The Impact of Warming on Assembly Processes and Diversity Patterns of Bacterial Communities in Mesocosms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mesocosm Experiment Setup
2.2. Samples Collection and Environmental Parameters
2.3. DNA Extraction, PCR, and Sequencing
2.4. Network Construction and Analysis
2.5. Bacterial Community Assembly Analysis
2.6. Statistical Analysis
3. Results
3.1. Warming Effects on Physical and Chemical Indicators
3.2. Bacterial Community Composition and Diversity in Water and Sediments
3.3. Co-Occurrence Patterns of Microbial Communities
3.4. Disentangling Community Assembly Processes in Water and Sediments
4. Discussion
4.1. Habitat Differences
4.2. Assembly Process Differences
4.3. The Impact of Seasonal Change and Warming
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPCC. Climate Change 2021: The Physical Science Basis; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2021. [Google Scholar]
- Woolway, R.I.; Albergel, C.; Frolicher, T.L.; Perroud, M. Severe Lake Heatwaves Attributable to Human-Induced Global Warming. Geophys. Res. Lett. 2022, 49, e2021GL097031. [Google Scholar] [CrossRef]
- Jassey, V.E.; Chiapusio, G.; Binet, P.; Buttler, A.; Laggoun-Defarge, F.; Delarue, F.; Bernard, N.; Mitchell, E.A.; Toussaint, M.L.; Francez, A.J.; et al. Above- and belowground linkages in Sphagnum peatland: Climate warming affects plant-microbial interactions. Glob. Chang. Biol. 2013, 19, 811–823. [Google Scholar] [CrossRef]
- Adrian, R.; O′Reilly, C.M.; Zagarese, H.; Baines, S.B.; Hessen, D.O.; Keller, W.; Livingstone, D.M.; Sommaruga, R.; Straile, D.; Van Donk, E.; et al. Lakes as sentinels of climate change. Limnol. Oceanogr. 2009, 54, 2283–2297. [Google Scholar] [CrossRef]
- Schmid, M.; Hunziker, S.; Wuest, A. Lake surface temperatures in a changing climate: A global sensitivity analysis. Clim. Chang. 2014, 124, 301–315. [Google Scholar] [CrossRef]
- Hunt, D.E.; Ward, C.S. A network-based approach to disturbance transmission through microbial interactions. Front. Microbiol. 2015, 6, 1182. [Google Scholar] [CrossRef]
- Yang, Y.; Dai, Y.; Li, N.; Li, B.; Xie, S.; Liu, Y. Temporal and Spatial Dynamics of Sediment Anaerobic Ammonium Oxidation (Anammox) Bacteria in Freshwater Lakes. Microb. Ecol. 2017, 73, 285–295. [Google Scholar] [CrossRef]
- Zhou, J.; Ning, D. Stochastic Community Assembly: Does It Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. MMBR 2017, 81, e00002-17. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O′Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. MMBR 2013, 77, 342–356. [Google Scholar] [CrossRef]
- Li, Y.; Gao, Y.; Zhang, W.; Wang, C.; Wang, P.; Niu, L.; Wu, H. Homogeneous selection dominates the microbial community assembly in the sediment of the Three Gorges Reservoir. Sci. Total Environ. 2019, 690, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D. Niche tradeoffs, neutrality, and community structure: A stochastic theory of resource competition, invasion, and community assembly. Proc. Natl. Acad. Sci. USA 2004, 101, 10854–10861. [Google Scholar] [CrossRef] [PubMed]
- Graham, E.B.; Stegen, J.C. Dispersal-Based Microbial Community Assembly Decreases Biogeochemical Function. Processes 2017, 5, 65. [Google Scholar] [CrossRef]
- Knelman, J.E.; Nemergut, D.R. Changes in community assembly may shift the relationship between biodiversity and ecosystem function. Front. Microbiol. 2014, 5, 424. [Google Scholar] [CrossRef]
- Ponisio, L.C.; Valdovinos, F.S.; Allhoff, K.T.; Gaiarsa, M.P.; Barner, A.; Guimarães, P.R.; Hembry, D.H.; Morrison, B.; Gillespie, R. A Network Perspective for Community Assembly. Front. Ecol. Evol. 2019, 7, 103. [Google Scholar] [CrossRef]
- Ernakovich, J.G.; Barbato, R.A.; Rich, V.I.; Schadel, C.; Hewitt, R.E.; Doherty, S.J.; Whalen, E.D.; Abbott, B.W.; Barta, J.; Biasi, C.; et al. Microbiome assembly in thawing permafrost and its feedbacks to climate. Glob. Chang. Biol. 2022, 28, 5007–5026. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Jiao, C.; Zhao, D.; Xu, H.; Huang, R.; Cao, X.; Yu, Z.; Wu, Q.L. Patterns and assembly processes of planktonic and sedimentary bacterial community differ along a trophic gradient in freshwater lakes. Ecol. Indic. 2019, 106, 105491. [Google Scholar] [CrossRef]
- Song, H.; Li, Z.; Du, B.; Wang, G.; Ding, Y. Bacterial communities in sediments of the shallow Lake Dongping in China. J. Appl. Microbiol. 2012, 112, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Ruan, X.; Zhang, Y.; Li, R. Illumina sequencing-based analysis of sediment bacteria community in different trophic status freshwater lakes. Microbiologyopen 2017, 6, e00450. [Google Scholar] [CrossRef]
- Parker, S.R.; West, R.F.; Boyd, E.S.; Feyhl-Buska, J.; Gammons, C.H.; Johnston, T.B.; Williams, G.P.; Poulson, S.R. Biogeochemical and microbial seasonal dynamics between water column and sediment processes in a productive mountain lake: Georgetown Lake, MT, USA. J. Geophys. Res.-Biogeosci. 2016, 121, 2064–2081. [Google Scholar] [CrossRef]
- Jiang, H.; Dong, H.; Zhang, G.; Yu, B.; Chapman, L.R.; Fields, M.W. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 2006, 72, 3832–3845. [Google Scholar] [CrossRef]
- Roske, K.; Sachse, R.; Scheerer, C.; Roske, I. Microbial diversity and composition of the sediment in the drinking water reservoir Saidenbach (Saxonia, Germany). Syst. Appl. Microbiol. 2012, 35, 35–44. [Google Scholar] [CrossRef]
- Lenaker, P.L.; Baldwin, A.K.; Corsi, S.R.; Mason, S.A.; Reneau, P.C.; Scott, J.W. Vertical Distribution of Microplastics in the Water Column and Surficial Sediment from the Milwaukee River Basin to Lake Michigan. Environ. Sci. Technol. 2019, 53, 12227–12237. [Google Scholar] [CrossRef] [PubMed]
- Pauer, J.J.; Auer, M.T. Nitrification in the water column and sediment of a hypereutrophic lake and adjoining river system. Water Res. 2000, 34, 1247–1254. [Google Scholar] [CrossRef]
- Dalu, T.; Wasserman, R.J.; Tonkin, J.D.; Mwedzi, T.; Magoro, M.L.; Weyl, O.L.F. Water or sediment? Partitioning the role of water column and sediment chemistry as drivers of macroinvertebrate communities in an austral South African stream. Sci. Total Environ. 2017, 607–608, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.L. Microorganisms and their roles in fundamental biogeochemical cycles. Curr. Opin. Biotechnol. 2011, 22, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.Q.; Bachand, S.C.; McIntyre, P.B.; Kraemer, B.M.; Vadeboncoeur, Y.; Kimirei, I.A.; Tamatamah, R.; McMahon, K.D.; Anantharaman, K. Depth-discrete metagenomics reveals the roles of microbes in biogeochemical cycling in the tropical freshwater Lake Tanganyika. ISME J. 2021, 15, 1971–1986. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, A.N.; Wang, J.; Liu, S.; Jiang, X.; Dang, C.; Ma, T.; Liu, S.; Chen, Q.; Xie, S.; et al. Integrated biogeography of planktonic and sedimentary bacterial communities in the Yangtze River. Microbiome 2018, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.A.; Lamanna, C.; Morlon, H.; Kerkhoff, A.J.; Enquist, B.J.; Green, J.L. Colloquium paper: Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. USA 2008, 105 (Suppl. S1), 11505–11511. [Google Scholar] [CrossRef]
- Ellegaard, M.; Clokie, M.R.J.; Czypionka, T.; Frisch, D.; Godhe, A.; Kremp, A.; Letarov, A.; McGenity, T.J.; Ribeiro, S.; John Anderson, N. Dead or alive: Sediment DNA archives as tools for tracking aquatic evolution and adaptation. Commun. Biol. 2020, 3, 169. [Google Scholar] [CrossRef]
- Langenheder, S.; Szekely, A.J. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 2011, 5, 1086–1094. [Google Scholar] [CrossRef]
- Classen, A.T.; Sundqvist, M.K.; Henning, J.A.; Newman, G.S.; Moore, J.A.M.; Cregger, M.A.; Moorhead, L.C.; Patterson, C.M. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere 2015, 6, 1–21. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Bruns, M.A.; Casadevall, A.; Criddle, C.S.; Eloe-Fadrosh, E.; Karl, D.M.; Nguyen, N.K.; Zhou, J. Microbes and Climate Change: A Research Prospectus for the Future. mBio 2022, 13, e0080022. [Google Scholar] [CrossRef] [PubMed]
- Aydogan, E.L.; Moser, G.; Müller, C.; Kämpfer, P.; Glaeser, S.P. Long-Term Warming Shifts the Composition of Bacterial Communities in the Phyllosphere of Galium album in a Permanent Grassland Field-Experiment. Front. Microbiol. 2018, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.C.; Tu, Q.Y. Investigation Specifications for Lake Eutrophication, 2nd ed.; China Environmental Science Press: Beijing, China, 1990. [Google Scholar]
- Liu, Y.; Ren, Z.; Qu, X.; Zhang, M.; Yu, Y.; Zhang, Y.; Peng, W. Microbial community structure and functional properties in permanently and seasonally flooded areas in Poyang Lake. Sci. Rep. 2020, 10, 4819. [Google Scholar] [CrossRef] [PubMed]
- Peter, H.; Sommaruga, R. Shifts in diversity and function of lake bacterial communities upon glacier retreat. ISME J. 2016, 10, 1545–1554. [Google Scholar] [CrossRef]
- Hall, E.K.; Neuhauser, C.; Cotner, J.B. Toward a mechanistic understanding of how natural bacterial communities respond to changes in temperature in aquatic ecosystems. ISME J. 2008, 2, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Adams, H.E.; Crump, B.C.; Kling, G.W. Temperature controls on aquatic bacterial production and community dynamics in arctic lakes and streams. Environ. Microbiol. 2010, 12, 1319–1333. [Google Scholar] [CrossRef]
- He, Q.; Wang, S.; Hou, W.; Feng, K.; Li, F.; Hai, W.; Zhang, Y.; Sun, Y.; Deng, Y. Temperature and microbial interactions drive the deterministic assembly processes in sediments of hot springs. Sci. Total Environ. 2021, 772, 145465. [Google Scholar] [CrossRef]
- Ren, L.; He, D.; Chen, Z.; Jeppesen, E.; Lauridsen, T.L.; Sondergaard, M.; Liu, Z.; Wu, Q.L. Warming and nutrient enrichment in combination increase stochasticity and beta diversity of bacterioplankton assemblages across freshwater mesocosms. ISME J. 2017, 11, 613–625. [Google Scholar] [CrossRef]
- Xu, J.; Wang, T.; Garcia Molinos, J.; Li, C.; Hu, B.; Pan, M.; Zhang, M. Effects of warming, climate extremes and phosphorus enrichment on the growth, sexual reproduction and propagule carbon and nitrogen stoichiometry of Potamogeton crispus L. Environ. Int. 2020, 137, 105502. [Google Scholar] [CrossRef]
- Wang, T.; Xu, J.; Molinos, J.G.; Li, C.; Hu, B.; Pan, M.; Zhang, M. A dynamic temperature difference control recording system in shallow lake mesocosm. MethodsX 2020, 7, 100930. [Google Scholar] [CrossRef]
- IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Stocker, T.F., Qin, D., Plattner, G.-K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., Midgley, P.M., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2013; p. 1535. [Google Scholar]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Rider, B.F.; Mellon, M.G. Colorimetric Determination of Nitrites. Ind. Eng. Chem. Anal. Ed. 1946, 18, 96–99. [Google Scholar] [CrossRef]
- Batten, J.J. Spectrophotometric Microdetermination of Nitrate with Chromotropic Acid Reagent. Anal. Chem. 1964, 36, 939–940. [Google Scholar] [CrossRef]
- Jespersen, A.M.; Christoffersen, K. Measurements of Chlorophyll-a from Phytoplankton Using Ethanol as Extraction Solvent. Arch. Hydrobiol. 1987, 109, 445–454. [Google Scholar] [CrossRef]
- Xu, N.; Tan, G.C.; Wang, H.Y.; Gai, X.P. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005, 71, 1501–1506. [Google Scholar] [CrossRef]
- Xiao, X.; Fan, M.; Wang, E.; Chen, W.; Wei, G. Interactions of plant growth-promoting rhizobacteria and soil factors in two leguminous plants. Appl. Microbiol. Biotechnol. 2017, 101, 8485–8497. [Google Scholar] [CrossRef] [PubMed]
- Barberan, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef]
- Zhao, D.; Shen, F.; Zeng, J.; Huang, R.; Yu, Z.; Wu, Q.L. Network analysis reveals seasonal variation of co-occurrence correlations between Cyanobacteria and other bacterioplankton. Sci. Total Environ. 2016, 573, 817–825. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B-Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Brislawn, C.J.; Graham, E.B.; Dana, K.; Ihardt, P.; Fansler, S.J.; Chrisler, W.B.; Cliff, J.B.; Stegen, J.C.; Moran, J.J.; Bernstein, H.C. Forfeiting the priority effect: Turnover defines biofilm community succession. ISME J. 2019, 13, 1865–1877. [Google Scholar] [CrossRef]
- Dini-Andreote, F.; Stegen, J.C.; van Elsas, J.D.; Salles, J.F. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 2015, 112, E1326–E1332. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef]
- Moroenyane, I.; Chimphango, S.B.; Wang, J.; Kim, H.K.; Adams, J.M. Deterministic assembly processes govern bacterial community structure in the Fynbos, South Africa. Microb. Ecol. 2016, 72, 313–323. [Google Scholar] [CrossRef]
- Meyerhof, M.S.; Wilson, J.M.; Dawson, M.N.; Michael Beman, J. Microbial community diversity, structure and assembly across oxygen gradients in meromictic marine lakes, Palau. Environ. Microbiol. 2016, 18, 4907–4919. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Konopka, A.E. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 2015, 6, 370. [Google Scholar] [CrossRef]
- Chase, J.M.; Myers, J.A. Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 2351–2363. [Google Scholar] [CrossRef] [PubMed]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar] [CrossRef]
- Anderson, M.J. PERMANOVA: A FORTRAN Computer Program for Permutational Multivariate Analysis of Variance Permutational Multivariate Analysis of Variance, a Computer Program; Department of Statistics, University of Auckland: Auckland, New Zealand, 2005. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R. Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Bucci, J.; Szempruch, A.; Caldwell, J.; Ellis, J.; Levine, J. Seasonal Changes in Microbial Community Structure in Freshwater Stream Sediment in a North Carolina River Basin. Diversity 2014, 6, 18–32. [Google Scholar] [CrossRef]
- Ren, L.J.; Liu, Y.Y.; Lauridsen, T.L.; Sondergaard, M.; Han, B.P.; Wang, J.J.; Jeppesen, E.; Zhou, J.Z.; Wu, Q.L.L. Warming exacerbates the impact of nutrient enrichment on microbial functional potentials important to the nutrient cycling in shallow lake mesocosms. Limnol. Oceanogr. 2021, 66, 2481–2495. [Google Scholar] [CrossRef]
- Mikhailov, I.S.; Zakharova, Y.R.; Bukin, Y.S.; Galachyants, Y.P.; Petrova, D.P.; Sakirko, M.V.; Likhoshway, Y.V. Co-occurrence Networks Among Bacteria and Microbial Eukaryotes of Lake Baikal During a Spring Phytoplankton Bloom. Microb. Ecol. 2019, 77, 96–109. [Google Scholar] [CrossRef]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Xun, W.; Li, W.; Xiong, W.; Ren, Y.; Liu, Y.; Miao, Y.; Xu, Z.; Zhang, N.; Shen, Q.; Zhang, R. Diversity-triggered deterministic bacterial assembly constrains community functions. Nat. Commun. 2019, 10, 3833. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.; Hugerth, L.W.; Logue, J.B.; Bruchert, V.; Andersson, A.F.; Holmstrom, S.J.M. Mineral Type Structures Soil Microbial Communities. Geomicrobiol. J. 2017, 34, 538–545. [Google Scholar] [CrossRef]
- Zinger, L.; Boetius, A.; Ramette, A. Bacterial taxa–area and distance–decay relationships in marine environments. Mol. Ecol. 2014, 23, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Z.; Qi, X.; Wang, Y.; Wang, Y.; Jiang, W.; He, F.; Wu, N. Environmental filtering, spatial processes and biotic interactions jointly shape different traits communities of stream macroinvertebrates. Front. Ecol. Evol. 2023, 11, 1196296. [Google Scholar] [CrossRef]
- Limberger, R.; Pitt, A.; Hahn, M.W.; Wickham, S.A. Spatial insurance in multi-trophic metacommunities. Ecol. Lett. 2019, 22, 1828–1837. [Google Scholar] [CrossRef]
- Yue, Y.; Tang, Y.; Cai, L.; Yang, Z.; Chen, X.; Ouyang, Y.; Dai, J.; Yang, M. Co-Occurrence Relationship and Stochastic Processes Affect Sedimentary Archaeal and Bacterial Community Assembly in Estuarine-Coastal Margins. Microorganisms 2022, 10, 1339. [Google Scholar] [CrossRef]
- Passy, S.I.; Larson, C.A. Succession in stream biofilms is an environmentally driven gradient of stress tolerance. Microb. Ecol. 2011, 62, 414–424. [Google Scholar] [CrossRef]
- Jäger, C.; Diehl, S. Resource competition across habitat boundaries: Asymmetric interactions between benthic and pelagic producers. Ecol. Monogr. 2016, 84, 287–302. [Google Scholar] [CrossRef]
- Fodelianakis, S.; Valenzuela-Cuevas, A.; Barozzi, A.; Daffonchio, D. Direct quantification of ecological drift at the population level in synthetic bacterial communities. ISME J. 2021, 15, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Bowers, R.M.; Sullivan, A.P.; Costello, E.K.; Collett, J.L., Jr.; Knight, R.; Fierer, N. Sources of bacteria in outdoor air across cities in the midwestern United States. Appl. Environ. Microbiol. 2011, 77, 6350–6356. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, E.S.; Ostman, O. The importance of dispersal for bacterial community composition and functioning. PLoS ONE 2011, 6, e25883. [Google Scholar] [CrossRef]
- Xia, M.; Xiong, F.; Li, X.; Li, D.; Wang, Z.; Zhai, D.; Liu, H.; Chen, Y.; Yu, J.; Wang, Y. Different Assembly Patterns of Planktonic and Sedimentary Bacterial Community in a Few Connected Eutrophic Lakes. Water 2022, 14, 723. [Google Scholar] [CrossRef]
- Ren, Z.; Ma, K.; Jia, X.; Wang, Q.; Zhang, C.; Li, X. Community Assembly and Co-Occurrence Patterns of Microeukaryotes in Thermokarst Lakes of the Yellow River Source Area. Microorganisms 2022, 10, 481. [Google Scholar] [CrossRef]
- Eiler, A.; Heinrich, F.; Bertilsson, S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2012, 6, 330–342. [Google Scholar] [CrossRef]
- Crump, B.C.; Peterson, B.J.; Raymond, P.A.; Amon, R.M.; Rinehart, A.; McClelland, J.W.; Holmes, R.M. Circumpolar synchrony in big river bacterioplankton. Proc. Natl. Acad. Sci. USA 2009, 106, 21208–21212. [Google Scholar] [CrossRef]
- Lambert, S.; Tragin, M.; Lozano, J.C.; Ghiglione, J.F.; Vaulot, D.; Bouget, F.Y.; Galand, P.E. Rhythmicity of coastal marine picoeukaryotes, bacteria and archaea despite irregular environmental perturbations. ISME J. 2019, 13, 388–401. [Google Scholar] [CrossRef]
- Fodelianakis, S.; Lorz, A.; Valenzuela-Cuevas, A.; Barozzi, A.; Booth, J.M.; Daffonchio, D. Dispersal homogenizes communities via immigration even at low rates in a simplified synthetic bacterial metacommunity. Nat. Commun. 2019, 10, 1314. [Google Scholar] [CrossRef]
- Phulpoto, I.A.; Jakhrani, B.A.; Panhyar, A.A.; Phulpoto, A.H.; Shaikh, S.H.; Channa, N.; Kanhar, N.A.; Qazi, M.A. Temperate hyper-saline ecosystems induce spatial distribution and halo-thermotolerance potential in indigenous cultivable bacterial community. Community Ecol. 2022, 23, 337–347. [Google Scholar] [CrossRef]
- Seidel, L.; Broman, E.; Nilsson, E.; Ståhle, M.; Ketzer, M.; Pérez-Martínez, C.; Turner, S.; Hylander, S.; Pinhassi, J.; Forsman, A.; et al. Climate change-related warming reduces thermal sensitivity and modifies metabolic activity of coastal benthic bacterial communities. ISME J. 2023, 17, 855–869. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Guo, W.; Zhang, J.; Lou, X. Influence of Heat Events on the Composition of Airborne Bacterial Communities in Urban Ecosystems. Int. J. Environ. Res. Public Health 2018, 15, 2295. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Nunes, A.R. Exploring the interactions between vulnerability, resilience and adaptation to extreme temperatures. Nat. Hazards 2021, 109, 2261–2293. [Google Scholar] [CrossRef]
- Ren, L.; Lu, Z.; Xia, X.; Peng, Y.; Gong, S.; Song, X.; Jeppesen, E.; Han, B.-P.; Wu, Q.L. Metagenomics reveals bacterioplankton community adaptation to long-term thermal pollution through the strategy of functional regulation in a subtropical bay. Water Res. 2022, 216, 118298. [Google Scholar] [CrossRef]
- Polazzo, F.; Hermann, M.; Crettaz-Minaglia, M.; Rico, A. Impacts of extreme climatic events on trophic network complexity and multidimensional stability. Ecology 2023, 104, e3951. [Google Scholar] [CrossRef]
- Jiao, C.; Zhao, D.; Zeng, J.; Guo, L.; Yu, Z. Disentangling the seasonal co-occurrence patterns and ecological stochasticity of planktonic and benthic bacterial communities within multiple lakes. Sci. Total Environ. 2020, 740, 140010. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Zhang, P.; Xue, K.; Liang, Y.; Van Nostrand, J.D.; Yang, Y.; He, Z.; Wu, L.; Stahl, D.A.; et al. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc. Natl. Acad. Sci. USA 2014, 111, E836–E845. [Google Scholar] [CrossRef] [PubMed]
- Zingel, P.; Cremona, F.; Nõges, T.; Cao, Y.; Neif, É.M.; Coppens, J.; Işkın, U.; Lauridsen, T.L.; Davidson, T.A.; Søndergaard, M.; et al. Effects of warming and nutrients on the microbial food web in shallow lake mesocosms. Eur. J. Protistol. 2018, 64, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Luo, Y.; Xia, J.; Jiang, L.; Zhou, X.; Lu, M.; Liang, J.; Shi, Z.; Shelton, S.; Cao, J. Stronger warming effects on microbial abundances in colder regions. Sci. Rep. 2015, 5, 18032. [Google Scholar] [CrossRef] [PubMed]
- Dutta, H.; Dutta, A. The microbial aspect of climate change. Energy Ecol. Environ. 2016, 1, 209–232. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Bei, Q.; Reitz, T.; Schnabel, B.; Eisenhauer, N.; Schädler, M.; Buscot, F.; Heintz-Buschart, A. Extreme summers impact cropland and grassland soil microbiomes. ISME J. 2023, 17, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Yu, M.; Chen, H.; Zhao, H.; Huang, Y.; Su, W.; Xia, F.; Chang, S.X.; Brookes, P.C.; Dahlgren, R.A.; et al. Elevated temperature shifts soil N cycling from microbial immobilization to enhanced mineralization, nitrification and denitrification across global terrestrial ecosystems. Glob. Chang. Biol. 2020, 26, 5267–5276. [Google Scholar] [CrossRef]
- Ye, W.; Liu, X.; Lin, S.; Tan, J.; Pan, J.; Li, D.; Yang, H. The vertical distribution of bacterial and archaeal communities in the water and sediment of Lake Taihu. FEMS Microbiol. Ecol. 2009, 70, 107–120. [Google Scholar] [CrossRef]
- He, D.; Ren, L.; Wu, Q.L. Contrasting diversity of epibiotic bacteria and surrounding bacterioplankton of a common submerged macrophyte, Potamogeton crispus, in freshwater lakes. FEMS Microbiol. Ecol. 2014, 90, 551–562. [Google Scholar] [CrossRef] [PubMed]
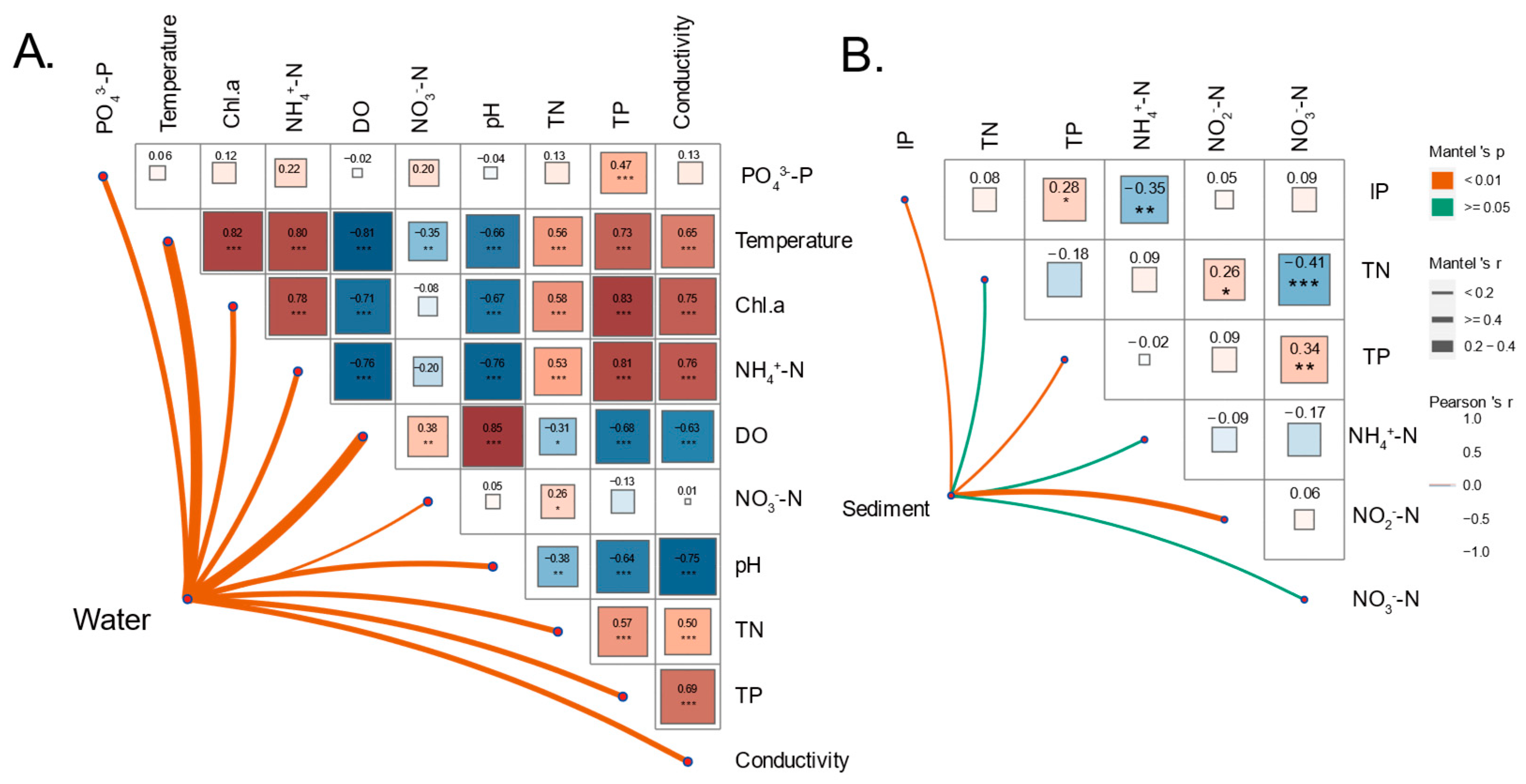


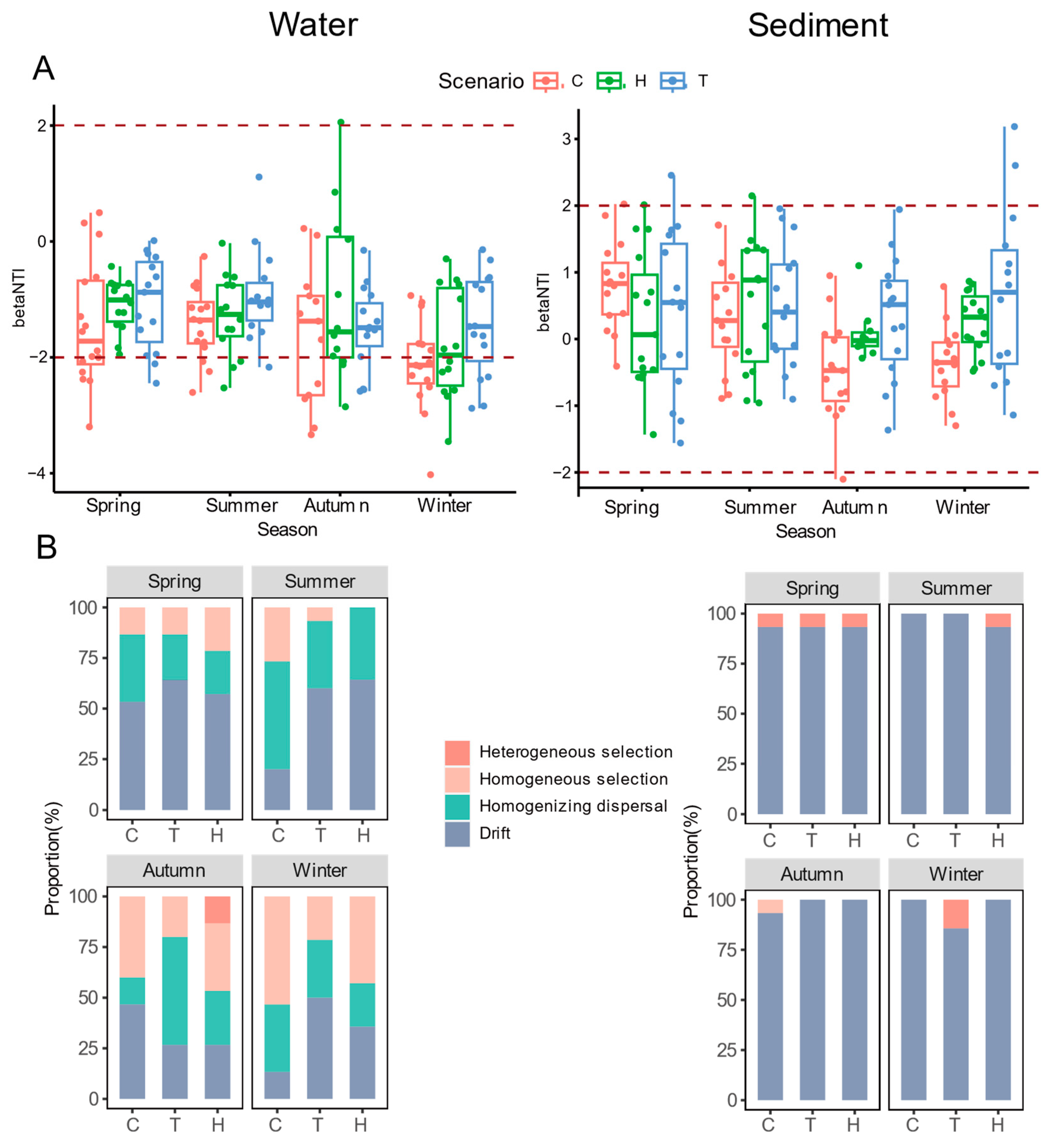
| Habitat | Scenario | Nodes | Edges | Average Degree | Average Transitivity | Average Clustering Coefficient | Average Path Length | Diameter | Modularity | Graph Density | Positive Relationship (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Water | Total | 316 | 892 | 5.646 | 0.390 | 0.458 | 3.614 | 10.816 | 0.053 | 0.018 | 97.646 |
| C | 325 | 2012 | 12.382 | 0.487 | 0.502 | 1.230 | 6.838 | 0.009 | 0.038 | 82.604 | |
| T | 351 | 1720 | 9.801 | 0.461 | 0.491 | 2.096 | 8.295 | 0.021 | 0.028 | 93.953 | |
| H | 360 | 2962 | 16.456 | 0.520 | 0.550 | 0.994 | 5.170 | 0.003 | 0.046 | 86.124 | |
| Sediment | Total | 831 | 9531 | 22.939 | 0.507 | 0.610 | 1.501 | 7.148 | 0.005 | 0.028 | 87.483 |
| C | 655 | 8451 | 25.805 | 0.456 | 0.472 | 0.471 | 4.438 | 0.0005 | 0.039 | 71.459 | |
| T | 704 | 10,006 | 28.426 | 0.508 | 0.479 | 0.459 | 6.127 | 0.001 | 0.040 | 77.294 | |
| H | 621 | 12,642 | 40.715 | 0.526 | 0.520 | 0.456 | 5.804 | 0.001 | 0.066 | 57.475 |
| Water | Sediment | ||||
|---|---|---|---|---|---|
| r | p | r | p | ||
| Temperature | 0.0369 | 0.219 | TN | 0.1322 | 0.014 * |
| pH | 0.2039 | 0.001 *** | NH4+-N | 0.0020 | 0.432 |
| DO | 0.1067 | 0.008 * | NO2−-N | 0.1195 | 0.029 * |
| Conductivity | 0.0259 | 0.001 *** | NO3−-N | −0.0110 | 0.536 |
| NO3−-N | −0.0429 | 0.759 | TP | 0.0853 | 0.063 |
| NH4+-N | 0.1620 | 0.002 ** | IP | 0.1180 | 0.015 * |
| TN | 0.0146 | 0.019 * | |||
| PO43−-P | 0.0663 | 0.194 | |||
| TP | 0.1160 | 0.035 * | |||
| Chl-a | 0.2213 | 0.003 ** | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Yan, Y.; Huang, J.; Wang, Z.; Feng, M.; Cheng, H.; Zhang, P.; Zhang, H.; Xu, J.; Zhang, M. The Impact of Warming on Assembly Processes and Diversity Patterns of Bacterial Communities in Mesocosms. Microorganisms 2023, 11, 2807. https://doi.org/10.3390/microorganisms11112807
Yang Q, Yan Y, Huang J, Wang Z, Feng M, Cheng H, Zhang P, Zhang H, Xu J, Zhang M. The Impact of Warming on Assembly Processes and Diversity Patterns of Bacterial Communities in Mesocosms. Microorganisms. 2023; 11(11):2807. https://doi.org/10.3390/microorganisms11112807
Chicago/Turabian StyleYang, Qian, Yifeng Yan, Jinhe Huang, Zhaolei Wang, Mingjun Feng, Haowu Cheng, Peiyu Zhang, Huan Zhang, Jun Xu, and Min Zhang. 2023. "The Impact of Warming on Assembly Processes and Diversity Patterns of Bacterial Communities in Mesocosms" Microorganisms 11, no. 11: 2807. https://doi.org/10.3390/microorganisms11112807
APA StyleYang, Q., Yan, Y., Huang, J., Wang, Z., Feng, M., Cheng, H., Zhang, P., Zhang, H., Xu, J., & Zhang, M. (2023). The Impact of Warming on Assembly Processes and Diversity Patterns of Bacterial Communities in Mesocosms. Microorganisms, 11(11), 2807. https://doi.org/10.3390/microorganisms11112807






