Solitary Bees Host More Bacteria and Fungi on Their Cuticle than Social Bees
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cuticle Preparation
2.3. Nucleic Acid Extraction
2.4. Quantitative PCR
2.5. Next-Generation Sequencing
2.6. Statistics
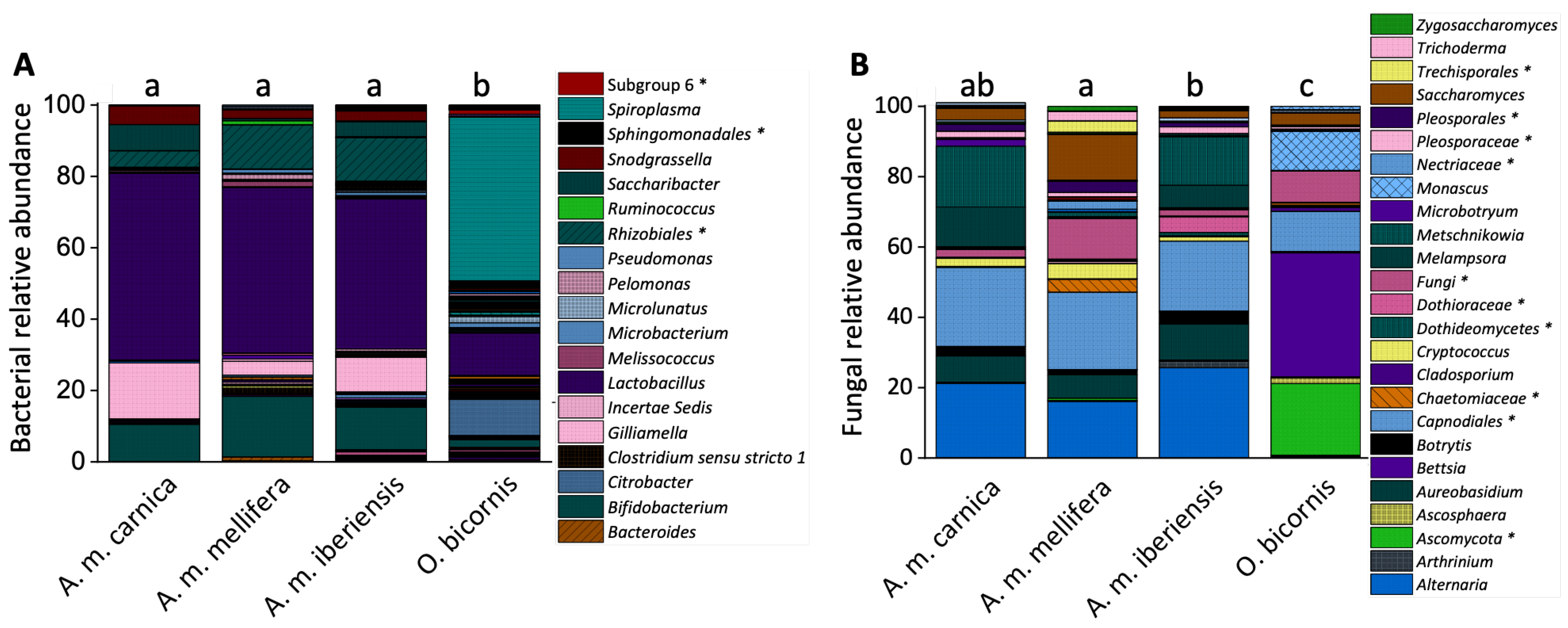
3. Results
3.1. Bacterial and Fungal Gene Copy Numbers
3.2. Cuticular Microbiome Composition
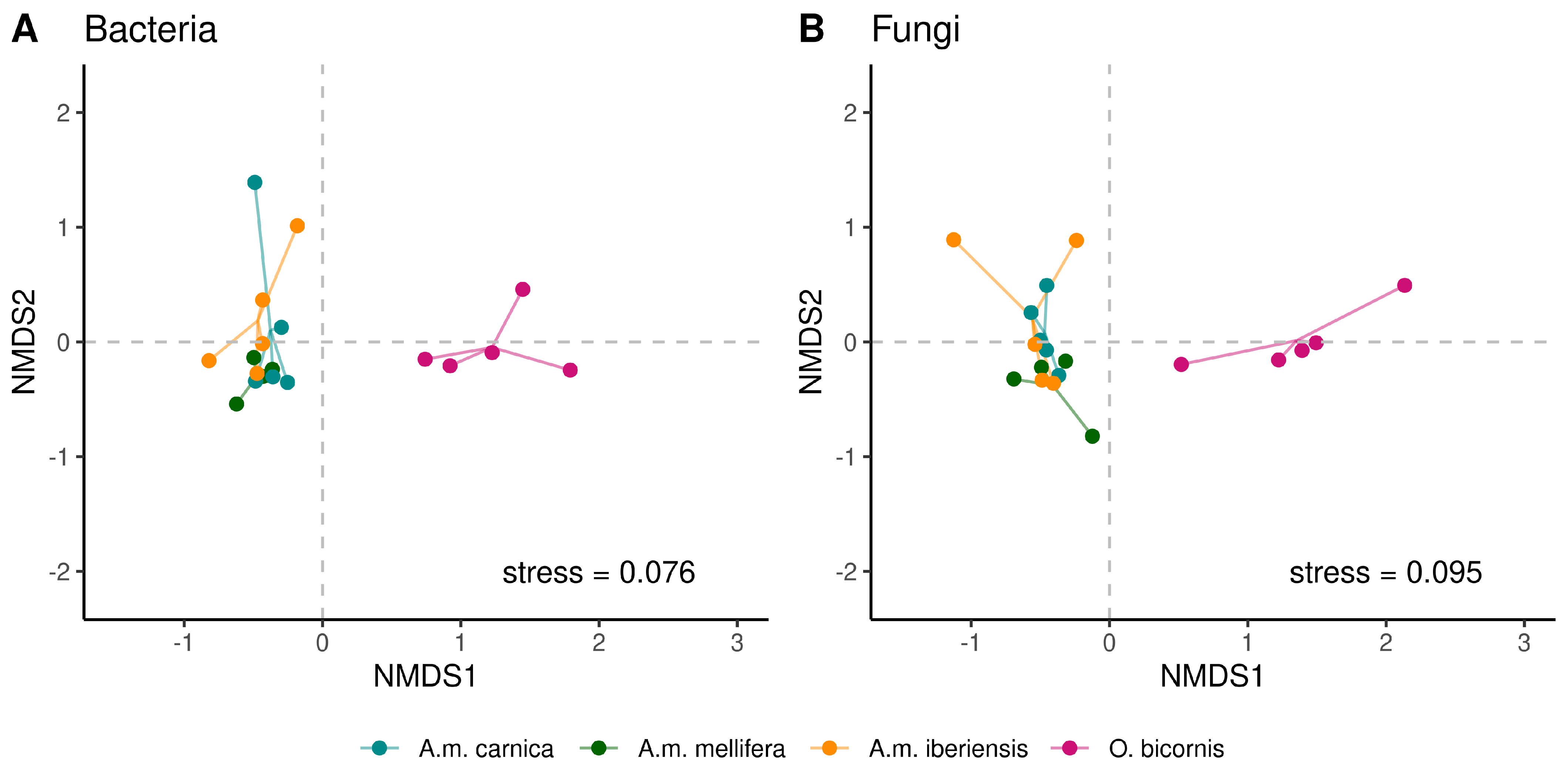
3.3. Cuticular Microbiome Diversity
4. Discussion
4.1. Social Bees Display a Reduced Cuticular Microbiome
4.2. Honeybee Subspecies from the Same Environment Resemble Similar Cuticular Microbiomes
4.3. The Cuticular Microbiome Differs from the Gut Microbiome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hein, L. The economic value of the pollination service, a review across scales. Open Ecol. J. 2009, 2, 74–82. [Google Scholar] [CrossRef]
- Winston, M.L. The Biology of the Honey Bee; Harvard University Press: Cambridge, MA, USA; London, UK, 1987. [Google Scholar]
- Ruttner, F. Biogeography and Taxonomy of Honeybees; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- De La Rúa, P.; Galián, J.; Serrano, J.; Moritz, R.F. Genetic structure and distinctness of Apis mellifera L. populations from the Canary Islands. Mol. Ecol. 2001, 10, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Radloff, S.; Hepburn, H.; Hepburn, C.; De la Rúa, P. Morphometric affinities and population structure of honey bees of the Balearic Islands (Spain). J. Apic. Res. 2001, 40, 97–103. [Google Scholar] [CrossRef]
- Danforth, B.N.; Cardinal, S.; Praz, C.; Almeida, E.A.; Michez, D. The impact of molecular data on our understanding of bee phylogeny and evolution. Annu. Rev. Entomol. 2013, 58, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Westrich, P. Die Wildbienen Baden-Württembergs. Bände I+II; Ulmer: Stuttgart, Germany, 1990. [Google Scholar]
- Strohm, E.; Daniels, H.; Warmers, C.; Stoll, C. Nest provisioning and a possible cost of reproduction in the megachilid bee Osmia rufa studied by a new observation method. Ethol. Ecol. Evol. 2002, 14, 255–268. [Google Scholar] [CrossRef]
- Fanning, S.; Mitchell, A.P. Fungal biofilms. PLoS Pathog. 2012, 8, e1002585. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Lederberg, J.; McCray, A.T. Ome SweetOmics–A genealogical treasury of words. Science 2001, 15, 8. [Google Scholar]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Miller, D.L.; Parish, A.J.; Newton, I.L. Transitions and transmission: Behavior and physiology as drivers of honey bee-associated microbial communities. Curr. Opin. Microbiol. 2019, 50, 1–7. [Google Scholar] [CrossRef]
- Corby-Harris, V.; Maes, P.; Anderson, K.E. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE 2014, 9, e95056. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Rao, V.D.; Yeoman, C.J.; Wilson, B.A.; White, B.A.; Goldenfeld, N.; Robinson, G.E. Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera). PLoS ONE 2015, 10, e0123911. [Google Scholar] [CrossRef] [PubMed]
- Tarpy, D.R.; Mattila, H.R.; Newton, I.L. Development of the honey bee gut microbiome throughout the queen-rearing process. Appl. Environ. Microbiol. 2015, 81, 3182–3191. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Fruciano, C.; Hildebrand, F.; Al Toufalilia, H.; Balfour, N.J.; Bork, P.; Engel, P.; Ratnieks, F.L.; Hughes, W.O. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evol. 2018, 8, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef] [PubMed]
- Kolmes, S.A. Grooming specialists among worker honey bees, Apis mellifera. Anim. Behav. 1989, 37, 1048–1049. [Google Scholar] [CrossRef]
- Simone-Finstrom, M. Social immunity and the superorganism: Behavioral defenses protecting honey bee colonies from pathogens and parasites. Bee World 2017, 94, 21–29. [Google Scholar] [CrossRef]
- Keller, A.; Grimmer, G.; Steffan-Dewenter, I. Diverse microbiota identified in whole intact nest chambers of the red mason bee Osmia bicornis (Linnaeus 1758). PLoS ONE 2013, 8, e78296. [Google Scholar] [CrossRef]
- McFrederick, Q.S.; Rehan, S.M. Characterization of pollen and bacterial community composition in brood provisions of a small carpenter bee. Mol. Ecol. 2016, 25, 2302–2311. [Google Scholar] [CrossRef]
- McFrederick, Q.S.; Thomas, J.M.; Neff, J.L.; Vuong, H.Q.; Russell, K.A.; Hale, A.R.; Mueller, U.G. Flowers and wild megachilid bees share microbes. Microb. Ecol. 2017, 73, 188–200. [Google Scholar] [CrossRef]
- Voulgari-Kokota, A.; Grimmer, G.; Steffan-Dewenter, I.; Keller, A. Bacterial community structure and succession in nests of two megachilid bee genera. FEMS Microbiol. Ecol. 2019, 95, fiy218. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Kikuchi, Y.; Fukatsu, T. How many symbionts are provided by mothers, acquired by offspring, and needed for successful vertical transmission in an obligate insect–bacterium mutualism? Mol. Ecol. 2007, 16, 5316–5325. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Brandel, A.; Becker, M.C.; Balles, R.; Abdelmohsen, U.R.; Ankenbrand, M.J.; Sickel, W. Wild bees and their nests host Paenibacillus bacteria with functional potential of avail. Microbiome 2018, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ushio, M.; Yamasaki, E.; Takasu, H.; Nagano, A.J.; Fujinaga, S.; Honjo, M.N.; Ikemoto, M.; Sakai, S.; Kudoh, H. Microbial communities on flower surfaces act as signatures of pollinator visitation. Sci. Rep. 2015, 5, 8695. [Google Scholar] [CrossRef] [PubMed]
- Ożarowski, M.; Karpiński, T.M.; Alam, R.; Lochyńska, M. Antifungal Properties of Chemically Defined Propolis from Various Geographical Regions. Microorganisms 2022, 10, 364. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Peng, A.H.; Lu, D.D.; Song, Y.J.; Wang, C.L.; Xie, W.D. Manumycin-type derivatives from a Streptomyces sp. associated with meson bee Osmia cornifrons. Nat. Prod. Commun. 2018, 13, 1934578X1801300605. [Google Scholar] [CrossRef]
- Scheiner, R.; Lim, K.; Meixner, M.D.; Gabel, M.S. Comparing the appetitive learning performance of six European honeybee subspecies in a common apiary. Insects 2021, 12, 768. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef]
- Porras-Alfaro, A.; Liu, K.L.; Kuske, C.R.; Xie, G. From genus to phylum: Large-subunit and internal transcribed spacer rRNA operon regions show similar classification accuracies influenced by database composition. Appl. Environ. Microbiol. 2014, 80, 829–840. [Google Scholar] [CrossRef]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge–accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Whelan, R.A.; Doranalli, K.; Rinttilä, T.; Vienola, K.; Jurgens, G.; Apajalahti, J. The impact of Bacillus subtilis DSM 32315 on the pathology, performance, and intestinal microbiome of broiler chickens in a necrotic enteritis challenge. Poult. Sci. 2019, 98, 3450–3463. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Buettner, C.; Noll, M. Differences in microbial key players in anaerobic degradation between biogas and sewage treatment plants. Int. Biodeterior. Biodegrad. 2018, 133, 124–132. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, H.R.; Schigel, D.; Tedersoo, L.; Larsson, K.H.; May, T.W.; Taylor, A.F.; Jeppesen, T.S.; Frøslev, T.G.; Lindahl, B.D.; et al. The taxon hypothesis paradigm—On the unambiguous detection and communication of taxa. Microorganisms 2020, 8, 1910. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F.; et al. The Biological Observation Matrix (BIOM) format or: How I learned to stop worrying and love the ome-ome. Gigascience 2012, 1, 7. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjöller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Martinson, V.G.; Danforth, B.N.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2011, 20, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Lozo, J.; Berić, T.; Terzić-Vidojević, A.; Stanković, S.; Fira, D.; Stanisavljević, L. Microbiota associated with pollen, bee bread, larvae and adults of solitary bee Osmia cornuta (Hymenoptera: Megachilidae). Bull. Entomol. Res. 2015, 105, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Fünfhaus, A.; Ebeling, J.; Genersch, E. Bacterial pathogens of bees. Curr. Opin. Insect Sci. 2018, 26, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Blomquist, G.J.; Chu, A.J.; Remaley, S. Biosynthesis of wax in the honeybee, Apis mellifera L. Insect Biochem. 1980, 10, 313–321. [Google Scholar] [CrossRef]
- Pinilla-Gallego, M.S.; Crum, J.; Schaetzl, R.; Isaacs, R. Soil textures of nest partitions made by the mason bees Osmia lignaria and O. cornifrons (Hymenoptera: Megachilidae). Apidologie 2018, 49, 464–472. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Przybyłek, I.; Karpiński, T.M. Antibacterial properties of propolis. Molecules 2019, 24, 2047. [Google Scholar] [CrossRef]
- Drescher, N.; Klein, A.M.; Schmitt, T.; Leonhardt, S.D. A clue on bee glue: New insight into the sources and factors driving resin intake in honeybees (Apis mellifera). PLoS ONE 2019, 14, e0210594. [Google Scholar] [CrossRef]
- Cini, A.; Bordoni, A.; Cappa, F.; Petrocelli, I.; Pitzalis, M.; Iovinella, I.; Dani, F.R.; Turillazzi, S.; Cervo, R. Increased immunocompetence and network centrality of allogroomer workers suggest a link between individual and social immunity in honeybees. Sci. Rep. 2020, 10, 8928. [Google Scholar] [CrossRef]
- Kunkel, H.; Kloft, W. Fortschritte auf dem Gebiet der Honigtauforschung. Apidologie 1977, 8, 369–391. [Google Scholar] [CrossRef]
- Konrad, R.; Wäckers, F.L.; Romeis, J.; Babendreier, D. Honeydew feeding in the solitary bee Osmia bicornis as affected by aphid species and nectar availability. J. Insect Physiol. 2009, 55, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Rering, C.C.; Rudolph, A.B.; Beck, J.J. Pollen and yeast change nectar aroma and nutritional content alone and together, but honey bee foraging reflects only the avoidance of yeast. Environ. Microbiol. 2021, 23, 4141–4150. [Google Scholar] [CrossRef] [PubMed]
- Scheiner, R.; Strauß, S.; Thamm, M.; Farré-Armengol, G.; Junker, R.R. The bacterium Pantoea ananatis modifies behavioral responses to sugar solutions in honeybees. Insects 2020, 11, 692. [Google Scholar] [CrossRef] [PubMed]
- Bugarova, V.; Godocikova, J.; Bucekova, M.; Brodschneider, R.; Majtan, J. Effects of the carbohydrate sources nectar, sucrose and invert sugar on antibacterial activity of honey and bee-processed syrups. Antibiotics 2021, 10, 985. [Google Scholar] [CrossRef] [PubMed]
- Shackleton, K.; Balfour, N.J.; Al Toufailia, H.; Gaioski Jr, R.; de Matos Barbosa, M.; Silva, C.A.d.S.; Bento, J.M.; Alves, D.A.; Ratnieks, F.L. Quality versus quantity: Foraging decisions in the honeybee (Apis mellifera scutellata) feeding on wildflower nectar and fruit juice. Ecol. Evol. 2016, 6, 7156–7165. [Google Scholar] [CrossRef]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef]
- Moran, N.A.; Hansen, A.K.; Powell, J.E.; Sabree, Z.L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE 2012, 7, e36393. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.; Eckholm, B.; Mott, B.; DeGrandi-Hoffman, G. An emerging paradigm of colony health: Microbial balance of the honey bee and hive (Apis mellifera). Insectes Sociaux 2011, 58, 431–444. [Google Scholar] [CrossRef]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef]
- Taylor, M.A.; Robertson, A.W.; Biggs, P.J.; Richards, K.K.; Jones, D.F.; Parkar, S.G. The effect of carbohydrate sources: Sucrose, invert sugar and components of mānuka honey, on core bacteria in the digestive tract of adult honey bees (Apis mellifera). PLoS ONE 2019, 14, e0225845. [Google Scholar] [CrossRef]
- Palmer-Young, E.C.; Raffel, T.R.; McFrederick, Q.S. pH-mediated inhibition of a bumble bee parasite by an intestinal symbiont. Parasitology 2019, 146, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Schmid-Hempel, P. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol. Lett. 2012, 15, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Cariveau, D.P.; Elijah Powell, J.; Koch, H.; Winfree, R.; Moran, N.A. Variation in gut microbial communities and its association with pathogen infection in wild bumble bees (Bombus). ISME J. 2014, 8, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, J.; Chen, Y.; Evans, J.D.; Dai, R.; Luo, W.; Li, J. Characterization of gut bacteria at different developmental stages of Asian honey bees, Apis cerana. J. Invertebr. Pathol. 2015, 127, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Erban, T.; Ledvinka, O.; Kamler, M.; Nesvorna, M.; Hortova, B.; Tyl, J.; Titera, D.; Markovic, M.; Hubert, J. Honeybee (Apis mellifera)-associated bacterial community affected by American foulbrood: Detection of Paenibacillus larvae via microbiome analysis. Sci. Rep. 2017, 7, 5084. [Google Scholar] [CrossRef]
- Erban, T.; Ledvinka, O.; Kamler, M.; Hortova, B.; Nesvorna, M.; Tyl, J.; Titera, D.; Markovic, M.; Hubert, J. Bacterial community associated with worker honeybees (Apis mellifera) affected by European foulbrood. PeerJ 2017, 5, e3816. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef]
- Walker, A.W.; Duncan, S.H.; McWilliam Leitch, E.C.; Child, M.W.; Flint, H.J. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl. Environ. Microbiol. 2005, 71, 3692–3700. [Google Scholar] [CrossRef]
- Hammer, T.J.; Le, E.; Moran, N.A. Thermal niches of specialized gut symbionts: The case of social bees. Proc. R. Soc. B 2021, 288, 20201480. [Google Scholar] [CrossRef]
- Fahimipour, A.K.; Hartmann, E.M.; Siemens, A.; Kline, J.; Levin, D.A.; Wilson, H.; Betancourt-Román, C.M.; Brown, G.; Fretz, M.; Northcutt, D.; et al. Daylight exposure modulates bacterial communities associated with household dust. Microbiome 2018, 6, 175. [Google Scholar] [CrossRef]
- Ludvigsen, J.; Andersen, Å.; Hjeljord, L.; Rudi, K. The honeybee gut mycobiota cluster by season versus the microbiota which cluster by gut segment. Vet. Sci. 2020, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Callegari, M.; Crotti, E.; Fusi, M.; Marasco, R.; Gonella, E.; De Noni, I.; Romano, D.; Borin, S.; Tsiamis, G.; Cherif, A.; et al. Compartmentalization of bacterial and fungal microbiomes in the gut of adult honeybees. Npj Biofilms Microbiomes 2021, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.H.; Jung, M.J.; Kim, P.S.; Bae, J.W. Social status shapes the bacterial and fungal gut communities of the honey bee. Sci. Rep. 2018, 8, 2019. [Google Scholar] [CrossRef] [PubMed]
- Tauber, J.P.; Nguyen, V.; Lopez, D.; Evans, J.D. Effects of a resident yeast from the honeybee gut on immunity, microbiota, and Nosema disease. Insects 2019, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Prasongsuk, S.; Lotrakul, P.; Ali, I.; Bankeeree, W.; Punnapayak, H. The current status of Aureobasidium pullulans in biotechnology. Folia Microbiol. 2018, 63, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, Y.; Luo, Z.; Gao, L.; Li, R.; Zhang, Y.; Kalaji, H.M.; Qiang, S.; Chen, S. Recent Advances in Alternaria Phytotoxins: A Review of Their Occurrence, Structure, Bioactivity, and Biosynthesis. J. Fungi 2022, 8, 168. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Jimenez, S.I.; Carroll, C.; Babcock, T.; Derstine, N.; Hadwin, A.; Moore, M.; Gries, G. Yeasts harbored by vespine wasps in the Pacific Northwest. Environ. Entomol. 2017, 46, 217–225. [Google Scholar] [CrossRef]
- Colda, A.; Bossaert, S.; Verreth, C.; Vanhoutte, B.; Honnay, O.; Keulemans, W.; Lievens, B. Inoculation of pear flowers with Metschnikowia reukaufii and Acinetobacter nectaris enhances attraction of honeybees and hoverflies, but does not increase fruit and seed set. PLoS ONE 2021, 16, e0250203. [Google Scholar] [CrossRef]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]



| Bee Species/Subspecies | Stat | p-Value | Family | Genus | Function | ||
|---|---|---|---|---|---|---|---|
| Bacteria | A.m. iberiensis | 0.695 | 0.0048 | ** | Lactobacillales uncl. | Lactobacillales uncl. | NA |
| 0.669 | 0.0322 | * | Elev-16S-1332 | Elev-16S-1332 uncl. | NA | ||
| 0.665 | 0.0322 | * | Chitinophagaceae | Chitinophagaceae uncl. | aerobic chemoheterotrophy chemoheterotrophy | ||
| 0.614 | 0.0367 | * | Rhodospirillales Incertae Sedis | Candidatus Alysiosphaera | NA | ||
| 0.456 | 0.0355 | * | Solirubrobacteraceae | Solirubrobacter | aerobic chemoheterotrophy chemoheterotrophy | ||
| 0.423 | 0.0177 | * | Sphingomonadaceae | Sphingomonas | aerobic chemoheterotrophy chemoheterotrophy | ||
| O. bicornis | 0.833 | 0.0030 | ** | Xanthomonadales Incertae Sedis | Steroidobacter | aerobic chemoheterotrophy | |
| 0.782 | 0.0003 | *** | Hyphomicrobiaceae | Rhodoplanes | phototrophy | ||
| 0.747 | 0.0003 | *** | Actinobacteria uncl. | Actinobacteria uncl. | NA | ||
| 0.744 | 0.0003 | *** | Rhodobiaceae | Rhodobium | aerobic chemoheterotrophy photoheterotrophy phototrophy | ||
| 0.707 | 0.0303 | * | Planctomycetaceae | Pir4 lineage | chemoorganotrophy | ||
| 0.701 | 0.0342 | * | Nocardiaceae | Nocardia | aerobic chemoheterotrophy chemoheterotrophy | ||
| 0.661 | 0.0003 | *** | Spiroplasmataceae | Spiroplasma | parasitic | ||
| 0.657 | 0.0347 | * | Nocardioidaceae | Kribbella | chemoorganotrophy | ||
| 0.629 | 0.0303 | * | Verrucomicrobiaceae | Verrucomicrobiaceae uncl. | aerobic chemoheterotrophy chemoheterotrophy | ||
| Fungi | A.m. mellifera | 0.659 | 0.0368 | * | Phaeosphaeriaceae | Phaeosphaeriaceae uncl. | litter saprotroph |
| 0.608 | 0.0406 | * | Tremellales uncl. | Cryptococcus | unspecified saprotroph | ||
| 0.600 | 0.0368 | * | Eremascaceae | Eremascus | unspecified saprotroph | ||
| 0.512 | 0.0083 | ** | Nectriaceae | Nectriaceae uncl. | plant pathogen animal parasite | ||
| A.m. iberiensis | 0.736 | 0.0008 | *** | Xylariaceae | Xylaria | wood saprotroph | |
| 0.682 | 0.0383 | * | Ascomycota uncl. | Torula | foliar endophyte | ||
| 0.678 | 0.0383 | * | Tremellomycetes uncl. | Tremellomycetes uncl. | NA | ||
| 0.610 | 0.0269 | * | Pleosporaceae | Alternaria | plant pathogen | ||
| 0.593 | 0.0383 | * | Xylariales uncl. | Dinemasporium | litter saprotroph | ||
| 0.526 | 0.0424 | * | Metschnikowiaceae | Metschnikowia | nectar tap saprotroph | ||
| 0.440 | 0.0436 | * | Herpotrichiellaceae | Exophiala | animalparasite | ||
| 0.434 | 0.0438 | * | Amphisphaeriaceae | Amphisphaeriaceae uncl. | plant pathogen | ||
| A.m. carnica | 0.7 | 0.0087 | ** | Microbotryaceae | Microbotryum | plant pathogen | |
| O. bicornis 1 | 0.900 | 0.0001 | *** | Davidiellaceae | Cladosporium | litter saprotroph | |
| 0.889 | 0.0001 | *** | Polyporaceae | Fomes | wood saprotroph | ||
| 0.877 | 0.0001 | *** | Mycosphaerellaceae | Mycosphaerellaceae uncl. | plant pathogen | ||
| 0.873 | 0.0001 | *** | Capnodiales uncl. | Toxicocladosporium | plant pathogen | ||
| 0.845 | 0.0005 | *** | Capnodiales uncl. | Pseudotaeniolina | litter saprotroph | ||
| 0.827 | 0.0037 | ** | Cystofilobasidiaceae | Guehomyces | NA | ||
| 0.816 | 0.0001 | *** | Saccharomycetaceae | Saccharomycetaceae uncl. | nectar tap saprotroph | ||
| 0.805 | 0.0001 | *** | Cordycipitaceae | Cordycipitaceae uncl. | animal parasite | ||
| 0.802 | 0.0001 | *** | Ascosphaeraceae | Bettsia | animal parasite | ||
| 0.799 | 0.0003 | *** | Trichocomaceae | Aspergillus | unspecifiedsaprotroph |
| Index | Contrast | p-Value | ||
|---|---|---|---|---|
| Richness | R2 = 0.58 F(3,16) = 9.696 p = 0.0007 | A. mellifera vs. O. bicornis A.m. carnica vs. A.m. mellifera A.m. carnica vs. A.m. iberiensis A.m. mellifera vs. A.m. iberiensis | 0.0014 0.8306 0.0249 0.0076 | ** ns * ** |
| Chao1 | R2 = 0.52 F(3,16) = 7.849 p = 0.0002 | A. mellifera vs. O. bicornis A.m. carnica vs. A.m. mellifera A.m. carnica vs. A.m. iberiensis A.m. mellifera vs. A.m. iberiensis | 0.005 0.6236 0.0544 0.0083 | ** ns ns ** |
| Evenness | R2 = 0.42 F(3,16) = 5.514 p = 0.0085 | A. mellifera vs. O. bicornis A.m. carnica vs. A.m. mellifera A.m. carnica vs. A.m. iberiensis A.m. mellifera vs. A.m. iberiensis | 0.0036 0.9508 0.2180 0.1318 | ** ns ns ns |
| ACE | R2 = 0.55 F(3,16) = 8.634 p = 0.0012 | A. mellifera vs. O. bicornis A.m. carnica vs. A.m. mellifera A.m. carnica vs. A.m. iberiensis A.m. mellifera vs. A.m. iberiensis | 0.003 0.5574 0.0538 0.0065 | ** ns ns ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thamm, M.; Reiß, F.; Sohl, L.; Gabel, M.; Noll, M.; Scheiner, R. Solitary Bees Host More Bacteria and Fungi on Their Cuticle than Social Bees. Microorganisms 2023, 11, 2780. https://doi.org/10.3390/microorganisms11112780
Thamm M, Reiß F, Sohl L, Gabel M, Noll M, Scheiner R. Solitary Bees Host More Bacteria and Fungi on Their Cuticle than Social Bees. Microorganisms. 2023; 11(11):2780. https://doi.org/10.3390/microorganisms11112780
Chicago/Turabian StyleThamm, Markus, Fabienne Reiß, Leon Sohl, Martin Gabel, Matthias Noll, and Ricarda Scheiner. 2023. "Solitary Bees Host More Bacteria and Fungi on Their Cuticle than Social Bees" Microorganisms 11, no. 11: 2780. https://doi.org/10.3390/microorganisms11112780
APA StyleThamm, M., Reiß, F., Sohl, L., Gabel, M., Noll, M., & Scheiner, R. (2023). Solitary Bees Host More Bacteria and Fungi on Their Cuticle than Social Bees. Microorganisms, 11(11), 2780. https://doi.org/10.3390/microorganisms11112780








