial Spectrum of Bacillus brevis. Mar. Drugs 2019, 17, 128. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Shi, J.; Zhao, Z.; Ran, F.; Mo, F.; Long, Y.; Yin, X.; Li, W.; Chen, T.; Chen, J. First Report of Crown Gall of Kiwifruit (Actinidia deliciosa) Caused by Agrobacterium fabacearum in China and the Establishment of Loop-Mediated Isothermal Amplification Technique. Int. J. Mol. Sci. 2021, 23, 207. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Li, R. Detection and Control of Fusarium oxysporum from Soft Rot in Dendrobium officinale by Loop-Mediated Isothermal Amplification Assays. Biology 2021, 10, 1136. [Google Scholar] [CrossRef]
- Herpe, G.; Lederlin, M.; Naudin, M.; Ohana, M.; Chaumoitre, K.; Gregory, J.; Vilgrain, V.; Freitag, C.A.; De Margerie-Mellon, C.; Flory, V.; et al. Efficacy of Chest CT for COVID-19 Pneumonia Diagnosis in France. Radiology 2021, 298, E81–E87. [Google Scholar] [CrossRef]
- Patanita, M.; Campos, M.D.; Félix, M.D.R.; Carvalho, M.; Brito, I. Effect of Tillage System and Cover Crop on Maize Mycorrhization and Presence of Magnaporthiopsis maydis. Biology 2020, 9, 46. [Google Scholar] [CrossRef]
- Román-Reyna, V.; Dupas, E.; Cesbron, S.; Marchi, G.; Campigli, S.; Hansen, M.A.; Bush, E.; Prarat, M.; Shiplett, K.; Ivey, M.L.L.; et al. Metagenomic Sequencing for Identification of Xylella fastidiosa from Leaf Samples. mSystems 2021, 6, e0059121. [Google Scholar] [CrossRef]
- Abdolahzadeh, A.; Dolgosheina, E.V.; Unrau, P.J. RNA detection with high specificity and sensitivity using nested fluorogenic Mango NASBA. RNA 2019, 25, 1806–1813. [Google Scholar] [CrossRef]
- Krttzman, G. A method for detection of seedborne bacterial diseases in tomato seeds. Phytoparasitica 1991, 19, 133–141. [Google Scholar] [CrossRef]
- Aono, Y.; Nakayama, T.; Ozawa, T.; Ushio, Y.; Yasuoka, S.; Fujimoto, T.; Ohki, T.; Oka, N.; Maoka, T. Simple and sensitive BIO-PCR detection of potato blackleg pathogens from stem, tuber, and soil samples. J. Gen. Plant Pathol. 2021, 87, 209–218. [Google Scholar] [CrossRef]
- Li, X.; Nie, J.; Ward, L.J.; Nickerson, J.; De Boer, S.H. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection and identification of Pectobacterium atrosepticum. Can. J. Plant Pathol. 2011, 33, 447–457. [Google Scholar] [CrossRef]
- Duarte, V.; Boer, S.; Ward, L.J.; Oliveira, A. Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. J. Appl. Microbiol. 2010, 96, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Gillis, A.; Santana, M.A.; Rodríguez, M.; Romay, G. First Report of Bell Pepper Soft-Rot Caused by Pectobacterium carotovorum subsp. brasiliense in Venezuela. Plant Dis. 2017, 101, 1671. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, J.-B.; Lim, J.-A.; Han, S.-W.; Heu, S. Genetic Diversity of Pectobacterium carotovorum subsp. brasiliensis Isolated in Korea. Plant Pathol. J. 2014, 30, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Cheung, C.-Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef]
- Immanuel, T.; Taylor, R.; Keeling, S.; Brosnahan, C.; Alexander, B. Discrimination between viable and dead Xanthomonas fragariae in strawberry using viability PCR. J. Phytopathol. 2020, 168, 363–373. [Google Scholar] [CrossRef]
- Stulberg, M.; Santillana, G.; Studholme, D.J.; Kasiborski, B.; Rascoe, J. Genomics-informed molecular detection of Xanthomonas vasicola pv. vasculorum strains causing severe bacterial leaf streak of corn. Phytopathology 2019, 110, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Santander, R.D.; Meredith, C.L.; Aimovi, S.G. Publisher Correction: Development of a viability digital PCR protocol for the selective detection and quantification of live Erwinia amylovora cells in cankers. Sci. Rep. 2019, 9, 15796. [Google Scholar] [CrossRef]
- Meng, X.; Chai, A.; Chen, L.; Shi, Y.; Xie, X.; Ma, Z.; Li, B. Rapid detection and quantification of viable Pseudomonas syringae pv. lachrymans cells in contaminated cucumber seeds using propidium monoazide and a real-time PCR assay. Can. J. Plant Pathol. 2016, 38, 296–306. [Google Scholar] [CrossRef]
- Fronhoffs, S.; Totzke, G.; Stier, S.; Wernert, N.; Rothe, M.; Brüning, T.; Koch, B.; Sachinidis, A.; Vetter, H.; Ko, Y. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol. Cell. Probes 2002, 16, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-L.; Qi, X.-H.; Han, Z.-Y.; Guo, Y.-B.; Wang, Y.-N.; Hu, T.-L.; Wang, L.-M.; Cao, K.-Q.; Wang, S.-T. Latent Infection of Valsa mali in the Seeds, Seedlings and Twigs of Crabapple and Apple Trees is a Potential Inoculum Source of Valsa Canker. Sci. Rep. 2019, 9, 7738. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Ma, L.; Zhao, C.; Yan, J.; Che, S.; Zhou, Z.; Wang, H.; Yang, L.; Hu, B. Transcriptome of Pectobacterium carotovorum subsp. carotovorum PccS1 infected in calla plants in vivo highlights a spatiotemporal expression pattern of genes related to virulence, adaptation, and host response. Mol. Plant Pathol. 2020, 21, 871–891. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.Y.; Lee, M.; Park, S.K.; Lu, L.; Lee, G.; Kim, S.G.; Kang, S.Y.; Lim, Y.P. Jasmonate regulates plant resistance to Pectobacterium brasiliense by inducing indole glucosinolate biosynthesis. Front. Plant Sci. 2022, 13, 964092. [Google Scholar] [CrossRef]
- Jin, Y.J.; Jo, D.; Kwon, S.-W.; Jee, S.; Kim, J.-S.; Raman, J.; Kim, S.-J. A New Approach Using the SYBR Green-Based Real-Time PCR Method for Detection of Soft Rot Pectobacterium odoriferum Associated with Kimchi Cabbage. Plant Pathol. J. 2022, 38, 656–664. [Google Scholar] [CrossRef]
- Jeong, S.-G.; Lee, J.Y.; Yoon, S.-R.; Moon, E.W.; Ha, J.-H. A quantitative PCR based method using propidium monoazide for specific and sensitive detection of Pectobacterium carotovorum ssp. carotovorum in kimchi cabbage (Brassica rapa L. subsp. pekinensis). LWT 2019, 113, 108327. [Google Scholar] [CrossRef]
- Muzhinji, N.; Dube, J.P.; de Haan, E.G.; Woodhall, J.W.; van der Waals, J.E. Development of a TaqMan PCR assay for specific detection and quantification of Pectobacterium brasiliense in potato tubers and soil. Eur. J. Plant Pathol. 2020, 158, 521–532. [Google Scholar] [CrossRef]
- van der Wolf, J.M.; de Haan, E.G.; Kastelein, P.; Krijger, M.; de Haas, B.H.; Velvis, H.; Mendes, O.; Kooman-Gersmann, M.; van der Zouwen, P.S. Virulence of Pectobacterium carotovorum subsp. brasiliense on potato compared with that of other Pectobacterium and Dickeya species under climatic conditions prevailing in The Netherlands. Plant Pathol. 2017, 66, 571–583. [Google Scholar] [CrossRef]
- Bellieny-Rabelo, D.; Nkomo, N.P.; Shyntum, D.Y.; Moleleki, L.N. Horizontally Acquired Quorum-Sensing Regulators Recruited by the PhoP Regulatory Network Expand the Host Adaptation Repertoire in the Phytopathogen Pectobacterium brasiliense. mSystems 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- Rivière, A.; Gagnon, M.; Weckx, S.; Roy, D.; Vuyst, L.D. Mutual Cross-Feeding Interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 Explain the Bifidogenic and Butyrogenic Effects of Arabinoxylan Oligosaccharides. Appl. Environ. Microbiol. 2015, 81, 7767–7781. [Google Scholar] [CrossRef]
- Okada, A.; Tsuchida, M.; Rahman, M.M.; Inoshima, Y. Two-Round Treatment With Propidium Monoazide Completely Inhibits the Detection of Dead Campylobacter spp. Cells by Quantitative PCR. Front. Microbiol. 2022, 13, 801961. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Mazza, A.; Masson, L.; Camper, A.K.; Brousseau, R. Selective detection of live bacteria combining propidium monoazide sample treatment with microarray technology. J. Microbiol. Methods 2009, 76, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wagnon, R.; Moreno, D.; Timilsina, S.; Jones, J.; Vallad, G.; Turechek, W.W. A Long-Amplicon Viability-qPCR Test for Quantifying Living Pathogens that Cause Bacterial Spot in Tomato Seed. Plant Dis. 2021, 106, 1474–1485. [Google Scholar] [CrossRef] [PubMed]
- Elizaquível, P.; Sánchez, G.; Selma, M.V.; Aznar, R. Application of propidium monoazide-qPCR to evaluate the ultrasonic inactivation of Escherichia coli O157:H7 in fresh-cut vegetable wash water. Food Microbiol. 2012, 30, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Vesper, S.; McKinstry, C.; Hartmann, C.; Neace, M.; Yoder, S.; Vesper, A. Quantifying fungal viability in air and water samples using quantitative PCR after treatment with propidium monoazide (PMA). J. Microbiol. Methods 2008, 72, 180–184. [Google Scholar] [CrossRef]
- Crespo-Sempere, A.; Estiarte, N.; Marín, S.; Sanchis, V.; Ramos, A.J. Propidium monoazide combined with real-time quantitative PCR to quantify viable Alternaria spp. contamination in tomato products. Int. J. Food Microbiol. 2013, 165, 214–220. [Google Scholar] [CrossRef]
- Zhang, Z.; Fan, J.; Feng, M.; Qiu, H.; Hu, A. Polymerase Chain Reaction-Assisted Evaluation of the Efficacy of Seed-Treatment Prevention of Sporisorium reilianum Infection in Sorghum Seedlings. Front. Microbiol. 2021, 12, 745144. [Google Scholar] [CrossRef]
- Dersch, L.M.; Beckers, V.; Rasch, D.; Melzer, G.; Bolten, C.; Kiep, K.; Becker, H.; Bläsing, O.E.; Fuchs, R.; Ehrhardt, T.; et al. Novel Approach for High-Throughput Metabolic Screening of Whole Plants by Stable Isotopes. Plant Physiol. 2016, 171, 25–41. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, S.; Cao, Z.; Ouyang, W.; Zhang, Q.; Xie, L.; Zheng, R.; Guo, M.; Ma, M.; Hu, Z.; et al. Chromatin loops associated with active genes and heterochromatin shape rice genome architecture for transcriptional regulation. Nat. Commun. 2019, 10, 3640. [Google Scholar] [CrossRef]
- Wang, S.; Jin, N.; Jin, L.; Xiao, X.; Hu, L.; Liu, Z.; Wu, Y.; Xie, Y.; Zhu, W.; Lyu, J.; et al. Response of Tomato Fruit Quality Depends on Period of LED Supplementary Light. Front. Nutr. 2022, 9, 833723. [Google Scholar] [CrossRef]
- Saito, H.; Sasaki, M.; Nonaka, Y.; Tanaka, J.; Tokunaga, T.; Kato, A.; Thuy, T.T.T.; Vang, L.V.; Tuong, L.M.; Kanematsu, S.; et al. Spray Application of Nonpathogenic Fusaria onto Rice Flowers Controls Bakanae Disease (Caused by Fusarium fujikuroi) in the Next Plant Generation. Appl. Environ. Microbiol. 2021, 87, e01959-20. [Google Scholar] [CrossRef] [PubMed]


| Strain No. | Species | Isolate Code | Host | Geographic Origin | qPCR Test a |
|---|---|---|---|---|---|
| 1 | Pectobacterium brasiliense | SX309 | Cucumber | Shanxi, China | + |
| 2 | Pectobacterium brasiliense | B6 | Chinese cabbage | Beijing, China | + |
| 3 | Pectobacterium brasiliense | KC08 | Chinese cabbage | Beijing, China | + |
| 4 | Pectobacterium brasiliense | WBC12 | Chinese cabbage | Beijing, China | + |
| 5 | Pectobacterium brasiliense | WQC3 | Celery | Beijing, China | + |
| 6 | Pectobacterium brasiliense | Y2 | Bok choy | Beijing, China | + |
| 7 | Pectobacterium brasiliense | WYC1 | Bok choy | Beijing, China | + |
| 8 | Pectobacterium brasiliense | HG1501290301 | Cucumber | Shanxi, China | + |
| 9 | Pectobacterium brasiliense | HG1501290302 | Cucumber | Shanxi, China | + |
| 10 | Pectobacterium brasiliense | HG1501290303 | Cucumber | Shanxi, China | + |
| 11 | Pectobacterium brasiliense | HG1501290304 | Cucumber | Shanxi, China | + |
| 12 | Pectobacterium brasiliense | HG1501290305 | Cucumber | Shanxi, China | + |
| 13 | Pectobacterium brasiliense | HG1501290306 | Cucumber | Shanxi, China | + |
| 14 | Pectobacterium brasiliense | HG1501290307 | Cucumber | Shanxi, China | + |
| 15 | Pectobacterium brasiliense | HG1501290308 | Cucumber | Shanxi, China | + |
| 16 | Pectobacterium aroidearum | MTL1911110302 | Common calla-lily rhizome | China | − |
| 17 | Pectobacterium aroidearum | MY2 | Konjac | Yunnan, China | − |
| 18 | Pectobacterium aroidearum | MY7 | Konjac | Yunnan, China | − |
| 19 | Pectobacterium aroidearum | MY10 | Konjac | Yunnan, China | − |
| 20 | Pectobacterium aroidearum | MY10 | Konjac | Sichuan, China | − |
| 21 | Pectobacterium carotovorum | ATCC 15713 | Potato | Denmark | − |
| 22 | Pectobacterium carotovorum | ATCC 39048 | Potato | UK | − |
| 23 | Pectobacterium carotovorum | B2 | Chinese cabbage | Beijing, China | − |
| 24 | Pectobacterium carotovorum | Y13 | Bok choy | Beijing, China | − |
| 25 | Pectobacterium carotovorum | WBC8 | Bok choy | Beijing, China | − |
| 26 | Pectobacterium Polaris | NIBIO1006T | Potato | Norway | − |
| 27 | Pectobacterium Polaris | WBC1 | Chinese cabbage | Beijing, China | − |
| 28 | Pectobacterium Polaris | WBC6 | Chinese cabbage | Beijing, China | − |
| 29 | Pectobacterium Polaris | WBC9 | Chinese cabbage | Beijing, China | − |
| 30 | Pectobacterium Polaris | HYC22041801 | Broccoli | Yunnan, China | − |
| 31 | Pectobacterium Polaris | SC22041801 | Lettuce | Yunnan, China | − |
| 23 | Pectobacterium odoriferum | ATCC 25272 | Potato | Unknown | − |
| 33 | Pectobacterium odoriferum | T4 | Chinese cabbage | Beijing, China | − |
| 34 | Pectobacterium odoriferum | WBC30 | Chinese cabbage | Beijing, China | − |
| 35 | Pectobacterium odoriferum | Q3 | Celery | Beijing, China | − |
| 36 | Pectobacterium odoriferum | WQC17 | Celery | Beijing, China | − |
| 37 | Pectobacterium versatile | BXH21032402 | Hydrangea | Yunnan, China | − |
| 38 | Pectobacterium versatile | Y6 | Bok choy | Beijing, China | − |
| 39 | Pectobacterium versatile | KC-01 | Chinese cabbage | Beijing, China | − |
| 40 | Pectobacterium versatile | KC-03 | Chinese cabbage | Beijing, China | − |
| 41 | Pectobacterium versatile | KC-06 | Chinese cabbage | Beijing, China | − |
| 42 | Pseudomonas syringae tomato | ATCC BAA-871 | Tomato | UK | − |
| 43 | Pseudomonas syringae lachrymans | NM002 | Cucumber | Beijing, China | − |
| 44 | Xanthomonas campestris | ATCC 33913 | Cabbage | UK | − |
| 45 | Ralstonia solanacearum | FQ1112080402 | Tomato | Unknown | − |
| 46 | Acidovorax citrulli | ATCC 29625 | Watermelon | USA | − |
| 47 | Clavibacter michiganensis subsp. Michiganensis | ATCC 10202 | Tomato | Unknown | − |
| 48 | Erwinia amylovora | XL2104240301 | Pear | Xinjiang, China | − |
| Ratio of Viable Cells a | qPCR | qPCR-PMA | Colony Counting | ||
|---|---|---|---|---|---|
| Ct Values | Log10 (copies·mL−1) b | Ct values | Log10 (copies·mL−1) b | Log10 (CFU·mL−1) c | |
| 100% | 21.71 ± 0.12 a | 6.27 | 20.96 ± 0.17 a | 6.49 | 7.04 |
| 50% | 22.98 ± 0.11 a | 5.89 | 22.87 ± 0.16 b | 5.92 | 6.79 |
| 10% | 24.11 ± 0.07 b | 5.56 | 25.43 ± 0.08 c | 5.20 | 6.01 |
| 1% | 22.34 ± 0.15 a | 6.08 | 27.98 ± 0.21 d | 4.42 | 5.08 |
| 0.1% | 22.79 ± 0.35 a | 5.95 | 32.21 ± 0.15 e | 3.17 | 4.24 |
| 0.01% | 25.65 ± 0.13 b | 5.11 | 34.73 ± 0.16 f | 2.34 | 3.31 |
| Inoculating Concentration of Pbr (CFU·mL−1) | Ct Values | Number of Live Bacteria Per Gram of Cucumber Seeds (CFU·g−1) a | Number of Live Bacteria Per Gram of Cucumber Seeds (CFU·g−1) b | Bioassay |
|---|---|---|---|---|
| DI (%) | ||||
| 1 × 108 | 23.89 ± 0.32 a | 8.49 × 105 | 9.12 × 105 | 78.34 ± 1.34 |
| 1 × 107 | 27.13 ± 0.14 b | 1.86 × 105 | 1.62 × 105 | 53.73 ± 2.81 |
| 1 × 106 | 29.64 ± 0.26 c | 1.71 × 104 | 2.18 × 104 | 38.58 ± 2.54 |
| 1 × 105 | 32.28 ± 0.17 d | 2.85 × 103 | 4.02 × 103 | 0.00 |
| 1 × 104 | 34.59 ± 0.28 e | 5.94 × 102 | 4.43 × 102 | 0.00 |
| 1 × 103 | >35 | - | - | 0.00 |
| 1 × 102 | >35 | - | - | 0.00 |
| 1 × 101 | >35 | - | - | 0.00 |
| Naturally Infested Seed Lots | Ct Values | Concentration (CFU·g−1) | Colony Counting (CFU·g−1) | |
|---|---|---|---|---|
| Sample No. | Location | |||
| 1 | Xinjiang, China | 31.73 ± 0.45 a | 4.14 × 103 | 5.21 × 104 |
| 2 | Xinjiang, China | 31.41 ± 0.91 a | 5.15 × 103 | 4.42 × 104 |
| 3 | Xinjiang, China | 33.37 ± 1.12 b | 1.36 × 103 | 3.23 × 104 |
| 4 | Inner Mongolia, China | 34.31 ± 0.33 bc | 7.19 × 102 | 1.54 × 103 |
| 5 | Inner Mongolia, China | 33.41 ± 0.59 b | 1.32 × 103 | 4.58 × 103 |
| 6 | Inner Mongolia, China | 33.91 ± 0.13 b | 9.43 × 102 | 2.14 × 103 |
| 7 | Heilongjiang, China | 33.22 ± 0.47 b | 1.51 × 103 | 7.33 × 103 |
| 8 | Heilongjiang, China | 32.85 ± 0.36 ab | 1.94 × 103 | 7.29 × 103 |
| 9 | Xinjiang, China | >35 | - | - |
| 10 | Xinjiang, China | >35 | - | - |
| 11 | Xinjiang, China | >35 | - | - |
| 12 | Inner Mongolia, China | >35 | - | - |
| 13 | Inner Mongolia, China | >35 | - | - |
| 14 | Heilongjiang, China | >35 | - | - |
| 15 | Heilongjiang, China | >35 | - | - |
| Naturally Infested Seed Lots | Ct Values | Concentration (CFU·g−1) | Colony Counting (CFU·g−1) | |
|---|---|---|---|---|
| Sample No. | Location | |||
| 1 | Shandong, China | 28.17 ± 0.32 ab | 4.64 × 104 | 1.33 × 105 |
| 2 | Shandong, China | 28.89 ± 0.21 b | 2.85 × 104 | 4.83 × 105 |
| 3 | Shandong, China | 27.39 ± 0.42 a | 7.89 × 103 | 4.31 × 104 |
| 4 | Liaoning, China | 27.32 ± 0.33 a | 8.27 × 104 | 2.93 × 105 |
| 5 | Liaoning, China | 28.55 ± 0.16 ab | 3.59 × 104 | 1.81 × 105 |
| 6 | Liaoning, China | 29.03 ± 0.19 b | 2.59 × 103 | 3.23 × 104 |
| 7 | Shanxi, China | 30.11 ± 0.23 c | 1.24 × 103 | 1.67 × 104 |
| 8 | Shanxi, China | 28.75 ± 0.31 ab | 3.13 × 104 | 5.24 × 105 |
| 9 | Shanxi, China | 28.14 ± 0.28 a | 4.74 × 104 | 4.23 × 105 |
| Other 31 samples | >35 | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Chen, R.; Yang, R.; Wei, X.; Xie, H.; Shi, Y.; Xie, X.; Chai, A.; Fan, T.; Li, B.; et al. Rapid Detection and Quantification of Viable Cells of Pectobacterium brasiliense Using Propidium Monoazide Combined with Real-Time PCR. Microorganisms 2023, 11, 2808. https://doi.org/10.3390/microorganisms11112808
Li J, Chen R, Yang R, Wei X, Xie H, Shi Y, Xie X, Chai A, Fan T, Li B, et al. Rapid Detection and Quantification of Viable Cells of Pectobacterium brasiliense Using Propidium Monoazide Combined with Real-Time PCR. Microorganisms. 2023; 11(11):2808. https://doi.org/10.3390/microorganisms11112808
Chicago/Turabian StyleLi, Junhui, Ruxing Chen, Ruwei Yang, Xinchen Wei, Hua Xie, Yanxia Shi, Xuewen Xie, Ali Chai, Tengfei Fan, Baoju Li, and et al. 2023. "Rapid Detection and Quantification of Viable Cells of Pectobacterium brasiliense Using Propidium Monoazide Combined with Real-Time PCR" Microorganisms 11, no. 11: 2808. https://doi.org/10.3390/microorganisms11112808
APA StyleLi, J., Chen, R., Yang, R., Wei, X., Xie, H., Shi, Y., Xie, X., Chai, A., Fan, T., Li, B., & Li, L. (2023). Rapid Detection and Quantification of Viable Cells of Pectobacterium brasiliense Using Propidium Monoazide Combined with Real-Time PCR. Microorganisms, 11(11), 2808. https://doi.org/10.3390/microorganisms11112808







