Aquiluscidin, a Cathelicidin from Crotalus aquilus, and the Vcn-23 Derivative Peptide, Have Anti-Microbial Activity against Gram-Negative and Gram-Positive Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Sample Collection
2.3. cDNA Cloning and Sequencing
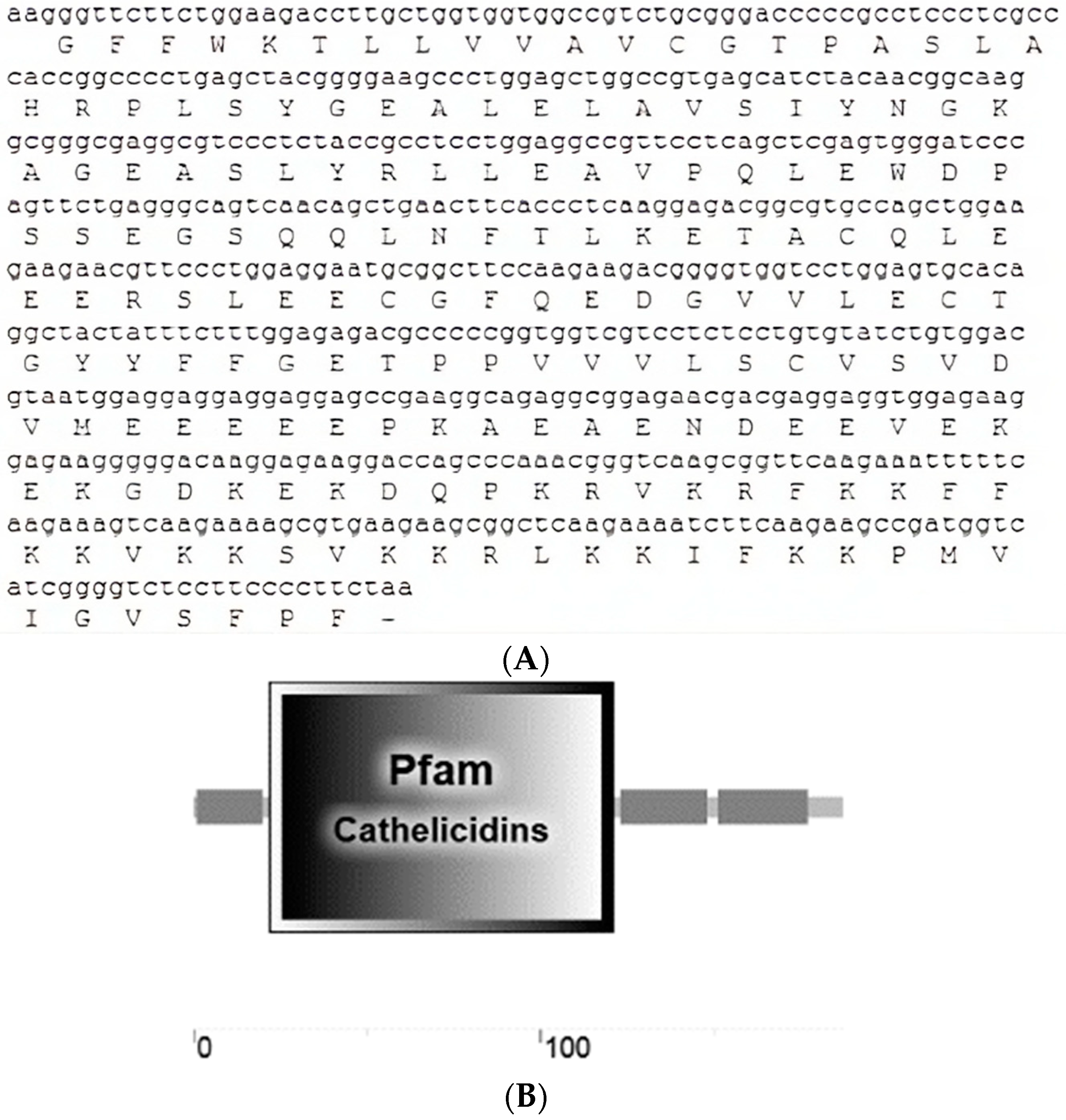
2.4. Bioinformatics Analysis
2.5. Synthetic Peptides
2.6. Cytotoxicity Analysis
2.7. Hemolytic Activity Analysis
2.8. Antibacterial Activity Analysis
3. Results
3.1. Crotalus aquilus Expressed a Cathelicidin Gene
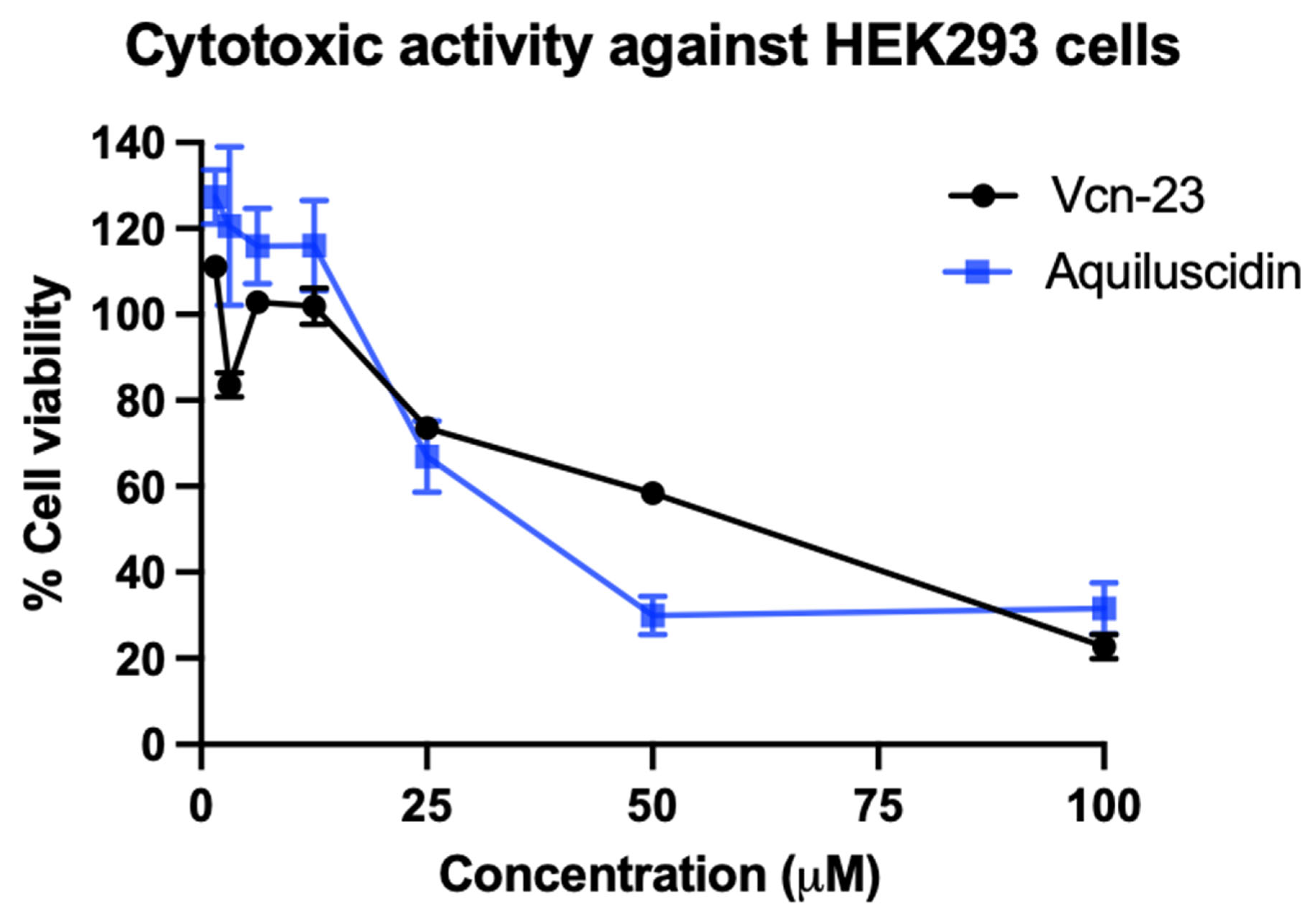
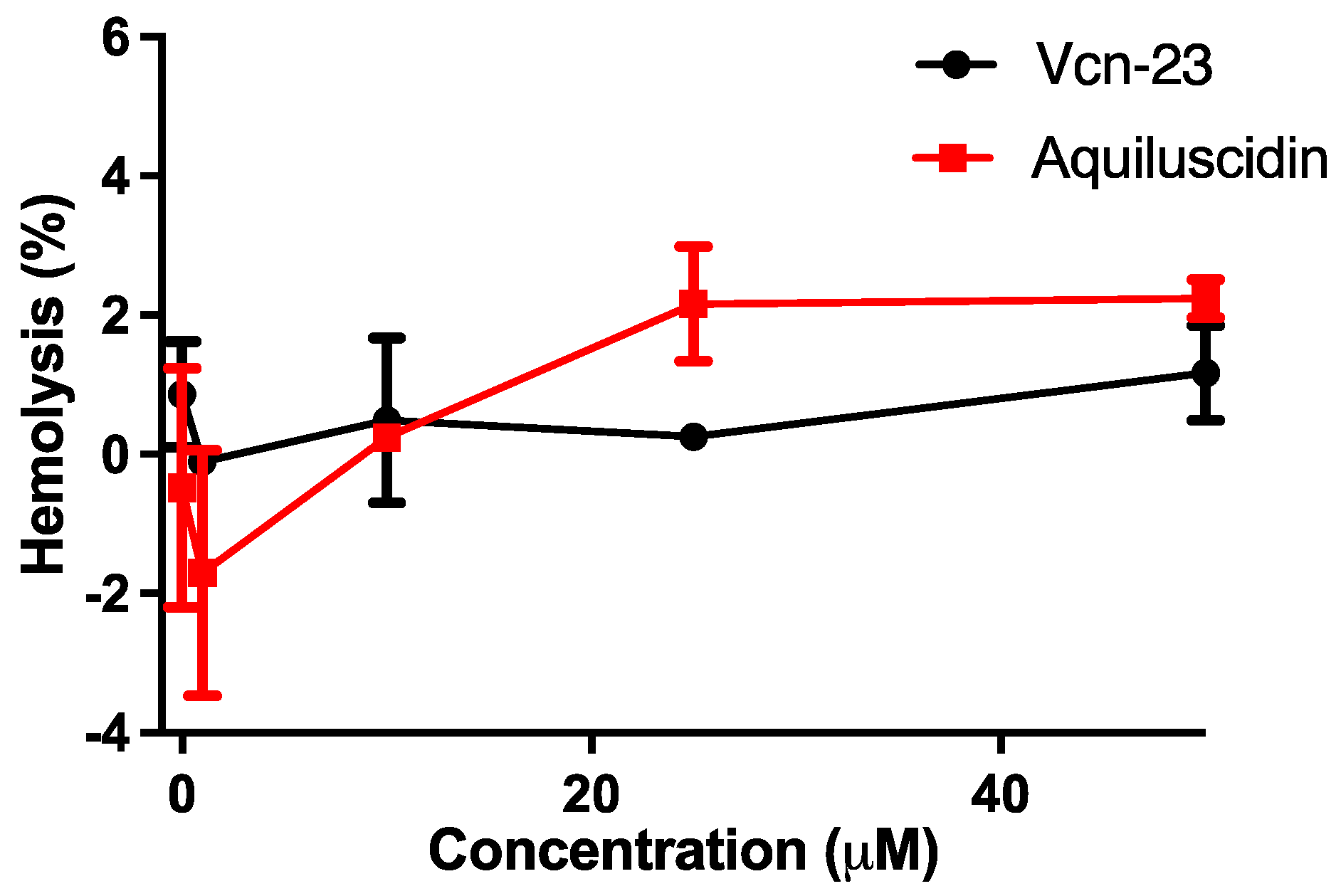
3.2. Cytotoxic and Hemolytic Effects
3.3. Antibacterial Activity
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Anti-Microbial Resistance. 2016. Available online: https://www.who.int/es/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 1 April 2019).
- Wang, G.; Mishra, B.; Lau, K.; Lushnikova, T.; Golla, R.; Wang, X. Anti-microbial peptides in 2014. Pharmaceuticals 2015, 8, 123–150. [Google Scholar] [CrossRef]
- O’Neill, J. Review on Anti-Microbial Resistance Anti-Microbial Resistance: Tackling a Crisis for the Health and Wealth of Nations. London: Review on Anti-Microbial Resistance. 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 3 May 2023).
- Greber, K.E.; Dawgul, M. Anti-microbial Peptides under Clinical Trials. Curr. Top. Med. Chem. 2017, 17, 620–628. [Google Scholar] [CrossRef]
- Wu, R.; Patocka, J.; Nepovimova, E.; Oleksak, P.; Valis, M.; Wu, W.; Kuca, K. Marine Invertebrate Peptides: Antimicrobial Peptides. Front. Microbiol. 2021, 12, 785085. [Google Scholar] [CrossRef]
- Chen, P.; Ye, T.; Li, C.; Praveen, P.; Hu, Z.; Li, W.; Shang, C. Embracing the era of antimicrobial peptides with marine organisms. Nat. Prod. Rep. 2023. Advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fan, L.; Pan, H.; Li, Y.; Qiu, Y.; Lu, Y. Antimicrobial peptides from marine animals: Sources, structures, mechanisms and the potential for drug development. Front. Mar. Sci. 2023, 9, 1112595. [Google Scholar] [CrossRef]
- Kesting, M.R.; Stoeckelhuber, M.; Hölzle, F.; Mücke, T.; Neumann, K.; Woermann, K.; Jacobsen, F.; Steinstraesser, L.; Wolff, K.D.; Loeffelbein, D.J.; et al. Expression of anti-microbial peptides in cutaneous infections after skin surgery. Br. J. Dermatol. 2010, 163, 121–127. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Anti-microbial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, I.; Lehrer, R.I. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 1996, 396, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Boparai, J.K.; Sharma, P.K. Mini Review on Anti-microbial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2019, 27, 4–16. [Google Scholar] [CrossRef]
- van Hoek, M.L. Anti-microbial peptides in reptiles. Pharmaceuticals 2014, 7, 723–753. [Google Scholar] [CrossRef]
- Xiao, Y.; Cai, Y.; Bommineni, Y.R.; Fernando, S.C.; Prakash, O.; Gilliland, S.E.; Zhang, G. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 2006, 281, 2858–2867. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsson, G.H.; Agerberth, B.; Odeberg, J.; Bergman, T.; Olsson, B.; Salcedo, R. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 1996, 238, 325–332. [Google Scholar] [CrossRef]
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta Biomembr. 2015, 1858, 546–566. [Google Scholar] [CrossRef]
- Chen, W.; Yang, B.; Zhou, H.; Sun, L.; Dou, J.; Qian, H.; Huang, W.; Mei, Y.; Han, J. Structure-activity relationships of a snake cathelicidin-related peptide, BF-15. Peptides 2011, 32, 2497–2503. [Google Scholar] [CrossRef]
- De Latour, F.A.; Amer, L.S.; Papanstasiou, E.A.; Bishop, B.M.; Van Hoek, M.L. Biochemical and Biophysical Research Communications Anti-microbial activity of the Naja atra cathelicidin and related small peptides. Biochem. Biophys. Res. Commun. 2010, 396, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Falcao, C.B.; Pérez-Peinado, C.; De La Torre, B.G.; Mayol, X. Structural Dissection of Crotalicidin, a Rattlesnake Venom Cathelicidin, Retrieves a Fragment with Anti-microbial and Antitumor Activity. J. Med. Chem. 2015, 58, 8553–8563. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hong, J.; Liu, X.; Yang, H.; Liu, R.; Wu, J.; Wang, A.; Lin, D.; Lai, R. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PLoS ONE 2008, 3, e3217. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, H.; Yu, G.; Liu, X.; Shen, J. Peptides Structure—Function relationship of king cobra cathelicidin. Peptides 2010, 31, 1488–1493. [Google Scholar] [CrossRef]
- Falcao, C.B.; De La Torre, B.G.; Pérez-Peinado, C.; Barron, A.E.; Andreu, D.; Rádis-Baptista, G. Vipericidins: A novel family of cathelicidin-related peptides from the venom gland of South American pit vipers. Amino Acids 2014, 46, 2561–2571. [Google Scholar] [CrossRef]
- Wei, L.; Gao, J.; Zhang, S.; Wu, S.; Xie, Z.; Ling, G.; Kuang, Y.Q.; Yang, Y.; Yu, H.; Wang, Y. Identification and characterization of the first cathelicidin from sea snakes with potent anti-microbial and antiinflammatory activity and special mechanism. J. Biol. Chem. 2015, 290, 16633–16652. [Google Scholar] [CrossRef]
- Zhao, H.; Gan, T.X.; Liu, X.D.; Jin, Y.; Lee, W.H.; Shen, J.H.; Zhang, Y. Identification and characterization of novel reptile cathelicidins from elapid snakes. Peptides 2008, 29, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- de Barros, E.; Gonçalves, R.M.; Cardoso, M.H.; Santos, N.C.; Franco, O.L.; Cândido, E.S. Snake Venom Cathelicidins as Natural Anti-microbial Peptides. Front. Pharmacol. 2019, 10, 1415. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, C.S.; Falcão, C.B.; Fontenelle, R.O.; Andreu, D.; Rádis-Baptista, G. Anti-fungal activity of Ctn[15-34], the C-terminal peptide fragment of crotalicidin, a rattlesnake venom gland cathelicidin. J. Antibiot. 2017, 70, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Mello, C.P.; Lima, D.B.; de Menezes, R.R.P.P.B.; Bandeira, I.C.J.; Tessarolo, L.D.; Sampaio, T.L.; Falcão, C.B.; Rádis-Baptista, G.; Martins, A.M.C. Evaluation of the antichagasic activity of batroxicidin, a cathelicidin-related anti-microbial peptide found in Bothrops atrox venom gland. Toxicon 2017, 130, 56–62. [Google Scholar] [CrossRef]
- Fang, Y.; He, X.; Zhang, P.; Shen, C.; Mwangi, J.; Xu, C.; Mo, G.; Lai, R.; Zhang, Z. In vitro and In Vivo Antimalarial Activity of LZ1, a Peptide Derived from Snake Cathelicidin. Toxins 2019, 11, 379. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.; Lemos-Espinal, J. Amphibians and Reptiles of the State of Queretaro, Mexico; National Commission for the Knowledge and Use of Biodiversity (CONABIO): Mexico City, Mexico, 2010; Volume 428, pp. 149–150. [Google Scholar]
- Jepson, L. Exotic Animal Medicine a Quick Reference Guide; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Birchard, S.J.; Sherding, R.G. Saunders Manual of Small Animal Practice, 2nd ed.; W.B. Saunders: Philadelphia, PA, USA, 2000. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2017, 46, D493–D496. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Finn Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Brown, N.P.; Leroy, C.; Sander, C. MView: A web-compatible database search or multiple alignment viewer. Bioinformatics 1998, 14, 380–381. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Torrent, M.; Di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An automated web server for prediction of protein anti-microbial regions. Bioinformatics 2012, 28, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Nogués, V.M.; Boix, E. A theoretical approach to spot active regions in anti-microbial proteins. BMC Bioinform. 2009, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Lazcano-Pérez, F.; Zavala-Moreno, A.; Rufino-González, Y.; Ponce-Macotela, M.; García-Arredondo, A.; Cuevas-Cruz, M.; Gómez-Manzo, S.; Marcial-Quino, J.; Arreguín-Lozano, B.; Arreguín-Espinosa, R. Hemolytic, anticancer and antigiardial activity of Palythoa caribaeorum venom. J. Venom. Anim. Toxins Incl. Trop. Dis. 2018, 24, 12. [Google Scholar] [CrossRef]
- Alsever, J.B.; Ainslie, R.B. A new method for the preparation of dilute blood plasma and the operation of a complete transfusion service. NY State J. Med. 1941, 41, 126–131. [Google Scholar]
- M07-A9; Methods for Dilution Anti-Microbial Susceptibility Tests for Bacteria That Grow Aerobically, Approved Standard, 9th ed., CLSI document M07-A9. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012.
- M100; Performance Standards for Anti-Microbial Susceptibility Testing, 26th ed. CLSI document M100. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016.
- Brogden, K.A.; Bates, A.M.; Fischer, C.L. Anti-Microbial Peptides in Host Defense: Functions beyond Anti-Microbial Activity. In Anti-Microbial Peptides; Harder, J., Schröder, J.M., Eds.; Birkhäuser Advances in Infectious Diseases; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Hancock, R.E.; Diamond, G. The role of cationic anti-microbial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Anti-microbial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Dürr, U.H.; Sudheendra, U.S.; Ramamoorthy, A. LL-37, the only human member of the cathelicidin family of anti-microbial peptides. Biochim. Biophys. Acta 2006, 1758, 1408–1425. [Google Scholar] [CrossRef]
- Lu, Y.; Ma, Y.; Wang, X.; Liang, J.; Zhang, C.; Zhang, K.; Lin, G.; Lai, R. The first anti-microbial peptide from sea amphibian. Mol. Immunol. 2008, 45, 678–681. [Google Scholar] [CrossRef]
- Tomasinsig, L.; Zanetti, M. The cathelicidins–structure, function and evolution. Curr. Protein Pept. Sci. 2005, 6, 23–34. [Google Scholar] [CrossRef]
- Shinnar, A.E.; Butler, K.L.; Park, H.J. Cathelicidin family of antimicrobial peptides: Proteolytic processing and protease resistance. Bioorganic Chem. 2003, 31, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Peinado, C.; Dias, S.A.; Mendonça, D.A.; Castanho, M.A.R.B.; Veiga, A.S.; Andreu, D. Structural determinants conferring unusual long life in human serum to rattlesnake-derived antimicrobial peptide Ctn[15-34]. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2019, 25, e3195. [Google Scholar] [CrossRef] [PubMed]
- Rivas, L.; Luque-Ortega, J.R.; Andreu, D. Amphibian antimicrobial peptides and Protozoa: Lessons from parasites. Biochim. Biophys. Acta 2009, 1788, 1570–1581. [Google Scholar] [CrossRef]
- Dathe, M.; Wieprecht, T. Structural features of helical anti-microbial peptides: Their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1999, 1462, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Li, S.A.; Lee, W.H.; Zhang, Y. Efficacy of OH-CATH30 and its analogs against drug-resistant bacteria in vitro and in mouse models. Anti-Microb. Agents Chemother. 2012, 56, 3309–3317. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 2009, 1788, 1687–1692. [Google Scholar] [CrossRef]


| Anti-Microbial MIC + µg/mL (µM) | ||||
|---|---|---|---|---|
| Microorganism | Aquiluscidin | Vcn-23 | Ampicillin | Gentamicin |
| E. coli TOP10 | 2 (0.47) | 2 (0.70) | 1 (2.86) | 0.125 (0.26) |
| S. aureus ATCC6538 | 8 (1.91) | 4 (1.40) | 0.5 (1.43) | 0.125 (0.26) |
| P. aeruginosa | 4 (0.95) | 4 (1.40) | 1 (2.86) | 0.125 (0.26) |
| E. coli CI * | 4 (0.95) | 4 (1.40) | 256 (732.67) | 0.125 (0.26) |
| S. aureus CI * | 8 (1.91) | 4 (1.40) | 0.125 (0.35) | 0.125 (0.26) |
| P. aeruginosa CI * | 4 (0.95) | 4 (1.40) | 256 (732.67) | 0.25 (0.52) |
| S. saprophyticus ATCC BAA-750 | 4 (0.95) | 2 (0.70) | 1 (2.86) | 0.125 (0.26) |
| S. typhymurium ATCC 14028 | 8 (1.91) | 2 (0.70) | 0.5 (1.43) | 0.25 (0.52) |
| E. casseliflavus ATCC 700327 | 4 (0.95) | 2 (0.70) | 0.125 (0.35) | 1 (2.09) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Arvizu, E.E.; Silis-Moreno, T.M.; García-Arredondo, J.A.; Rodríguez-Torres, A.; Cervantes-Chávez, J.A.; Mosqueda, J. Aquiluscidin, a Cathelicidin from Crotalus aquilus, and the Vcn-23 Derivative Peptide, Have Anti-Microbial Activity against Gram-Negative and Gram-Positive Bacteria. Microorganisms 2023, 11, 2778. https://doi.org/10.3390/microorganisms11112778
Hernández-Arvizu EE, Silis-Moreno TM, García-Arredondo JA, Rodríguez-Torres A, Cervantes-Chávez JA, Mosqueda J. Aquiluscidin, a Cathelicidin from Crotalus aquilus, and the Vcn-23 Derivative Peptide, Have Anti-Microbial Activity against Gram-Negative and Gram-Positive Bacteria. Microorganisms. 2023; 11(11):2778. https://doi.org/10.3390/microorganisms11112778
Chicago/Turabian StyleHernández-Arvizu, Edwin Esaú, Teresa Monserrat Silis-Moreno, José Alejandro García-Arredondo, Angelina Rodríguez-Torres, José Antonio Cervantes-Chávez, and Juan Mosqueda. 2023. "Aquiluscidin, a Cathelicidin from Crotalus aquilus, and the Vcn-23 Derivative Peptide, Have Anti-Microbial Activity against Gram-Negative and Gram-Positive Bacteria" Microorganisms 11, no. 11: 2778. https://doi.org/10.3390/microorganisms11112778
APA StyleHernández-Arvizu, E. E., Silis-Moreno, T. M., García-Arredondo, J. A., Rodríguez-Torres, A., Cervantes-Chávez, J. A., & Mosqueda, J. (2023). Aquiluscidin, a Cathelicidin from Crotalus aquilus, and the Vcn-23 Derivative Peptide, Have Anti-Microbial Activity against Gram-Negative and Gram-Positive Bacteria. Microorganisms, 11(11), 2778. https://doi.org/10.3390/microorganisms11112778







